Abstract
PEN is an abundant peptide in the brain that has been implicated in the regulation of feeding. We identified a receptor for PEN in mouse hypothalamus and Neuro2A cells. PEN bound to and activated GPR83, a G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptor (GPCR). Reduction of GPR83 expression in mouse brain and Neuro2A cells reduced PEN binding and signaling, consistent with GPR83 functioning as the major receptor for PEN. In some brain regions, GPR83 colocalized with GPR171, a GPCR that binds the neuropeptide bigLEN, another neuropeptide that is involved in feeding and is generated from the same precursor protein as is PEN. Coexpression of these two receptors in cell lines altered the signaling properties of each receptor, suggesting a functional interaction. Our data established PEN as a neuropeptide that binds GPR83 and suggested that these two ligand-receptor systems—PEN-GPR83 and bigLEN-GPR171—may be functionally coupled in the regulation of feeding.
INTRODUCTION
Neuropeptides play important roles in cell-cell signaling, and many neuropeptide receptors are potential therapeutic targets. The peptides SAAS, PEN, and LEN, which are so-named because of the presence of these amino acid residues in their sequences, are among the most abundant peptides present in mouse hypothalamus and are all produced from the same precursor protein, proSAAS (1, 2). Peptides containing the SAAS and LEN sequences are produced as big and little (longer and shorter) peptides, whereas only a single PEN peptide has been identified (Fig. 1A). The enzymes responsible for the cleavage of proSAAS into SAAS, PEN, LEN (bigLEN and littleLEN), and other peptides are the same enzymes that produce most neuropeptides: prohormone convertases and carboxy-peptidase E (3–6). Furthermore, differential cleavage by various peptidases leads to a range of big and little forms of the proSAAS-derived peptides, many of which may be functional neuropeptides (7).
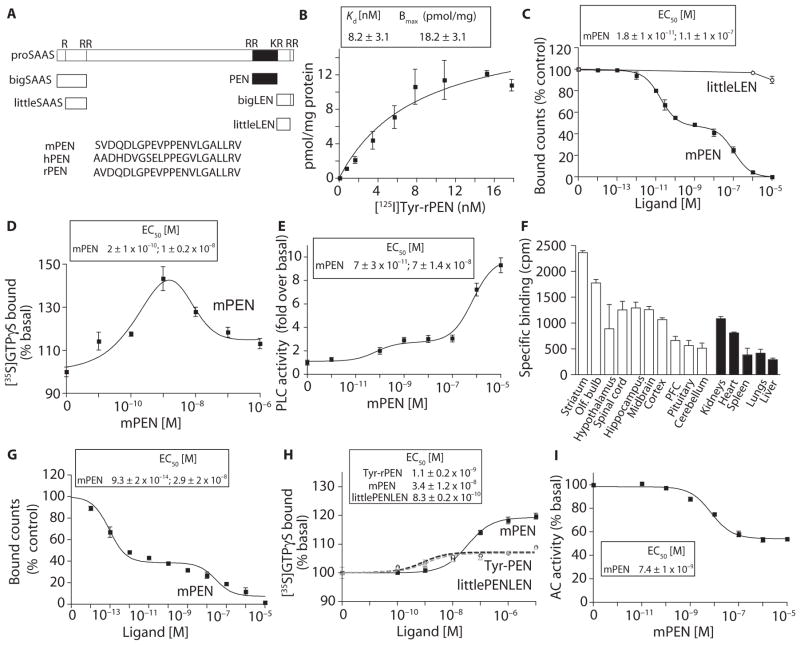
Fig. 1. PEN binds and activates a GPCR in the brain.
(A) Schematic representation of peptides derived from proSAAS processing. PEN sequences of mouse (mPEN), rat (rPEN), and human (hPEN) are in single-letter amino acid code. (B) Saturation binding with [125I]Tyr-rPEN in mouse hypothalamic membranes (30 μg). (C) The ability of mPEN and rlittleLEN to displace [125I]Tyr-rPEN (3 nM) binding in mouse hypothalamic membranes (30 μg). (D) The effect of mPEN on [35S]GTPγS binding in mouse hypothalamic membranes (20 μg). (E) The effect of mPEN on phospholipase C (PLC) activity in mouse hypothalamic membranes (10 μg). (F) Specific binding of [125I]Tyr-rPEN in different mouse brain regions and peripheral tissues. (G) Displacement by mPEN of [125I]Tyr-rPEN (3 nM) binding to mouse hippocampal membranes (30 μg). (H) The effect of mPEN, Tyr-rPEN, or rlittlePENLEN on [35S]GTPγS binding in mouse hippocampal membranes (20 μg). (I) The effect of mPEN on adenylyl cyclase (AC) activity in mouse hippocampal membranes (2 μg). Data represent means ± SE (n = 3 to 8 individual experiments).
Both PEN and LEN peptides are present in neuropeptide Y (NPY) neurons in the arcuate nucleus of the mouse hypothalamus (8). These cells also contain agouti-related peptide (AgRP) and function in the stimulation of feeding (9, 10). Consistent with a role in feeding and body weight regulation, transgenic mice overexpressing PCSK1N (the gene encoding proSAAS) are slightly overweight (11), and mice with a disruption in the PCSK1N gene, which eliminates the production of proSAAS, are underweight (12). In addition, intracerebroventricular injection of antibodies to either bigLEN or PEN blocks feeding (8), suggesting that these peptides stimulate feeding.
GPR171, a G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptor (GPCR), binds and is activated by bigLEN (13). GPR171 is present in the hypothalamus and other areas involved in feeding and body weight regulation (13). GPR171 does not bind PEN; therefore, we tested for the existence of a PEN receptor. Using various binding and signaling assays, we found evidence for PEN receptors in mouse hypothalamus and in the Neuro2A cell line. We tested candidate orphan GPCRs by individually expressing them in a heterologous cell line and found that PEN activated GPR83. Furthermore, knockdown of GPR83 in Neuro2A cells or knockout of GPR83 in mouse brain substantially reduced PEN binding and PEN-induced signaling. We also explored interactions between GPR83 and GPR171; these two receptors are colocalized in some brain regions. We found that PEN signaling is modulated by bigLEN and vice versa in cell lines expressing both receptors. Furthermore, these two receptors colocalized and were in close enough proximity for direct interactions in the ventral hypothalamus. These results indicated that crosstalk between the GPR83 and GPR171 signaling pathways may occur in a region of the brain that controls feeding and other reward behaviors.
RESULTS
PEN binds and activates a GPCR in the brain
PEN is an abundant peptide derived from the neuropeptide precursor proSAAS. Mouse PEN (mPEN) and rat PEN (rPEN) only differ by one residue at the N-terminal end, whereas human PEN (hPEN) is more divergent and has the sequence PEG instead of PEN (Fig. 1A). To determine whether a receptor for PEN is present in mouse brain, we performed ligand-binding studies with N-terminally tyrosinated radioidodinated rPEN (Tyr-rPEN). We detected saturable radioligand binding with a high affinity of ~8 nM in mouse hypothalamic membrane preparations (Fig. 1B). mPEN dose-dependently displaced the radioligand (Fig. 1C), revealing a high-affinity site (18 pM) and a lower-affinity site (110 nM). The peptide littleLEN from rat (rlittleLEN) did not compete for binding (Fig. 1C).
