Abstract
The specificity of nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) for the RT of human immunodeficiency virus type 1 (HIV-1) has prevented the use of simian immunodeficiency virus (SIV) in the study of NNRTIs and NNRTI-based highly active antiretroviral therapy. However, a SIV-HIV-1 chimera (RT-SHIV), in which the RT from SIVmac239 was replaced with the RT-encoding region from HIV-1, is susceptible to NNRTIs and is infectious to rhesus macaques. We have evaluated the antiviral activity of efavirenz against RT-SHIV and the emergence of efavirenz-resistant mutants in vitro and in vivo. RT-SHIV was susceptible to efavirenz with a mean effective concentration of 5.9 ± 4.5 nM, and RT-SHIV variants selected with efavirenz in cell culture displayed 600-fold-reduced susceptibility. The efavirenz-resistant mutants of RT-SHIV had mutations in RT similar to those of HIV-1 variants that were selected under similar conditions. Efavirenz monotherapy of RT-SHIV-infected macaques produced a 1.82-log-unit decrease in plasma viral-RNA levels after 1 week. The virus load rebounded within 3 weeks in one treated animal and more slowly in a second animal. Virus isolated from these two animals contained the K103N and Y188C or Y188L mutations. The RT-SHIV-rhesus macaque model may prove useful for studies of antiretroviral drug combinations that include efavirenz.
Highly active antiretroviral therapy (HAART) has been a significant advance in the treatment of AIDS. HAART has enabled the long-term suppression of human immunodeficiency virus type 1 (HIV-1) loads to low or undetectable levels in many patients, lowered mortality rates, and improved quality of life (8, 22). The successes of HAART, however, are tempered by the presence of reservoirs of latent or persistent virus and residual viral replication (22). Another major problem is the emergence of drug-resistant variants, which may lead to virus load rebound and treatment failure (7, 8, 17, 19, 22).
Three main classes of antiretroviral drugs are widely used in various combinations for HAART: nucleoside analog reverse transcriptase (RT) inhibitors (NRTIs), nonnucleoside RT inhibitors (NNRTIs), and protease inhibitors (PIs). Early HAART regimens included one PI and two NRTIs (10, 12). More recently, NNRTI use in HAART has increased due, at least in part, to toxicities associated with the use of PIs. One of the most widely used NNRTIs is efavirenz (Sustiva). Efavirenz is used in HAART regimens that contain PIs and in PI-sparing regimens (11, 25). The combination of efavirenz with two NRTIs, lamuvidine and zidovudine, was shown to be more effective in lowering plasma virus loads and delaying the onset of virological failure and was better tolerated than a similar PI-based regimen (25). Efavirenz, in contrast to other approved NNRTIs, nevirapine and delavirdine, has a more favorable resistance profile, often requiring two or more mutations in reverse trascriptase (RT) to generate high-level drug resistance.
In order to study in more detail the problems associated with viral resistance, persistence, and residual replication within the treated host, a suitable animal model for studies of HAART is needed. Simian immunodeficiency virus (SIV) infection of rhesus macaques has proven to be a useful animal model for studies of AIDS pathogenesis, but its usefulness as a model for HAART has been limited by the inability of commonly used NNRTIs to inhibit SIV replication. However, a chimera of SIV (RT-SHIV) in which the RT from SIVmac239 was replaced with the RT from an HIV-1 clone (HXBc2) is infectious in rhesus macaques (26) and susceptible to several NRTIs and NNRTIs (3, 4). In this study, the RT-SHIV-rhesus macaque model has been used to evaluate the efficacy of efavirenz and the emergence of efavirenz-resistant mutants.
MATERIALS AND METHODS
Chemicals and drugs.
Efavirenz (Sustiva) was provided by M. Nasr, Division of AIDS, National Institute of Allergy and Infectious Diseases, Bethesda, Md., and T. Evans, University of California, Davis. Nevirapine was provided by M. Nasr. All other chemicals were reagent grade or better.
Cells and virus.
HeLa H1-JC.37 cells (21) are an H1-J clone of HeLa cells that naturally express CXCR4 and were engineered to stably express CD4 and CCR5. These cells are permissive to infection by all isolates and tropisms of HIV-1 that have been examined and are also permissive to SIV (14). HeLa H1-JC.37 cells were grown in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, Calif.) supplemented with 10% fetal bovine serum (FBS) (Gemini Bio-Products, Woodland, Calif.) that was heat inactivated for 30 min at 56°C, 100 U of penicillin/ml, 0.1 mg of streptomycin/ml, and 2.0 mM l-glutamine. CEMx174 cells, which are permissive to both HIV-1 and SIV (23), were grown in RPMI 1640 (Invitrogen) supplemented with 10% FBS, 100 U of penicillin/ml, 0.1 mg of streptomycin/ml, and 2.0 mM l-glutamine. All cells were maintained at 37°C with a humidified 5% CO2 atmosphere.
Stocks of RT-SHIV and SIVmac239 were prepared by transfection of cells with the appropriate 5′- and 3′-half clones. The 5′-half clone of RT-SHIV, containing the RT-encoding region from HIV-1 HXBc2 (26), was provided by J. Sodroski, Harvard Medical School, Boston, Mass.. The two half clones of SIVmac239, containing a nef open reading frame in the 3′-half clone, have been described (18). Infectious stocks of SIVmac239 or RT-SHIV were prepared following the transfection of appropriate clones into CEMx174 cells by electroporation, as previously described (20). Virus stocks were prepared from culture supernatants of infected CEMx174 cells. Cells were removed from the culture medium by centrifugation at 600 × g for 5 min. Aliquots of supernatants were stored frozen at −80°C. These stocks were thawed and used to infect cells for subsequent studies.