Because most neuropeptides activate GPCRs, which signal through heterotrimeric G proteins containing an α subunit activated by guanine nucleotide exchange of GDP (guanosine diphosphate) for GTP (guanosine 5′-triphosphate) and are inactivated by autocatalytic GTPase (guanosine triphosphatase) activity and reassociation with the βγ heterodimer, we examined the ability of mPEN to activate G proteins, as assessed by GTPγS binding to mouse hypothalamic membranes. Subnanomolar concentrations of mPEN increased GTPγS binding [EC50 (median effective concentration), ~0.2 nM], whereas concentrations above 1 nM were less effective, implying desensitization of the response (Fig. 1D). To assess the G protein activated by mPEN, we measured adenylyl cyclase activity (to assess Gαs- or Gαi-mediated signaling) and PLC activity (to assess Gαq-mediated signaling) in the presence of a protease inhibitor cocktail to prevent peptide degradation. The adenylyl cyclase activity assay primarily measures the activity of the Gαi or Gαs subunit, whereas the PLC assay measures the activity that could be mediated either by the Gαq or Gβγ subunits. Studies assaying adenylyl cyclase activity in mouse hypothalamic membranes gave inconsistent results, with some samples exhibiting bell-shaped dose-response curves and some exhibiting a small decrease (~21 ± 2%) in cAMP (adenosine 3′,5′-monophosphate) levels (fig. S1), whereas those measuring PLC activity were consistent and reproducible (Fig. 1E). Therefore, we focused on PLC activity. We detected a robust and dose-dependent increase in PLC activity in response to mPEN (Fig. 1E), suggesting that the hypothalamic receptor for PEN is a Gαq-coupled GPCR that activates the PLC-mediated signaling cascade.
During the studies with hypothalamic membranes, we observed substantial variation in the total amount of mPEN binding between individual animals. To directly examine this, we compared the relative amount of mPEN binding in 18 individual hypothalami from male mice and found them to vary by two- to fivefold (fig. S2). A comparison of the relative amounts of mPEN binding in several brain regions and peripheral organs revealed the highest amount of mPEN binding in the striatum (Fig. 1F). The olfactory bulb, spinal cord, hypothalamus, hippocampus, midbrain, and cortex also had high amounts of mPEN binding (Fig. 1F). We also detected mPEN binding in the kidney, heart, spleen, lung, and liver (Fig. 1F). Thus, these results indicated that mPEN binds to one or more receptors in the brain and periphery.
As found for the hypothalamic mPEN receptor, the mPEN receptor in hippocampal membranes showed both high- and low-affinity sites when radiolabeled Tyr-rPEN was displaced with mPEN (Fig. 1G). In contrast to the hypothalamic response, the activation profile of hippocampal membranes with GTPγS had a monophasic dose-response curve (EC50 of 34 nM) with no detectable desensitization at high concentrations (Fig. 1H). Tyr-rPEN and rlittlePENLEN, another proSAAS-derived peptide (14), exhibited lower efficacies and higher affinities, compared with mPEN (Fig. 1H). An additional difference between the hypothalamic and hippocampal response was that mPEN dose-dependently inhibited adenylyl cyclase activity (Fig. 1I), whereas there was no consistent and reproducible effect on PLC activity (fig. S3). Together, these results indicated that PEN receptors are present in the hippocampus and hypothalamus, and the receptors couple to different G proteins in the different regions, Gαq in the hypothalamus and Gαi in the hippocampus.
Because neuropeptides alter synaptic activity (15) and because PEN has been implicated in regulating feeding behaviors, we performed electrophysiological characterization of the effect of mPEN in slice preparations of the paraventricular nucleus (PVN) of the rat hypothalamus, a brain region involved in regulation of food intake and reward behaviors. The addition of mPEN inhibited presynaptic glutamate release (Fig. 2). We measured pharmacologically isolated α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR)–mediated spontaneous excitatory postsynaptic currents (sEPSCs) by voltage clamp recording of PVN neurons. The application of mPEN decreased the sEPSC frequency without changing the amplitude, suggesting a presynaptic effect on glutamate release (Fig. 2, A and B). Moreover, mPEN dose-dependently reduced the amplitude of evoked EPSCs with an EC50 of ~8 nM (Fig. 2C). Sequential application of 10- and 100-nM mPEN produced increasing inhibition of evoked EPSC amplitude, and the inhibition was reversible after washout (fig. S4). Additionally, in most samples, application of 100 nM mPEN induced an increase in paired-pulse ratio (Fig. 1D), which is another indication of a presynaptic effect.
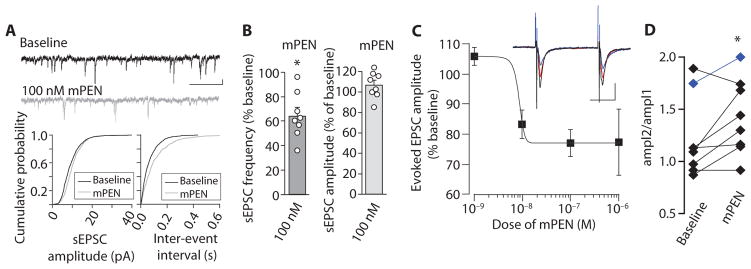
Fig. 2. Effect of mPEN on synaptic activity of PVN neurons in rat hypothalamic slices.
(A) Spontaneous excitatory postsynaptic current sEPSC traces (top) in a PVN neuron after bath application of 100 nM mPEN (scale bar, 5 pA and 200 ms). Cumulative plots for this neuron demonstrate the effect of mPEN on sEPSC amplitudes (left) and inter-sEPSC intervals (right). (B) Graphical representation of frequency (left) and mean amplitudes (right) of sEPSCs in PVN neurons after mPEN application. Each circle represents an individual neuron. (C) Dose response for change in evoked EPSC amplitude. Frequency (baseline, 5.8 ± 1.7 Hz; mPEN, 3.9 ± 1.3 Hz; P = 0.03; n = 8); amplitude (baseline, 11.8 ± 1.5 pA; mPEN, 12.5 ± 1.6 pA; P = 0.24; n = 8 individual neurons); and inset, example traces of evoked responses from one cell. Black, baseline; red, 10 nM mPEN; blue, 100 nM mPEN. Scale bars, 40 pA and 20 ms. n = 3 to 5 cells per dose. (D) The paired-pulse ratio in PVN neurons in response to 100 nM PEN. The data in blue are from the one neuron where the highest dose tested was 10 nM PEN. The data represent means ± SE (n = 3 to 8 individual experiments). *P < 0.05 [paired two-tailed t test for (B) and (D); for statistical analysis, see table S1].
A PEN receptor is present in Neuro2A cells
Multiple GPCR agonists induce neurite outgrowth in the mouse neuroblastoma cell line Neuro2A, indicating that various GPCRs are present in this cell line (13, 16, 17). Because receptor activation–mediated neurite outgrowth represents a functional outcome of GPCR activation in Neuro2A cells, this assay provides an easy and convenient test for the presence of a receptor (13, 16, 17). Exposing Neuro2A cells to mPEN, mouse bigLEN (mbigLEN), or rat SAAS (rSAAS) significantly increased the number of neurites, suggesting the presence of receptors for PEN and these other neuropeptides in these cells (Fig. 3A and fig. S5). Ligand-binding analysis revealed saturable binding of Tyr-rPEN (Fig. 3B). The affinity of rPEN for binding to the Neuro2A cells was 22 nM, which is lower than that detected for binding to mouse hypothalamic membranes, which had an affinity of ~8 nM (Fig. 1B). However, as seen with the mouse hypothalamus samples, dose-dependent displacement with mPEN revealed a high- and a lower-affinity binding site (Fig. 3C).
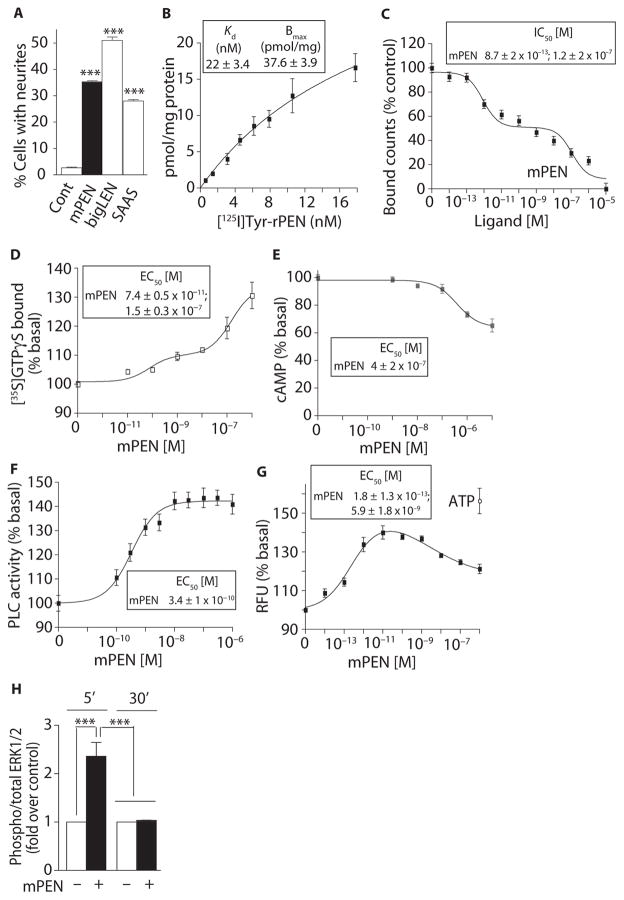
Fig. 3. PEN binds and activates a GPCR in Neuro2A cells.
(A) The effect of mPEN, mbigLEN, or rSAAS (1 μM) on neurite outgrowth in Neuro2A cells. (B) Saturation binding with [125I]Tyr-rPEN in Neuro2A cell membranes (30 μg). (C) The ability of mPEN to displace [125I]Tyr-rPEN (3 nM) binding from Neuro2A cell membranes (50 μg). (D) The effect of mPEN on [35S] GTPγS binding in Neuro2A membranes (20 μg). (E) The effect of mPEN on intracellular cAMP levels in Neuro2A cells (10,000 per well). (F) The effect of mPEN on PLC activity in Neuro2A membranes (10 μg). (G) The effect of mPEN on intracellular Ca2+ release in Neuro2A cells. ATP (adenosine 5′-triphosphate) (1 μM) was used as a positive control. RFU, relative fluorescence units. (H) The effect of mPEN (1 μM) on MAPK (mitogen-activated protein kinase) phosphorylation at 5 and 30 min in Neuro2A cells. Data represent means ± SE (n = 3 to 6 independent experiments). ***P < 0.0001 [one-way analysis of variance (ANOVA) for (A) and two-way ANOVA for (H)]; for details of statistical analysis, see table S1.
Analysis of G protein activation showed that GTPγS binding was biphasic in response to mPEN, exhibiting both a high and a low potency (Fig. 3D). As with the hippocampal samples, exposure of the Neuro2A cells to mPEN led to dose-dependent inhibition of adenylyl cyclase activity and a concomitant decrease in intracellular cAMP (Fig. 3E). As with the hypothalamic samples, mPEN also caused a dose-dependent increase in PLC activity in Neuro2A cells (Fig. 3F). Because PLC activity stimulates the production of IP3 (inositol 1,4,5-trisphosphate), which activates its receptor on the endoplasmic reticulum to release Ca2+, we also examined the effect of PEN on intracellular Ca2+ signals. In Neuro2A cells loaded with a Ca2+-sensitive fluorescent dye, we observed a dose-dependent increase in intracellular Ca2+ with a high potency of ~0.2 pM; mPEN concentrations greater than 1 nM led to a decreased Ca2+ response, suggesting desensitization (Fig. 3G). The different potencies for mPEN-mediated induction of the various signals may result from activities arising from the high- and low-affinity binding sites that we detected.
In Neuro2A cells, activation of the MAPK pathway occurs in response to GPCRs coupled to Gαi (13, 18). Consistent with this, mPEN activated the MAPK pathway, as evidenced by the increased phosphorylation of extracellular signal–regulated kinases 1 and 2 (ERK1/2) within 5 min of treatment; this effect was desensitized by 30 min (Fig. 3H). Together, these studies demonstrated that Neuro2A cells have one or more receptors for PEN with properties similar to those detected for PEN receptor(s) in the brain.
GPR83 is a PEN receptor: Heterologous expression of GPR83 mediates PEN binding and signaling
To identify a PEN receptor, we searched the literature for orphan GPCRs present in Neuro2A cells (19) and in the hypothalamus (20, 21). We selected hGPR83 (human GPR83), mGPR19, mGPR108, and mGPR165 as potential candidates. We also included mGPR171 because this receptor binds bigLEN, which is generated from the same precursor as PEN (1, 13). For these experiments, we used heterologous expression of the human and mouse GPCRs in Chinese hamster ovary (CHO) cells and human embryonic kidney (HEK) 293 cells because these cells are easy to transfect and contain many signaling effectors needed to study the properties of a GPCR in a heterologous system (22). We expressed the epitope-tagged versions of the receptors in CHO cells along with a chimeric promiscuous G protein [human Gα16/i3 (hGα16/i3)] and tested for hPEN-induced intracellular Ca2+ signals using the Ca2+-sensitive fluorescent dye. Cells expressing hGPR83 showed a robust response to hPEN, whereas cells expressing the other GPCRs did not (Fig. 4A and fig. S6). We also tested a scrambled peptide, rlittleLEN and mbigLEN, as negative controls, and ATP, which activates endogenous purinergic receptors to stimulate a Ca2+ signal, as a positive control.
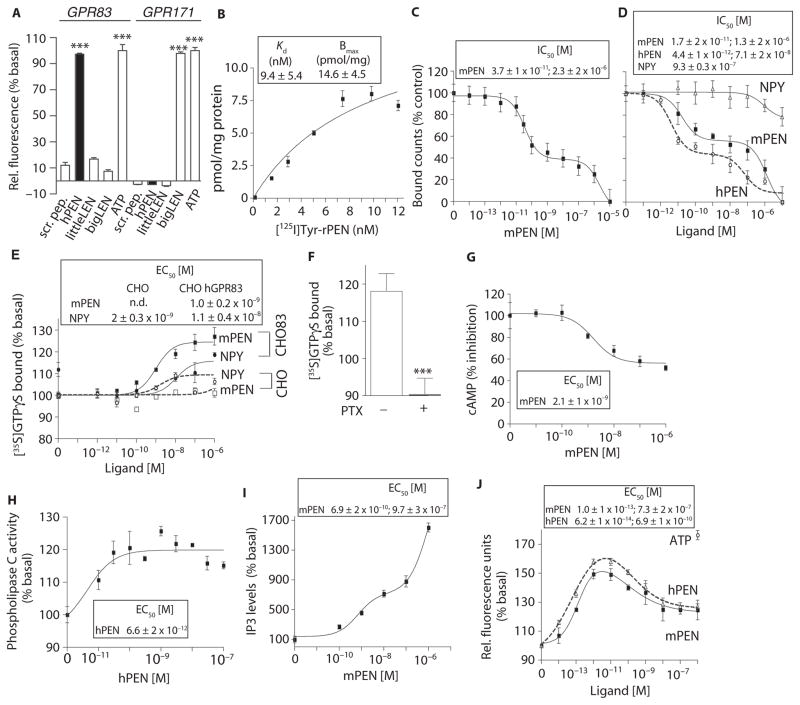
Fig. 4. Expression of GPR83 in heterologous cells confers PEN binding and signaling.