Uncloned HIV-IIIB was provided by J. Yee, California National Primate Research Center, University of California, Davis. Virus stocks were also prepared from infections of CEMx174 cells, as described above.
FIA.
Focal infectivity assays (FIAs) were used to determine titers of virus stocks and for drug-susceptibility assays. Assays with RT-SHIV, HIV-1, and SIV were all performed using HeLa H1-JC.37 indicator cells as previously described (9, 20). Immunostaining of RT-SHIV and SIV were performed with a 1/2,500 dilution of polyclonal antiserum that was obtained from rhesus macaques infected with SIV. For the HIV-1 assays, immunostaining was performed using the monoclonal antibody 22-6 (13) at a 1/800 dilution. Foci were counted under a dissecting microscope at 30 to 100× magnification. Data for drug susceptibility assays were plotted as a percentage of control foci (no drug) versus inhibitor concentrations. The concentrations required to inhibit focus formation by 50% (50% effective concentration [EC50]) were obtained from a best-fit line of the linear portions of those plots. EC50 values for each drug were determined from at least three separate experiments.
Selection of drug-resistant mutants.
Drug-resistant mutants of RT-SHIV or HIV-1 were selected by passage of cell-free virus in CEMx174 cells with stepwise increases in the drug concentration. Each selection was performed in triplicate. For the first passage, 106 cells were added to each of three 25-cm3 flasks containing RPMI supplemented with 0.1% FBS and 0.025 μM efavirenz. Cultures were inoculated with cell-free supernatant containing 1,000 focus-forming units of RT-SHIV or HIV-IIIB and incubated for 1 h at 37°C. FBS and drug were then added to adjust the final FBS concentration to 10% and the final drug concentration to the appropriate level. The medium and drug were replaced every 2 to 3 days, and cells were removed and subcultured as necessary. The cultures were monitored every 2 to 3 days by p27 (RT-SHIV) or p24 (HIV) antigen capture enzyme-linked immunosorbent assay (ELISA) as previously described (13, 16). When cultures tested positive by ELISA, they were centrifuged twice at 500 × g for 5 min each time to remove the cells. Aliquots of the cell-free supernatants were stored at −80°C as virus stocks. Pellets of infected CEMx174 cells were resuspended in phosphate-buffered saline and also stored at −80°C. For the second round of selection, 200 μl of cell-free supernatant was used to infect fresh CEMx174 cells in the presence of a twofold-higher drug concentration. The drug concentration was increased serially twofold at each subsequent round of selection.
Nucleic acid preparation and sequence analysis.
Total cellular DNA containing proviral DNA was extracted from infected cells using a DNeasy Tissue kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. DNA was eluted from the silica columns using 100 μl of buffer AE (Qiagen). A 2-μl aliquot of each DNA preparation was amplified by nested PCR using JumpStart REDTaq (Sigma-Aldrich, St. Louis, Mo.). In the first round of nested PCR with RT-SHIV, 0.4 μM (each) primers 239-2675 and 239-4751 (reverse [R]) was used, and in the second round, 0.4 μM (each) primers 239-2786 and 239-4615 (R) was added to each reaction mixture. Fragments of 2,129 and 1,875 bp were obtained from the first and second round, respectively. In the first round of HIV nested PCR, 0.4 μM (each) primers HXB2-2441 and HXB2-4384 (R) was added to each reaction mixture, and in the second round, 0.4 μM (each) primers HXB2-2493 and HXB2-4277 (R) was used. Fragments of 1,989 and 1,830 bp were obtained from the first and second round, respectively. All primers are shown in Table 1, and conditions for the PCRs were as described previously (20, 28). The amplicons were purified using either a PCR Purification kit (Qiagen) or Microcon YM-50 Centrifugal Filter Devices (Millipore, Bedford, Mass.). DNA sequence analyses were performed as previously described (20, 28) with the sequencing primers listed in Table 1.
TABLE 1.
Primers used for PCR and DNA sequence analyses
| Primer name | 3′ base | Sequence |
|---|---|---|
| SIV mac239 primersa | ||
| 239-2675 | 2675 | GTAACAGGAATAGAGTTAGGTCCAC |
| 239-2786 | 2786 | ATTAAAGGGACAATCATGACAG |
| 239-4615 (R) | 4615 | TCTGTCTGGCCACTATTCTG |
| 239-4751 (R) | 4751 | TCCACTAGCTACATGTACTGCAAC |
| HIV-1 HXB2 primersb | ||
| HXB2-2441 | 2441 | GTAAGACAGTATGATCAGATACTC |
| HXB2-2493 | 2493 | TACAGTATTAGTAGGACCTACAC |
| HXB2-EcoRV | 2987 | ACCAGGGATTAGATATCAGTAC |
| HXB2-3509 | 3509 | CCAGTACATGGAGTGTATTATGAC |
| HXB2-3837 | 3837 | GTGAAATTATGGTACCAGTTAG |
| HXB2-3039 (R) | 3039 | GGCTCTAAGATTTTTGTCAT |
| HXB2-3253 (R) | 3253 | GTACTGTCCATTTATCAGGAT |
| HXB2-4277 (R) | 4277 | AGCCATTGCTCTCCAATTACTG |
| HXB2-4384 (R) | 4384 | CTGGACTACAGTCTACTTGTC |
Viral RNA for RT-PCR was prepared from 140 μl of EDTA-anticoagulated cell-free plasma using the Viral RNA kit (Qiagen) according to the manufacturer's instructions. RNA was eluted from the silica columns in 50 μl of nuclease-free water. Nested RT-PCR was carried out under conditions previously described (20). Synthesis of cDNA was carried out using a 5-μl aliquot of RNA and primer HXB2-3253 (R). The first round of nested PCR was performed with primers 239-2675 and HXB2-3253 (R), and the second round was performed with primers 239-2786 and HXB2-3253 (R), yielding fragments of 927 and 813 bp, respectively.