(A) The effect of PEN (1 μM) on intracellular Ca2+ release in cells expressing hGPR83 along with a promiscuous chimeric hGα16/i3 protein. Scrambled peptide (1 μM), hPEN (1 μM), rlittleLEN (1 μM), mbigLEN (1 μM), or ATP (1 μM). (B) Saturation binding with [125I]Tyr-rPEN in CHO hGPR83 cell membranes (30 μg). (C) mPEN displacement of [125I]Tyr-rPEN (3 nM) binding to HEK-293 mGPR83 cells (50 μg). (D) The ability of mPEN, hPEN, and NPY to displace [125I]Tyr-rPEN (3 nM) binding to CHO hGPR83 cell membranes (50 μg). (E) The effect of mPEN on GTPγS binding to membranes (20 μg) from CHO hGPR83 cells (CHO83) or CHO alone. (F) The effect of pertussis toxin (PTX; 50 ng/ml) pretreatment on 1 μM PEN-mediated [35S]GTPγS binding in membranes (20 μg) from CHO hGPR83 cells versus absence of PTX. ***P < 0.0005 (t test). (G) The effect of mPEN on intracellular cAMP levels in CHO hGPR83 cells (10,000 cells per well). (H) The effect of PEN on PLC activity in membranes (10 μg) from CHO hGPR83 cells. (I) The effect of mPEN on IP3 levels in CHO hGPR83 cells (10,000 cells per well). (J) The effect of mPEN or hPEN on intracellular Ca2+ release in CHO hGPR83 cells expressing a promiscuous chimeric hGα16/i3 protein. ATP (1 μM) was used as a positive control. Data (A to J) represent means ± SE (n = 3 to 8 independent experiments). ***P < 0.001[one-way ANOVA for (A); t test for (F)]; details of the statistical analyses are in table S1.
To determine whether the pharmacological properties of GPR83 were like those observed for the PEN-binding site in mouse brain and Neuro2A cells, we examined the properties of GPR83 in HEK cells transiently expressing hemagglutinin A (HA)–tagged mouse GPR83 (mGPR83), tagged on the N terminus, or in CHO cells stably expressing untagged hGPR83. Ligand-binding analysis of hGPR83 revealed high-affinity binding for Tyr-rPEN with a dissociation constant (Kd) of 9.4 nM (Fig. 4B); this value is similar to that observed in mouse hypothalamus (~8 nM) (Fig. 1B). Ligand displacement analysis of mGPR83 revealed a dose-dependent displacement of [125I] Tyr-rPEN with mPEN with high- and low-affinity sites (Fig. 4C), as we observed for mouse hypothalamus and hippocampus (Fig. 1, C and G) and the Neuro2A cells (Fig. 3C). Ligand displacement analysis of untagged hGPR83 with hPEN or mPEN also revealed a dose-dependent displacement with high- and low-affinity sites (Fig. 4D). HA-tagged mGPR83 and untagged hGPR83 had similar binding profiles for mPEN (Fig. 4, C and D); thus, the HA tag did not appear to affect binding. A study examining rGPR83 reported binding of NPY (23). However, we found that NPY at concentrations below 1 μM did not displace [125I]Tyr-rPEN and exhibited only a small (~20%) and not significant (one-way ANOVA; P = 0.37, F7,40 = 1.109) displacement with 10 μM NPY (Fig. 4D).
Analysis of G protein activation showed that mPEN increased GTPγS binding in CHO cells expressing hGPR83 but had no effect in control CHO cells (Fig. 4E). In contrast, NPY induced a small increase in GTPγS binding both in control cells and in hGPR83-expressing cells (Fig. 4E). Pretreatment with pertussis toxin to block signaling through Gαi abolished the mPEN-mediated increases in GTPγS binding in hGPR83-expressing cells (Fig. 4F). In hGPR83-expressing cells, mPEN led to a dose-dependent reduction in cAMP (Fig. 4G) and a dose-dependent increase in PLC activity (Fig. 4H) and intracellular IP3 (Fig. 4I). As we observed with Neuro2A cells, mPEN or hPEN added to hGPR83-expressing CHO cells increased Ca2+ signals at low concentrations, and high concentrations reduced the response (Fig. 4J), suggesting desensitization of the receptor at high PEN concentrations. The results from ligand-binding and G protein activity studies showed that mGPR83 and hGPR83 exhibited properties like those of the PEN receptor in mouse brain and Neuro2A cells.
Many GPCRs undergo agonist-dependent endocytosis, and detecting such a cellular response would provide additional evidence that GPR83 is a receptor for PEN. Thus, we performed a receptor internalization assay in which the amount of HA-tagged mGPR83 at the cell surface was detected by enzyme-linked immunosorbent assay (ELISA) before and after exposure of the cells to either mPEN or hPEN. Analysis of the amount of receptor at the surface over time showed that the addition of either mPEN or hPEN resulted in rapid and robust internalization of mGPR83 (fig. S6B). These results support the hypothesis that GPR83 functions as a receptor for PEN.
Reducing GPR83 abundance impairs PEN signaling
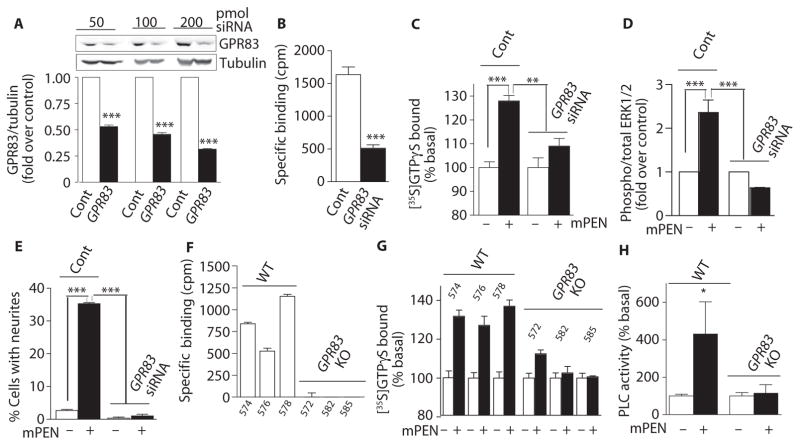
If GPR83 represents the PEN receptor that we detected in Neuro2A cells (Fig. 3), reduction or loss of GPR83 should diminish the response to PEN. We found that Neuro2A cells express GPR83 mRNA (fig. S7) and protein (Fig. 5A), and small interfering RNA (siRNA)–targeting GPR83 reduced the abundance of GPR83 protein (Fig. 5A). The GPR83 knockdown Neuro2A cells had significantly reduced PEN binding (Fig. 5B); the magnitude of the decrease in binding was consistent with the magnitude of the reduction in GPR83 protein. We also found that GPR83 knockdown significantly decreased PEN-stimulated GTPγS binding (Fig. 5C), ERK1/2 phosphorylation (Fig. 5D), and neurite outgrowth (Fig. 5E).