Animals and sample collection.
Juvenile rhesus macaques (Macaca mulatta) 6 to 8 months old (∼1.9 to 2.5 kg) were from the type D retrovirus- and SIV-free colony at the California National Primate Research Center. The facility operates according to the Guide for the Care and Use of Laboratory Animals prepared by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council. When necessary, animals were immobilized with ketamine-HCl (10 mg/kg of body weight; Parke-Davis, Morris Plains, N.J.) injected intramuscularly. EDTA-anticoagulated blood samples were collected regularly to measure viral and immunologic parameters. Complete blood counts were performed on EDTA-anticoagulated blood samples. The samples were analyzed by using an automated electronic cell counter (Baker 9000; Serano Baker Diagnostics), and differential counts were determined manually.
Virus inoculation.
Animals were inoculated intravenously with 1.0 ml of virus dilution containing 103 50% tissue culture infectious doses of cell-free RT-SHIV grown in CEMx174 cells. The titer of the RT-SHIV stock used for animal experiments was determined by limiting-dilution tissue culture methods that were previously described (27).
Preparation and administration of drug.
Initially, doses of efavirenz were prepared by mixing purified efavirenz powder with either fruit baby food (Gerber, Fremont, Mich.) or yogurt baby food with Tang (Kraft Foods, Northfield, Ill.) for flavor. Doses (60 mg/kg) were administered orally once daily using a 5-ml syringe. After the animals reached a body weight of 3 kg, they were fed 200 mg of efavirenz daily by mixing the contents of a 200-mg Sustiva capsule (DuPont Pharmaceutical, Wilmington, Del. [since acquired by Bristol-Myers Squibb]) into a peanut butter, peanut butter and jelly, or Nutella (Ferrero USA, Inc., Somerset, N.J.) sandwich.
Pharmacokinetics of efavirenz in rhesus macaques.
The safety and pharmacokinetics of efavirenz were analyzed in three juvenile macaques. These three animals were given efavirenz (60 mg/kg) orally once a day. Blood samples were taken at sequential times during a 24-h period on day 1 and on day 14 of drug dosing, and levels of efavirenz in plasma were quantified by using validated high-performance liquid chromatography (HPLC) assays.
The HPLC system consisted of a Beckman 126 pump, a Beckman 508 autosampler, and a Beckman 168 photo diode array detector. Detection and quantification of efavirenz were performed with a C18 precolumn and a Luna 5 μ C18 column (250 by 4.6 mm; Phenomenex, Torrance, Calif.). The mobile phase was comprised of 40% phosphate buffer (25 mM; pH 7.0) and 60% acetonitrile at a flow rate of 1.5 ml per min. Efavirenz was detected at a wavelength of 247 nm. The internal standard for assays was amprenavir, which was detected at 266 nm.
A standard curve of efavirenz was prepared by adding efavirenz to 100-μl aliquots of blank monkey plasma to yield concentrations of 0.5, 1, 5, 7.5, and 10 μM. After addition of the internal standard, plasma samples were extracted with 300 μl of acetonitrile. The supernatants were reduced to dryness under vacuum at room temperature. Residues were resuspended in 25 μl of the HPLC mobile phase and centrifuged at 7,500 × g for 5 min. Samples (23 μl) of the resulting supernatants were injected onto the HPLC column. The concentrations of efavirenz were determined from the slope of a standard curve of the peak ratio of efavirenz to the internal standard versus the concentration of efavirenz. The slope of the standard curve was determined by weighted (1/x2) least-squares regression analysis.
Pharmacokinetic parameters were determined by a noncompartmental method (WinNon; Scientific Consulting, Inc.). The area under the curve (AUC) and the first nonnormalized moment (AUMC) were determined by polynomial interpolation and integration from time zero to the last sample time, with extrapolation to time infinity using the least-squares terminal slope (λz). The half-life was calculated as 0.693/λz. Total clearance was calculated as dose/AUC, and the steady-state volume of distributon (Vss) was calculated as follows: Vss = (dose × AUMC)/AUC2.
Plasma viral RNA levels.
A real-time quantitative RT-PCR (TaqMan) assay with a sensitivity of 40 copies of viral RNA per ml of plasma was used to quantify RT-SHIV RNA as previously described (15).
Virus isolation (cell-free and cell-associated).
Cell-free and cell-associated infectious viruses were isolated by cocultivation of plasma or rhesus macaque peripheral blood mononuclear cells (PBMC) with 5 × 105 CEMx174 cells in 25-cm3 flasks. The medium was replaced every 2 to 4 days, and cells were removed and subcultured as necessary. The cultures were observed for cytopathic effect under a light microscope, and virus production was monitored by p27 capture ELISA. After testing positive for virus production and cytopathic effect, cultures were centrifuged twice at 600 × g for 5 min to remove the cells. Aliquots of the cell-free supernatants were stored at −80°C as virus stocks. Pellets of infected CEMx174 cells were resuspended in phosphate-buffered saline and also stored at −80°C.
Lymphocyte phenotyping by flow cytometry.
T-lymphocyte antigens were detected by direct labeling of whole blood with peridinin chlorophyll protein-conjugated anti-human CD8 (clone SK1; Becton Dickinson Immunocytometry Inc., San Jose, Calif.), phycoerythrin-conjugated anti-human CD4 (clone M-T477; Pharmingen), and fluorescein-conjugated anti-human CD3 (clone SP34; Pharmingen). A separate aliquot of blood was labeled with fluorescein-conjugated anti-human CD3 and PerCP-conjugated anti-human CD20 (clone L27; Becton Dickinson). Red blood cells were lysed, and the samples were fixed in paraformaldehyde using a Q-prep system (Coulter Corp., Hialeah, Fla.). Lymphocytes were gated by forward and side light scatter and were then analyzed with a FACSCAN flow cytometer (Becton Dickinson). CD4+ T lymphocytes and CD8+ T lymphocytes were defined as CD3+ CD4+ and CD3+ CD8+ lymphocyte populations, respectively. B lymphocytes were defined as CD3− CD20+ lymphocytes.
RESULTS
Inhibition of RT-SHIV by efavirenz.
The susceptibilities of RT-SHIV, HIV-1, and SIV to efavirenz were compared. The EC50 value obtained for inhibition of RT-SHIV by efavirenz was 5.9 ± 4.5 nM. This was similar to the values we obtained for inhibition of HIV-1 by efavirenz (8.7 ± 1.9 nM) and to previously published values for HIV-1 (6, 30). As expected, SIV was not inhibited by efavirenz (EC50 > 10 μM). RT activity obtained from lysed virus preparations of RT-SHIV was inhibited by efavirenz, whereas RT from SIV was not affected (data not shown).
Selection of RT-SHIV and HIV-1 variants resistant to efavirenz.
RT-SHIV variants resistant to efavirenz were selected by serial passage of virus in CEMx174 cells with stepwise increases in the drug concentration. Efavirenz-resistant variants of HIV-1 were selected under identical conditions. Two independent RT-SHIV variants, designated SE1 and SE2, were obtained following the seventh passage. A third independent RT-SHIV selection with efavirenz did not yield virus beyond the third passage. The efavirenz susceptibilities of SE1 and SE2 were determined by FIA, and the results are shown in Table 2. Both variants were >600-fold resistant to efavirenz.
TABLE 2.
Efavirenz susceptibilities of RT-SHIV and HIV-1 mutants selected in cell culture
| Virus | EC50 ± SD (nM)a | Fold resistanceb | Mutations in RT |
|---|---|---|---|
| Wild-type RT-SHIV | 5.9 ± 4.5 | 1.0 | None |
| RT-SHIV SE1 | 3,750 ± 250 | 636 | L100I, V179D, V35I, T386A |
| RT-SHIV SE2 | 3,600 ± 2,100 | 611 | K103N, V108I, G196R, V314L, Q547K |
| Wild-type HIV-1 | 8.7 ± 1.9 | 1.0 | None |
| HIV-1 HE1 | 920 ± 40 | 106 | L100I, L214F, M357T |
| HIV-1 HE2 | 830 ± 25 | 96 | L100I, E138K, G190A/E, L214F, M357T |
Within an experiment, each value represents three determinations. Results from three or more independent experiments were used to calculate the EC50 ± standard deviation.
EC50 value of mutant divided by EC50 value of wild-type virus.
Sequence analyses of the RT-encoding regions of the pol genes from SE1 and SE2 after the final passage are shown in Table 2. L100I and V179D were found in SE1, while K103N and V108I were found in SE2. Both genotypes are known to confer efavirenz resistance on HIV-1. Two additional mutations, V35I and T386A, were present in the RT of SE1, while three additional mutations, G196R, V314L, and Q547K, had emerged in the RT of SE2.
Three independent variants of HIV-1, designated HE1, HE2, and HE3, were isolated after the seventh round of selection (1.6 μM efavirenz), and two of these were characterized (HE1 and HE2). HE1 and HE2 each displayed ∼100-fold resistance to efavirenz (Table 2). Both of the mutants possessed the L100I mutation in the RT-encoding region, which is known to confer efavirenz resistance on HIV-1. L214F and M357T also emerged in both HIV-1 variants. E138K and G190A/E were present in the RT of HE2, and both of these mutations are associated with reduced susceptibility of HIV-1 to one or more NNRTIs (6, 24).
Efficacy of efavirenz in RT-SHIV-infected rhesus macaques.
In order to evaluate the suitability of the RT-SHIV-rhesus macaque model for in vivo studies of efavirenz, a preliminary study of efavirenz monotherapy was conducted. First, we evaluated the safety and pharmacokinetics of efavirenz in three juvenile macaques. The three animals were given efavirenz (60 mg/kg) orally in baby food once a day as described in Materials and Methods. Levels of efavirenz in the blood were determined during 24-h periods on day 1 and on day 14 of drug dosing (Fig. 1). The pharmacokinetic parameters were determined by noncompartmental methods, and the results are shown in Table 3. These parameters are comparable to previously published values (2). Peak levels of 1.9 to 5.7 μM were obtained 2 to 8 h after dosing. In all three animals and all times tested, the blood efavirenz concentrations were >35 times greater than the EC50 for inhibition of RT-SHIV. As in humans, the half-life of efavirenz in rhesus macaques was nearly a day. This dose of efavirenz was well tolerated by the three animals, and no adverse effects were noted.
FIG. 1.
Levels of efavirenz in blood of rhesus macaques. Efavirenz (60 mg/kg) was administered orally once daily, and samples were taken over a 24-h period on day 1 (A) and day 14 (B).
TABLE 3.