Fig. 5. Knockdown or knockout of GPR83 leads to reduced binding and signaling by PEN.
(A) The effect of expressing GPR83 siRNA (50 to 200 pmol) on the levels of GPR83 in Neuro2A cells. (B) The effect of expressing GPR83 siRNA (200 pmol) in Neuro2A cells on specific binding of [125I] Tyr-rPEN (3 nM) to membranes (50 μg). Specific binding is defined as the difference in [125I]Tyr-rPEN bound in the absence and presence of 10 μM mPEN. cpm, counts per minute. (C) The effect of mPEN (1 μM) on GTPγS binding in membranes (20 μg) from Neuro2A cells transfected with 200 pmol of GPR83 siRNA. (D) The effect of mPEN (1 μM; 5 min) on phosphorylation of MAPK in untransfected Neuro2A cells (Cont) and in cells expressing 200 pmol of GPR83 siRNA. (E) The effect of mPEN (1 μM) on neuritogenesis in untransfected Neuro2A cells (Cont) and cells expressing GPR83 siRNA (200 pmol). Data (A to E) represent means ± SE (n = 3 to 6 individual experiments). *P < 0.05; **P < 0.001; and ***P < 0.0001 [two-way ANOVA for (A), t test for (B), and two-way ANOVA for (C), (D), and (E)]. (F) [125I]Tyr-rPEN binding to membranes (30 μg) from individual GPR83 knockout and wild-type (WT) mice. The number on the x axis denotes the individual mouse number. (G) The effect of mPEN (1 μM) on GTPγS binding in hypothalamic membranes (20 μg) from individual GPR83 knockout mice. (H) The effect of mPEN (100 nM) on PLC activity in hypothalamic membranes (20 μg) from WT and GPR83 knockout (KO) mice. Data (H) represent means ± SE (n = 3 individual animal tissues); *P < 0.05 compared to WT without PEN (two-way ANOVA). In (H), the difference in the presence and absence of mPEN is not significant in the GPR83 knockout mouse tissue. For details of statistical analysis, see table S1.
mGPR83 is localized to chromosome 9, region A2-3, and hGPR83 to chromosome 11, region 11q21 (24). To date, four different isoforms of mGPR83 have been described that could arise from alternative splicing: isoform 1 encodes a 423–amino acid protein, isoform 2 represents a shorter form of 381 amino acids, isoform 3 has a 68–amino acid and isoform 4 a 20–amino acid insertion in the second cytoplasmic loop compared to isoform 1 (25). Transcript analysis of hGPR83 in human brain detected mostly the homolog of mouse isoform 1 and a minor amount of mouse isoform 2 (24). Mice with a global knockout of GPR83 (NM_010287) have been generated by replacing a 2374-kb genomic region containing exons 2 and 3 of GPR83 with a neomycin resistance cassette (26, 27). We used these mice and performed ligand-binding analysis of hypothalamic membranes from individual mice (wild-type mouse # 574, 576, and 578 or knockout mouse # 572, 582, and 585), which revealed that mice lacking GPR83 had no detectable PEN binding (Fig. 5F). Signaling analyses revealed that the PEN-mediated stimulation of G protein activity (GTPγS binding) (Fig. 5G) and PLC activity (Fig. 5H) was also absent with the exception of one mouse that exhibited some GTPγS binding. These results indicated that GPR83 expression is necessary to elicit PEN responses, which is further evidence that GPR83 functions as the PEN receptor in the hypothalamus.
GPR83 interacts with GPR171
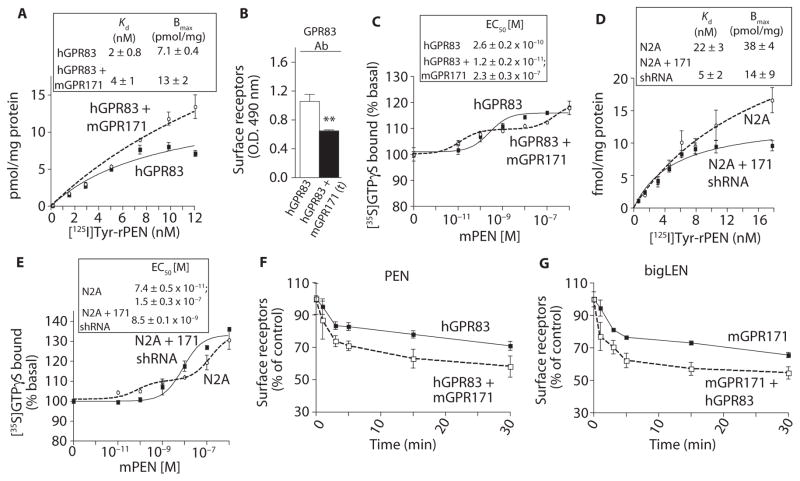
PEN is synthesized by posttranslational processing of proSAAS; this processing also generates other peptides, such as PENLEN that is further processed to bigLEN (1). Because PEN and bigLEN colocalize to NPY-positive neurons in the hypothalamus (28) and are both released upon stimulation (1), and because in AtT-20 cells (pituitary corticotrope tumor cells) both PEN and bigLEN are stored within the same vesicles (28), we examined whether the receptor for PEN (GPR83) and receptor for bigLEN (GPR171) were part of a functional complex. To assess potential functional interactions, we investigated the binding and signaling properties of hGPR83 in CHO cells in the absence or presence of mGPR171. CHO cells expressing both receptors bound more radiolabeled PEN than did cells expressing only hGPR83 (Fig. 6A). This was not due to an increase in the total amount of hGPR83 at the cell surface because the presence of mGPR171 decreased the amount of GPR83 at the cell surface based on ELISA using antibodies to GPR83 (Fig. 6B). The presence of mGPR171 also affected hGPR83-mediated signaling. In the absence of mGPR171, mPEN caused a monophasic dose-dependent increase in GTPγS binding (Figs. 4E and 6C). The presence of mGPR171 converted this monophasic curve into a biphasic curve; furthermore, in these cells, PEN exhibited higher potency for the first phase of the response when compared to that in cells expressing only hGPR83 (Fig. 6C).
Fig. 6. GPR83 functionally interacts with GPR171.
(A) The effect of expressing mGPR171 on [125I]Tyr-rPEN binding to membranes (30 μg) from CHO hGPR83 cells. (B) The effect of expressing mGPR171 on surface abundance of hGPR83 in CHO hGPR83 cells. (C) The effect of expressing mGPR171 on PEN-mediated GTPγS binding in CHO hGPR83 cells. (D) The effect of shRNA-mediated knockdown of GPR171 on [125I]Tyr-rPEN binding to Neuro2A cells. The GPR171 shRNA has been described previously (13). (E) The effect of shRNA-mediated knockdown of GPR171 in Neuro2A cells on PEN-mediated GTPγS binding. (F) The effect expressing mGPR171 in CHO hGPR83 cells (2 × 105 cells) on 100 nM mPEN-mediated hGPR83 internalization. (G) The effect of expressing hGPR83 in CHO mGPR171cells (2 × 105 cells) on bigLEN (100 nM)–mediated mGPR171 internalization. Data (A to G) represent means ± SE (n = 3 to 6 independent experiments). **P < 0.01; ***P < 0.001 [t test for (B); for details of statistical analysis, see table S1].