Pharmacokinetics of efavirenz in rhesus macaques
| Animal | Day | Parameter valuea
|
|||||
|---|---|---|---|---|---|---|---|
| Half-life (h) | AUC (μM · h) | ClT/F (liters/kg/h) | Cmax (μM) | Vss/F (liters/kg) | Tmax (h) | ||
| 32325 | 1 | 16.4 | 90.1 | 2.1 | 3.2 | 49.8 | 8 |
| 14 | 25.3 | 103.7 | 1.8 | 5.7 | 34.5 | 4 | |
| 32402 | 1 | 10.2 | 60.9 | 3.1 | 5.4 | 45.9 | 2 |
| 14 | 14.5 | 67.3 | 2.9 | 5.3 | 59.2 | 2 | |
| 32451 | 1 | 37 | 117.3 | 1.6 | 1.9 | 86.4 | 8 |
| 14 | 27 | 103.3 | 1.8 | 3.1 | 71.5 | 2 | |
| Avg | 1 | 21 ± 14 | 89 ± 28 | 2.3 ± 0.8 | 3.5 ± 1.7 | 61 ± 22 | |
| 14 | 22 ± 7 | 91 ± 21 | 2.2 ± 0.6 | 4.7 ± 1.4 | 55 ± 19 | ||
Pharmacokinetic parameters are area under the curve (AUC), systemic clearance (ClT), oral bioavailability (F), peak concentration (Cmax), steady-state volume of distribution (Vss), and time to peak concentration (Tmax).
Two groups of three juvenile rhesus macaques (groups A and B) were used for studies of efavirenz efficacy and the emergence of efavirenz resistance in RT-SHIV-infected macaques. All of the animals became persistently infected after inoculation with RT-SHIV, and peak virus loads were reached between 2 and 4 weeks postinfection, as shown in Fig. 2. Group B animals were treated with efavirenz (60 mg/kg orally once daily) beginning 6 weeks postinfection. Efavirenz monotherapy decreased plasma virus loads an average of 1.82 ± 0.17 log units for group B after 1 week of therapy. The difference in virus loads for control group A and the treated group B were statistically significant at 1 and 2 weeks postinitiation of therapy (P < 0.05; one-tailed t test). The virus load for animal 32325 began to rebound immediately after the initial decline and remained high (>106 RNA copies per ml of plasma) over the course of drug treatment. The virus load for animal 32402 decreased to <40 copies of viral RNA/ml and remained undetectable, except for a transient increase at week 26. Animal 32451 had intermediate to low virus loads over the course of drug treatment. Two of three control animals, 32014 and 32239, had consistently high virus loads (>104 RNA copies/ml) over the course of the study. However, the virus load for animal 32147 decreased steadily over time, and except for a transient increase at week 56, was undetectable after week 40 postinfection. (data not shown).
FIG. 2.
Viral-RNA levels in RT-SHIV-infected macaques. (A) Control animals (no drug). (B) Efavirenz-treated animals. The dashed lines represent the limit of detection of the assay. The arrows indicate the time that efavirenz treatment was started in the group B animals.
Efavirenz monotherapy was halted after 46 weeks (week 52 postinfection). Animal 32325 continued to exhibit high virus loads, in the range of 106 to 107 viral-RNA copies/ml, after cessation of treatment. No rebound in plasma viremia was evident for animal 32402. Plasma virus loads for animal 32451 slowly increased to >103 copies of RNA per ml of plasma after cessation of therapy.
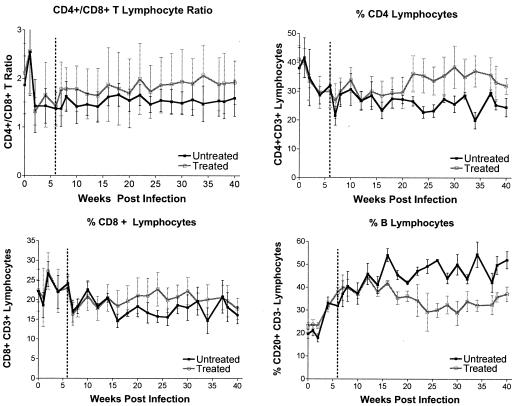
Absolute lymphocyte counts were highly variable, but all of the animals experienced transient lymphopenia 1 week postinfection. The percentages of lymphocytes were more reliable parameters to monitor infection. Prior to the start of treatment at week 6, there was no difference between the percentages of lymphocytes in groups A and B. However, after week 6, the efavirenz-treated animals (group B) had a higher CD4/CD8 T-lymphocyte ratio, higher percentages of CD4+ and CD8+ T lymphocytes, and a lower percentage of B lymphocytes than the untreated animals (Fig. 3) (two-way analysis of variance; treatment P values, <0.05 for each parameter).
FIG. 3.
Lymphocyte subsets in peripheral blood of RT-SHIV-infected animals. The percentages of lymphocyte subsets were determined by flow cytometry and are expressed as fractions of the total number of gated lymphocytes. The dashed lines indicate the start of efavirenz treatment 6 weeks after infection. The error bars indicate standard deviations.
In vivo emergence of efavirenz-resistant RT-SHIV variants.
Virus isolates obtained from PBMC and plasma of treated and untreated animals were cultured in CEMx174 cells and characterized by phenotypic and genotypic analyses. Virus from efavirenz-treated animal 32325, isolated 36 weeks postinfection (30 weeks of therapy), displayed >1,600-fold resistance to efavirenz and had the K103N and Y188L mutations in RT (Table 4). These mutations, alone or in combination, confer moderate to high-level resistance to efavirenz on HIV-1 (1). Several additional mutations, (W88S, T139V, G196R, I202V, K275R, M357T, T386A, and Q507H) were also present or emerged later, but their significance is not known. However, G196R and K275R emerged in viruses from multiple animals in both groups by 36 weeks postinfection. After 30 weeks of therapy, virus from efavirenz-treated animal 32451 had K103N and Y188C mutations in RT. Two additional mutations in RT, K275R and T376S, were also detected (Table 4).
TABLE 4.
Mutations inferred by genotypes of the RT-encoding regions of RT-SHIV isolates from untreated (group A) and efavirenz-treated (group B) animals
| Animal (group) | Mutation(s) at week:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 18 | 34 | 36 | 42 | 52 | 54 | 56 | 60 | |
| 32014 (A) | G196R | G196R | L74V | |||||
| L214F | G196R | |||||||
| V466A | T215A | |||||||
| K219E | ||||||||
| K275R | ||||||||
| V466A | ||||||||
| 32147 (A) | G196R | L74V | L74V | |||||
| K275R | G196R | G196R | ||||||
| K275R | L214F | |||||||
| K275R | ||||||||
| 32239 (A) | V75L | V75L | V75L | V75L | ||||
| K103N | K103N | W88C | W88C | |||||
| G196R | G196R | K103N | K103N | |||||
| K275R | K275R | G196R | G196R | |||||
| M357T | L214F | L214F | ||||||
| K275R | K275R | |||||||
| M357T | M357T | |||||||
| 32325 (B) | W88S | W88S | W88S | W88S | W88S | |||
| K103N | K103N | K103N | K103N | K103N | ||||
| Y188L | Y188L | T139V | T139V | D123G | ||||
| G196R | G196R | Y188L | Y188L | T139V | ||||
| K275R | K275R | G196R | G196R | Y188L | ||||
| M357T | M357T | I202V | I202V | G196R | ||||
| T386A | T386A | M357T | K275R | I202V | ||||
| Q507H | Q507H | T386A | M357T | K275R | ||||
| Q507H | T386A | M357T | ||||||
| Q507H | T386A | |||||||
| 32451 (B) | K103N | K103N | K103N | K103N | K103N | |||
| K275R | Y188C | Y188C | Y188C | T200A | ||||
| K275R | K275R | K275R | K275R | |||||
| T376S | T376S | T376S | R277K | |||||
| M348I | ||||||||
| E370Q | ||||||||
| T386S | ||||||||
After cessation of efavirenz therapy at 52 weeks postinfection, the Q507H mutation reverted to wild type in virus from animal 32325. The Y188C mutation in virus from 32451 reverted to wild type after 4 weeks without the drug.
Virus could not be isolated from PBMC or plasma of animal 32402 beyond 14 weeks postinfection. In addition, virus could not be isolated from lymph node mononuclear cells of the animal at week 52 postinfection, and the animal was negative by RT-PCR for plasma viremia at 44 weeks postinfection.
A mutation associated with efavirenz resistance in HIV-1 (K103N) occurred in RT-SHIV from one of the control animals. This mutation was detected at 18 weeks postinfection in isolates from animal 32239, and it remained present in virus isolates at 52 weeks postinfection.
Phenotypic analyses of viruses isolated from animals are shown in Table 5. RT-SHIV mutants isolated from the two efavirenz-treated animals were >1,600-fold resistant to efavirenz. Viruses from the two control animals displayed much lower levels of resistance.
TABLE 5.
Efavirenz susceptibilities of RT-SHIV mutants selected in vivoa
| Virus or source animal | EC50 (nM) | Fold resistancee |
|---|---|---|
| Wild-type RT-SHIV | 5.9 ± 4.5b | 1.0 |
| 32014 | 16c | 2.7 |
| 32239 | 1,130 ± 250d | 190 |
| 32325 | >10,000b | >1,600 |
| 32451 | >10,000c | >1,600 |
Within an experiment, each value represents three determinations. Viruses are from control animals 32014 and 32239 and from efavirenz-treated animals 32325 and 32451, as indicated.
Results from three or more independent experiments were used to calculate the EC50 ± standard deviation.
Results from one experiment were used to calculate the EC50.
Results from two independent experiments were used to calculate the EC50 ± standard deviation.
EC50 value of mutant divided by EC50 value of wild-type virus.
DISCUSSION
These results demonstrate that efavirenz inhibits RT-SHIV in vitro and in vivo. In cell culture, efavirenz was equally effective against RT-SHIV and HIV-1. Drug susceptibility assays with both viruses were performed with the same cell line and culture conditions in order to allow direct comparisons. Efavirenz treatment also resulted in a 1.8-log-unit reduction in virus loads after 1 week in RT-SHIV-infected rhesus macaques. Although the number of animals per group was small, this difference was statistically significant at 1 and 2 weeks after initiation of therapy. The course of efavirenz therapy in RT-SHIV-infected macaques was similar to that observed with NNRTI monotherapy of HIV-1 in humans. Following the initial decline in virus loads, there was a rapid rebound to levels that were not significantly different from virus loads in the no-drug control animals.
As expected, efavirenz-resistant mutants of RT-SHIV selected in vitro contained mutations known to confer efavirenz resistance on HIV-1. Despite identical in vitro selection conditions, the two RT-SHIV variants contained RT mutation profiles different from each other and from the HIV-1 variants. One of the mutants, SE1, had L100I and V179D mutations in RT. These mutations are associated with efavirenz-resistant HIV-1 selected in cell culture systems (29). The other mutant, SE2, had K103N and V108I mutations in RT. This combination of mutations has been reported for efavirenz-resistant HIV-1 selected in vitro or obtained from efavirenz-treated AIDS patients (1). Additional RT mutations in SE1 and SE2 (V35I and T386A for SE1 and G196R, V314L, and Q547K for SE2) were also different from each other and from the additional RT mutations in the HIV-1 variants. Most of the additional RT mutations in SE1 and SE2 have been seen in drug-resistant clinical HIV-1 isolates, but the effects of these mutations on efavirenz resistance or replication fitness are not known. Some of the mutations in RT, particularly those that also occur in the absence of drugs, may contribute to viral fitness by improving enzymatic function in the chimeric virus. The two RT-SHIV mutants selected in vitro also displayed a higher level of phenotypic resistance to efavirenz, (600-fold) than the HIV-1 mutants (100-fold) that were selected under similar conditions. These differences suggest that although efavirenz resistance in RT-SHIV is conferred by mutations known to confer efavirenz resistance on HIV-1, other viral determinants besides RT affect features of efavirenz resistance.