To study this further, we analyzed Neuro2A cells, which are positive for GPR171 and GPR83, and Neuro2A cells with a stable knockdown of GPR171 (13). GPR171 knockdown decreased the amount of radiolabeled rPEN binding (Fig. 6D) and resulted in a monophasic dose-dependent G protein activation (GTPγS binding) (Fig. 6E). Thus, the presence of GPR171 altered the pharmacological properties of GPR83, which could be due to direct receptor-receptor interactions or that the receptors are part of a multi-component signaling complex.
Heteromerization of GPCRs can sometimes enable the ligand of one receptor to promote the endocytosis of the other receptor. Therefore, we examined whether the presence of mGPR171 altered the ligand-mediated endocytic properties in CHO cells expressing one or both of the receptors. We measured receptor endocytosis by ELISA using antibodies that recognized either GPR83 or the myc tag at the N terminus of mGPR171. A time course of receptor endocytosis revealed that both the initial rate and extent of PEN-induced hGPR83 endocytosis were enhanced by the presence of mGPR171 (Fig. 6F). To examine whether the reciprocal also occurred, we performed a time course of bigLEN-induced mGPR171 endocytosis in CHO cells expressing mGPR171 alone or with hGPR83. Both the rate and extent of mGPR171 endocytosis were enhanced by the presence of GPR83 (Fig. 6G).
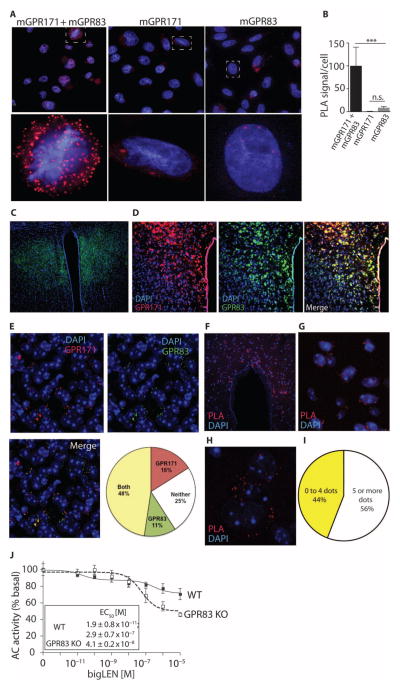
To test whether these two receptors were colocalized in the PVN and were in close enough proximity to interact, we performed colocalization analysis by immunohistochemistry in the PVN and PLAs (proximity ligation assays) in CHO cells expressing the receptors and in PVN using antibodies selective for the two receptors [for antibody selectivity data, see fig. S8 for GPR83 and Gomes et al. (13) for GPR171]. With the PLA in the CHO cells, we found a statistically significant increase in signal only in cells expressing both mGPR171 and mGPR83 (Fig. 7, A and B), consistent with the hypothesis that the two receptors interact to influence each other’s signaling. To confirm that these interactions could also occur in the brain, we first performed immunohistochemical analysis with the receptor-specific antibodies. We detected GPR83 in the PVN (Fig. 7C) and, furthermore, found that ~48% of the cells were positive for both GPR171 and GPR83 immunoreactivity, indicating these receptors are both present in a subset of the neurons in the PVN (Fig. 7, D and E). In contrast, in the lateral septum, a region with very low detectable GPR83 but high amounts of GPR171 immunoreactivity, very few cells (<5%) were positive for both GPR83 and GPR171 (fig. S9). To examine whether the receptors are close enough for physical interaction in the PVN, we performed PLA with the receptor-specific antibodies; many cells in the PVN had a positive signal (Fig. 7, F to H), and quantification showed that ~56% of cells exhibited close proximity between these two receptors (more than five red dots per cell; Fig. 7I).
Fig. 7. GPR83 and GPR171 are colocalized in the PVN and close enough to interact.
(A) PLA to determine the interaction of GPR83 and GPR171 in CHO cells coexpressing mGPR83 and mGPR171. (B) Quantification of the PLA signal, n = 30 cells. (C) Coronal section stained for GPR83 (green) in the PVN. (D) Immunohistochemical localization using the antibody recognizing GPR83 (green) and the antibody recognizing GPR171 (red) to determine colocalization of GPR83 and GPR171 in the PVN. Magnification, ×10. (E) Quantification of 63× images from the PVN shown as a pie chart. n = 50 cells per field (two fields per mice) from three independent animals. (F) PLA to determine the interaction of GPR83 and GPR171 in the PVN; 40× image. (G) A ×63 magnification of (F). (H) A zoomed-in image of (F) showing PLA signal in a single cell. (I) Quantification of data in (G). n = 50 cells per field (two fields per mice) from three independent animals. (J) The effect of knockdown of GPR83 (GPR83 knockout) on bigLEN-mediated adenylate cyclase activity. Data represent means ± SE (n = 3 independent animals). **P < 0.01; ***P < 0.001 [one-way ANOVA for (B); for details of statistical analysis, see table S1].
To further investigate the functional implications of receptor-receptor interactions, we examined the effect of GPR83 on GPR171 signaling in hypothalamic membranes from wild-type mice or mice lacking GPR83. In the hypothalamus, GPR171 couples to Gαi in response to bigLEN (13). We found that the profile of bigLEN-mediated signaling in membranes from GPR83 knockout animals was different from the profile in membranes from wild-type animals (Fig. 7J); the bigLEN dose-response curve was shifted from a biphasic dual-potency curve in the membranes from the wild-type mice to a monophasic curve with greater efficacy in the membranes from the GPR83 knockout mice (Fig. 7J). These data showed that GPR83 modulates GPR171 signaling in the hypothalamus.
DISCUSSION
One of the major findings of this study was that PEN binds and activates GPR83. PEN is produced from proSAAS along with other biologically active peptides, such as bigLEN and SAAS (1). SAAS has biological activity through an unidentified receptor (29), and bigLEN activates GPR171 (13). PEN and other proSAAS-derived peptides are among the most abundant peptides in the hypothalamus (7). These peptides are produced within the secretory pathway, stored in secretory granules, and secreted from cells (1). On the basis of the presence of PEN and other proSAAS peptides in AgRP neurons of the hypothalamus (8), as well as the results of transgenic mice overexpressing proSAAS and knockout mice lacking proSAAS (11, 12), proSAAS-derived peptides have been implicated in the regulation of body weight through an assumed effect on appetite and food intake. However, intracerebroventricular administration of proSAAS peptides, such as PEN or bigLEN or a combination of both these peptides, did not elicit a significant effect on food intake (8). This could be due to a high basal amount of proSAAS-derived peptides, because the peptides are rapidly degraded, because these peptides regulate body weight through mechanisms distinct from those affecting appetite, or a combination of these effects. Because we could only detect changes in signaling and in electrophysiological recordings in the presence of a protease inhibitor cocktail, we predict that their activity may be limited by degradation. Consistent with a role in appetite and feeding behavior, antibody-mediated neutralization of endogenous PEN reduces food intake for 1 to 14 hours (8), suggesting that PEN is an orexigenic neuropeptide. However, an important criteria for neuropeptides are that they bind to selective receptors and influence second messengers. The results of the present study support the hypothesis that PEN is a bona fide neuropeptide.