A concern with this animal model system is the appearance of the K103N mutation in one of the control RT-SHIV-infected macaques that were not treated with efavirenz. The appearance of this mutation in the absence of drug may make some studies of NNRTI resistance with RT-SHIV difficult to interpret. In a separate experiment, we found that one of six control animals (no drugs) harbored RT-SHIV with the K103N mutation (data not shown). It is unlikely that control animals were inadvertently given efavirenz, as treated and untreated animals were housed in separate cages with no possibility for direct contact. Contamination during virus isolation or PCR is also unlikely, because the genotype of RT-SHIV with K103N from the control animal is distinct from genotypes of viruses from any animals in the efavirenz-treated group. Moreover, the base change in codon 103 is A to T for isolates from the efavirenz-treated animals, whereas it is A to C in isolates from the control animal (32239). Finally, the level of efavirenz resistance for virus from control animal 32239 was not nearly as high as the levels for variants obtained from the drug-treated animals. Therefore, K103N appears to occur in some drug-naïve animals. Viruses from two control animals also had the L74V mutation, which has been associated with resistance to ddI and ddC (24) and to (−)-β-d-dioxolane-guanosine (5). Balzarini et al. also reported the emergence of L74V in RT-SHIV from drug-naive monkeys (3).
RT-SHIV isolates from each of the three control animals contained four or more mutations that resulted in amino acid changes in RT. The G196R mutation emerged in viruses from all three animals. These mutations may represent adaptation of the foreign (HIV-1) RT to the SIV genome, and we plan to evaluate the effects of these mutations on viral fitness. There may also be mutations in other SIV genes that interact with RT to compensate for differences between SIV and HIV-1 RTs. It is also possible that some of these mutations arise from selective pressures in the host macaques that are different from selective pressures in humans.
Our primary goal in this work is to establish an animal model that can be used to study details of HAART that are difficult or impossible to study in humans, such as tissue reservoirs of viral latency or persistence and sites of residual virus replication during optimal therapy. The in vivo efficacy of efavirenz that we have demonstrated suggests that RT-SHIV-infected macaques will be useful for studies with drug combinations that include this NNRTI. RT-SHIV is also similar to HIV-1 in susceptibility to PIs in vitro (9). To further evaluate this system as a HAART model, an experiment is in progress with a three-drug combination of efavirenz and two nucleoside analogs. The availability of a nonhuman primate model for HAART will allow invasive studies during successful HAART to more completely define sites of viral latency and residual virus replication. Many of these experiments would be impractical and/or unethical in humans.
Acknowledgments
We thank D. Bennett, T. Dearman, L. Hirst, W. von Morgenland, and the Veterinary Staff, Colony Services and Clinical Laboratory of the California National Primate Research Center, for expert technical assistance. We thank K. Van Rompay for useful discussions and suggestions.
This work was supported by NIH grant R01 AI47070 to T.W.N. R.F.S. is supported by an NIH-sponsored Center for AIDS Research grant (number 2P30 AI50409), NIH grant R 37 AI41980, and the Department of Veterans Affairs.
REFERENCES
- 1.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balani, S. K., L. R. Kauffman, F. A. deLuna, and J. H. Lin. 1999. Nonlinear pharmacokinetics of efavirenz (DMP-266), a potent HIV-1 reverse transcriptase inhibitor, in rats and monkeys. Drug Metab. Dispos. 27:41-45. [PubMed] [Google Scholar]
- 3.Balzarini, J., E. DeClercq, and K. Uberla. 1997. SIV/HIV-1 hybrid virus expressing the reverse transcriptase gene of HIV-1 remains sensitive to HIV-1 specific reverse transcriptase inhibitors after passage in rhesus macaques. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 15:1-4. [DOI] [PubMed] [Google Scholar]
- 4.Balzarini, J., M. Weeger, M. J. Camarasa, E. DeClercq, and K. Uberla. 1995. Sensitivity/resistance profile of simian immunodeficiency virus containing the reverse transcriptase gene of human immunodeficiency virus type 1 (HIV-1) toward the HIV-1-specific non-nucleoside reverse transcriptase inhibitors. Biochem. Biophys. Res. Commun. 211:850-856. [DOI] [PubMed] [Google Scholar]
- 5.Bazmi, H. Z., J. L. Hammond, S. C. Cavalcanti, C. K. Chu, R. F. Schinazi, and J. W. Mellors. 2000. In vitro selection of mutations in the human immunodeficiency virus type 1 reverse transcriptase that decrease susceptibility to (−)-β-d-dioxolane-guanosine and suppress resistance to 3′-azido-3′-deoxythymidine. Antimicrob. Agents Chemother. 44:1783-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Clercq, E. 1998. The role of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Antivir. Res. 38:153-179. [DOI] [PubMed] [Google Scholar]
- 7.Deeks, S. G. 2001. International perspectives on antiretroviral resistance. Nonnucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S25-S33. [DOI] [PubMed] [Google Scholar]
- 8.Fauci, A. S. 1999. The AIDS epidemic. N. Engl. J. Med. 341:1046-1050. [DOI] [PubMed] [Google Scholar]
- 9.Giuffre, A. C., J. Higgins, R. W. Buckheit, Jr., and T. W. North. 2003. Susceptibilities of simian immunodeficiency virus to protease inhibitors. Antimicrob. Agents Chemother. 47:1756-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, C. Gonzalez, D. McMahon, D. D. Richman, F. T. Valentine, L. Jonas, A. Meibohm, E. A. Emini, and J. A. Chodakewitz. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734-739. [DOI] [PubMed] [Google Scholar]
- 11.Haas, D. W., W. J. Fessel, R. A. Delapenha, H. Kessler, D. Seekins, M. Kaplan, N. M. Ruiz, L. M. Ploughman, D. F. Labriola, and D. J. Manion. 2001. Therapy with efavirenz plus indinavir in patients with extensive prior nucleoside reverse-transcriptase inhibitor experience: a randomized, double-blind, placebo-controlled trial. J. Infect. Dis. 183:392-400. [DOI] [PubMed] [Google Scholar]
- 12.Hammer, S. M., K. E. Squires, M. D. Hughes, J. M. Grimes, L. M. Demeter, J. S. Currier, J. J. Eron, Jr., J. E. Feinberg, H. H. Balfour, Jr., L. R. Deyton, J. A. Chodakewitz, and M. A. Fischl. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N. Engl. J. Med. 337:725-733. [DOI] [PubMed] [Google Scholar]
- 13.Higgins, J. R., N. C. Pedersen, and J. R. Carlson. 1986. Detection and differentiation by sandwich enzyme-linked immunosorbent assay of human T-cell lymphotropic virus type III/lymphadenopathy-associated virus- and acquired immunodeficiency syndrome-associated retroviruslike clinical isolates. J. Clin. Microbiol. 24:424-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 1997. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J. Virol. 71:8642-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leutenegger, C. M., J. Higgins, T. B. Matthews, A. F. Tarantal, P. A. Luciw, N. C. Pedersen, and T. W. North. 2001. Real-time TaqMan PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS Res. Hum. Retrovir. 17:243-251. [DOI] [PubMed] [Google Scholar]
- 16.Lohman, B. L., J. Higgins, M. L. Marthas, P. A. Marx, and N. C. Pedersen. 1991. Development of simian immunodeficiency virus isolation, titration, and neutralization assays which use whole blood from rhesus monkeys and an antigen capture enzyme-linked immunosorbent assay. J. Clin. Microbiol. 29:2187-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loveday, C. 2001. International perspectives on antiretroviral resistance. Nucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S10-S24. [DOI] [PubMed] [Google Scholar]
- 18.Luciw, P. A., E. Pratt-Lowe, K. E. Shaw, J. A. Levy, and C. Cheng-Mayer. 1995. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. USA 92:7490-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, V. 2001. International perspectives on antiretroviral resistance. Resistance to protease inhibitors. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S34-S50. [DOI] [PubMed] [Google Scholar]
- 20.Murry, J. P., J. Higgins, T. B. Matthews, V. Y. Huang, K. K. A. Van Rompay, N. C. Pedersen, and T. W. North. 2003. Reversion of the M184V mutation in reverse transcriptase of simian immunodeficiency virus is selected by tenofovir, even in the presence of lamivudine. J. Virol. 77:1120-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 23.Salter, R. D., D. N. Howell, and P. Cresswell. 1985. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics 21:235-246. [DOI] [PubMed] [Google Scholar]
- 24.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 1999. Mutations in retroviral genes associated with drug resistance: 1999-2000 update. Int. Antivir. News 7:46-69. [Google Scholar]
- 25.Staszewski, S., J. Morales-Ramirez, K. T. Tashima, A. Rachlis, D. Skiest, J. Stanford, R. Stryker, P. Johnson, D. F. Labriola, D. Farina, D. J. Manion, N. M. Ruiz, et al. 1999. Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir, and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. N. Engl. J. Med. 341:1865-1873. [DOI] [PubMed] [Google Scholar]
- 26.Uberla, K., C. Stahl-Hennig, D. Böttiger, K. Mätz-Rensing, F. J. Kaup, J. Li, W. A. Haseltine, B. Fleckenstein, G. Hunsmann, B. Öberg, and J. J. Sodroski. 1995. Animal model for therapy of acquired immunodeficiency syndrome with reverse transcriptase inhibitors. Proc. Natl. Acad. Sci. USA 92:8210-8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Rompay, K. K., M. L. Marthas, R. A. Ramos, C. P. Mandell, E. K. McGowan, S. M. Joye, and N. C. Pedersen. 1992. Simian immunodeficiency virus (SIV) infection of infant rhesus macaques as a model to test antiretroviral drug prophylaxis and therapy: oral 3′-azido-3′-deoxythymidine prevents SIV infection. Antimicrob. Agents Chemother. 36:2381-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Rompay, K. K., T. B. Matthews, J. Higgins, D. R. Canfield, R. P. Tarara, M. A. Wainberg, R. F. Schinazi, N. C. Pedersen, and T. W. North. 2002. Virulence and reduced fitness of simian immunodeficiency virus with the M184V mutation in reverse transcriptase. J. Virol. 76:6083-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winslow, D. L., S. Garber, C. Reid, H. Scarnati, D. Baker, M. M. Rayner, and E. D. Anton. 1996. Selection conditions affect the evolution of specific mutations in the reverse transcriptase gene associated with resistance to DMP 266. AIDS 10:1205-1209. [DOI] [PubMed] [Google Scholar]
- 30.Young, S. D., S. F. Britcher, L. O. Tran, L. S. Payne, W. C. Lumma, T. A. Lyle, J. R. Huff, P. S. Anderson, D. B. Olsen, S. S. Carroll, D. J. Pettibone, J. A. O'Brien, R. G. Ball, S. K. Balani, J. H. Lin, I. Chen, W. A. Schleif, V. V. Sardana, W. J. Long, V. W. Brynes, and E. A. Emini. 1995. L-743,726 (DMP-266): a novel, highly potent nonnucleoside inhibitor of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 39:2602-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]