GPR83 was a logical candidate to test for the PEN receptor for several reasons. This receptor is present at a relatively high abundance in regions of the brain involved in regulating energy metabolism (30–32). GPR83 is also known as JP05, GPR72, and the glucocorticoid-induced receptor, the latter because the mRNA for this receptor is increased in mouse thymocytes after treatment with either glucocorticoids or compounds that increase cAMP (33, 34). Although four isoforms of GPR83 have been reported for mouse (24, 25), the distribution of the isoforms in mouse brain has not been assessed. In humans, the most abundant GPR83-encoding transcript corresponds to the homolog of mouse isoform 1 (24). In mouse brain, GPR83 expression is increased after chronic amphetamine treatment (35) and reduced by short-term dexamethasone treatment (36). GPR83 knockout mice show a reduction in stress-evoked anxiety and delayed spatial learning in the Morris water maze (37). GPR83 knockout mice have normal body weight when fed a regular chow diet, yet they are resistant to obesity when placed on a high-fat diet (27).
Although GPR83 is currently referred to as an “orphan” GPCR, indicating that its ligand has not been definitively established, a report in 2007 claimed that NPY was a ligand for this receptor (23). We found that radiolabeled PEN binding to GPR83 was not affected by NPY concentrations below 1 μM, although 10 μM NPY caused a partial, nonsignificant, displacement of PEN binding. Another study reported that GPR83 is significantly activated by 0.1 mM zinc ions, although slightly lower or higher concentrations of zinc failed to significantly activate the receptor (38). The N-terminal extracellular domain of GPR83 has been reported to function as an intramolecular inverse agonist (39), raising the possibility that the receptor can be activated by proteolytic cleavage of the inhibitory domain. A study reported that GPR83 exhibits a high level of constitutive activity (40). These diverse results suggest that GPR83 may be activated by multiple modes (without ligands or with different ligands) in the brain as well as other tissues.
GPR83 transcripts are detected in the kidney, liver, spleen, and other peripheral tissues, with high levels in T cells (25, 26, 31, 32, 41–43). Although we did not test all of these tissues, in addition to detecting PEN binding in the brain, we detected PEN binding to the kidney, heart, spleen, lungs, and liver, tissues that have been reported as positive for GPR83. PEN is present in pancreatic endocrine cells, pituitary, and adrenal medulla and could act as a peptide hormone through peripheral GPR83 receptors. Alternatively, GPR83 may bind another ligand in the periphery. Our data indicated that in the hypothalamus and hippocampus, GPR83 is the PEN receptor. Whether GPR83 is the only receptor for PEN could be assessed by comparing PEN binding and PEN-dependent signaling in other tissues from wild-type and GPR83 knockout mice.
Another major finding was that GPR83 and GPR171 functionally interact, are close enough to directly interact in a subset of neurons in the PVN and in cultured cells, and thereby influence the signaling properties of each receptor. Although not all proSAAS-derived peptides appear colocalized in mouse brain, both PEN and bigLEN show substantial colocalization and are likely co-secreted (8). Thus, not only are both receptors likely to be exposed to ligand in response to the same synaptic activity, but their close proximity may enable the functional interaction of these receptors, which may have important physiological consequences.
GPR83 also interacts with the ghrelin receptor (encoded by Ghsr1a) when the two receptors are coexpressed in heterologous cells (COS7 and HEK-293) (27). GPR83 and GHSR1A are both present in specific subsets of mouse arcuate nucleus neurons, and the orexigenic effect of ghrelin is potentiated in GPR83 knockout mice (27). Similarly, we found that bigLEN-mediated GPR171 inhibition of adenylyl cyclase was greater in hypothalamus from GPR83 knockout mice compared to the inhibition in tissue from wild-type mice. However, the interaction between GPR83 and GPR171 that we identified was complex; cells expressing a combination of the two receptors showed biphasic dose-response curves with high- and low-affinity sites for PEN and bigLEN, whereas cells expressing only one of the receptors had a single site with an intermediate affinity. Similarly, brain tissue from wild-type mice and Neuro2A cells had biphasic dose-response curves consistent with receptor interactions. We expect that for PEN and bigLEN, both the high- and low-affinity sites could have important biological functions. In some brain regions, the amounts of these peptides are very high (~0.5 to 2 nmol per gram of tissue) (44, 45), which is comparable to the amounts of other abundant brain peptides, such as enkephalin (46). Thus, if 1 nmol of peptide is evenly distributed in 1 g of tissue, its concentration would be 1 μM, which is sufficient to stimulate the lower-affinity site. Neuropeptides are stored within secretory granules where the concentration can reach 5 to 10 mM. Receptors close to the sites of peptide release could be exposed to micromolar concentrations of peptides, whereas receptors on cells located farther from the site of neuropeptide release would be exposed to substantially lower concentrations. Therefore, both high- and low-affinity sites on GPR83 are likely physiologically important.
MATERIALS AND METHODS
Materials
Tyr-PEN, hPEN, mPEN, rlittlePENLEN, rSAAS, mbigLEN, and rlittleLEN were from Phoenix Pharmaceuticals Inc, and mPEN was custom-synthesized by GenScript. Neuro2A, a mouse neuroblastoma cell line, CHO cells, and HEK-293 cells were from ATCC. Dulbecco’s modified Eagle’s medium (DMEM) and penicillin-streptomycin were from Corning cellgro. F12 was from Gibco. Lipofectamine and Fluo-4 NW calcium dye were from Invitrogen. Fetal bovine serum (FBS) was from Biowest. Mouse GPR19, GPR108, GPR165, and GPR171 cDNAs (complementary DNAs) were obtained from Open Biosystems and subcloned into pCMV-myc-N-terminal epitope tagging mammalian expression vector (Stratagene) according to the manufacturer’s protocol (13). The untagged hGPR83 plasmid cDNA was obtained from cdna.org. The generation and characterization of mGPR83 (HA-tagged) plasmid have been described previously (38). Control siRNA, mGPR83 siRNA, and myc antibodies (cat. # sc-40) were from Santa Cruz Biotechnology. The siRNA to GPR83 (cat. # sc-75191) comprises three sequences as follows: #1, CAUCACCAAGGGUGUCAUAtt (sense) and UAUGACACCCUUGGUGAUGtt (antisense); #2, CCAUGAGCAGUACUUGUUAtt (sense) and UAACAAGUACUGCUCAUGGtt (antisense); and #3, GUUCAGCCCUCAAUUUGUAtt (sense) and UACAAAUUGAGGGCUGAACtt (antisense). mGPR83 short hairpin RNAs (shRNAs) were purchased from Sigma-Aldrich (clone ID: TRCN0000026837, sequence CCGGCCATCACCAAGGGTGTCATATCTCGAGATATGACACCCTTGGTGATGGTTTTT; clone ID: TRCN0000026857, sequence CCGGCCATGAGCAGTACTTGTTATACTCGAGTATAACAAGTACTGCTCATGGTTTTT). Antibodies recognizing tubulin (cat. # T5168), protease inhibitor cocktail (cat. # P2714), and phosphatase inhibitor cocktail (cat. # P0044) were from Sigma-Aldrich. NPY was obtained from Tocris. Pierce Iodination Reagent was from Thermo Scientific. [125I] (cat. # NEZ033L001MC) and [35S]GTPγS (cat. # NEG030H250UC) were from PerkinElmer. Antibodies to phospho-ERK1/2 (cat. # 4370S) and total ERK1/2 (cat. # 4696S) were from Cell Signaling Technology. Antibodies to GPR83 (cat. # ab72175) were from Abcam. Antibodies to GPR171 were generated in rat against the amino acid sequence MTNSSFFCPVY, and tested for specificity using a standard protocol [fig. S8 and (47)]. Rabbit (cat. # PI-1000) and mouse (cat. # PI-2000) secondary antibodies coupled to horseradish peroxidase were from Vector Laboratories. Rabbit IRDye 800 (cat. # 926-32211) and mouse IRDye 680 (cat. # 926-68070) secondary antibodies were from LI-COR. GPR83 knockout [genotyped by quantitative polymerase chain reaction (PCR) analysis] and control wild-type mouse brains were generated as described (27).
Cell culture and transfections
HEK-293 and Neuro2A cells were grown in DMEM containing 4.5 g/liter glucose (cat. # 10-013-CV, Corning cellgro), 10% FBS, and 1× penicillin-streptomycin solution (cat. # 30-002-CI, Corning) at 37°C and 10% CO2/O2. CHO cells were grown in F12 medium containing 1.802 g/liter glucose (cat. # 11765-054, Gibco Life Sciences), 10% FBS, and 1× penicillin-streptomycin solution at 37°C and 10% CO2/O2. Cells transiently expressing each of the myc-tagged receptors (mGPR19, mGPR108, mGPR165, and mGPR171), untagged hGPR83, or HA-tagged mGPR83 alone or with a chimeric hGα16/i3 protein were generated using Lipofectamine 2000 (cat. # 11668-019, Invitrogen) according to the manufacturer’s protocol. Stable cell lines expressing either mGPR171, untagged hGPR83, or HA-tagged mGPR83 were maintained in growth media containing G418 (500 μg/ml). Cells coexpressing hGPR83 (untagged) and mGPR171 (myc-tagged) were generated by transiently transfecting CHO cells stably expressing hGPR83 or mGPR171 with mGPR171 or hGPR83, respectively, using Lipofectamine 2000 as described in the manufacturer’s protocol. Neuro2A cells stably expressing GPR171 or GPR83 shRNA were generated as described by the shRNA manufacturer (Sigma-Aldrich). Neuro2A cells were transfected with mGPR83 siRNA using Lipofectamine 2000 as described in the manufacturer’s protocol.
Membrane preparation
Membranes were prepared as described previously (13) from wild-type male mouse hypothalamus, olfactory bulb, midbrain, striatum, cortex, pituitary, prefrontal cortex, cerebellum, hippocampus, kidney, heart, spleen, lungs, GPR83 knockout mouse hypothalamus or hippocampus (27), CHO cells alone or expressing untagged hGPR83, HEK cells expressing HA-tagged mGPR83, and Neuro2A cells alone or expressing either GPR83 siRNA, GPR83 shRNA, or GPR171 shRNA. Briefly, tissues/cells were homogenized in 25 volumes (1 g wet weight/25 ml) of ice-cold 20 mM tris-Cl buffer containing 250 mM sucrose, 2 mM EGTA, and 1 mM MgCl2 (pH 7.4) followed by centrifugation at 27,000g for 15 min at 4°C. The pellet was resuspended in 25 ml of the same buffer, and the centrifugation step was repeated. The resulting membrane pellet was resuspended in 40 volumes (of original wet weight) of 2 mM tris-Cl buffer containing 2 mM EGTA and 10% glycerol (pH 7.4). The protein content of the homogenates was determined using the Pierce BCA Protein Assay Reagent, after which homogenates were stored in aliquots at −80°C until use.
Binding assays
Tyr-rPEN (200 μg) was radioiodinated using [125I] and Pierce Iodination Reagent as described in the manufacturer’s protocol (Thermo Scientific). The specific activity of the iodinated peptide was 54.1 Ci/mmol at the time of iodination (the radiolabeled peptide was used within 60 days of iodination). Binding assays were carried out using membranes (30 to 50 μg protein) from hypothalamus, hippocampus, Neuro2A cells, CHO cells expressing hGPR83, or HEK cells expressing HA-tagged mGPR83 as described previously (18, 48, 49). Briefly, saturation binding assays were carried out for 1 hour at 37°C in 50 mM tris-Cl (pH 7.4) containing protease inhibitor cocktail (Sigma-Aldrich) and [125I]Tyr-rPEN (0 to 10 nM). Nonspecific binding was determined using 10 μM unlabeled Tyr-rPEN. Displacement binding assays were carried out for 1 hour at 37°C using 50 mM tris-Cl (pH 7.4) containing protease inhibitor cocktail, 3 nM of [125I]Tyr-rPEN, and different concentrations (0 to 10 μM) of mPEN, hPEN, rlittleLEN (LENPSPQAPA), and NPY. At the end of the incubation period, samples were filtered using a Brandel filtration system and GF/B filters. Filters were washed three times with 3 ml of ice-cold 50 mM tris-Cl (pH 7.4) and bound radioactivity was measured using a scintillation counter.
[35S]GTPγS binding
[35S]GTPγS binding assays were carried out as described previously (13, 49). Briefly, membranes (20 μg) from hypothalamus, Neuro2A cells, or CHO cells expressing hGPR83 were incubated for 1 hour at 30°C with Tyr-rPEN, mPEN, and rlittlePENLEN (0 to 1 μM final concentration) in the presence of 2 mM GDP and 0.5 nM [35S]GTPγS in 50 mM Hepes containing 5 mM MgCl2, 100 mM NaCl, 1 mM EDTA (pH 7.4), and a protease inhibitor cocktail. Nonspecific binding was determined in the presence of 10 μM cold [35S]GTPγS. Basal values represent values obtained in the presence of GDP and in the absence of ligand. At the end of the incubation period, samples were filtered using a Brandel filtration system and GF/B filters. Filters were washed three times with 3 ml of ice-cold 50 mM tris-Cl (pH 7.4), and bound radioactivity was measured using a scintillation counter. For experiments involving pertussis toxin, membranes were pretreated with toxin (50 ng/ml) for 30 min on ice before carrying out the assay.
PLC activity assay
PLC activity was measured by incubating membranes (10 μg) from hypothalamus, Neuro2A cells, or CHO cells expressing hGPR83 with mPEN (0 to 10 μM final concentration) for 1 hour at 37°C in 0.1 M tris-Cl buffer (pH 7.4) containing 10 mM CaCl2, 80 mM p-nitrophenylphosphorylcholine, and protease inhibitor cocktail (50). The amount of p-nitrophenol released was measured at 410 nm.
IP3 measurement
CHO cells expressing hGPR83 (10,000 per well) in Hepes-buffered Hanks’ balanced salt solution (HBSS) lacking CaCl2 and MgCl2 and containing protease inhibitor cocktail were treated with mPEN (0 to 10 μM final concentration) for 3 min at room temperature in the presence of protease inhibitor cocktail, and the amount of IP3 formed was measured using the IP3 kit from DiscoveRx.
Adenylyl cyclase activity
Membranes (2 μg per well) from hippocampus or Neuro2A cells or CHO cells expressing hGPR83 cells (10,000 per well) were pretreated with 40 μM forskolin in the absence or presence of mPEN or mbigLEN (0 to 10 μM final concentration) in the presence of 1× protease inhibitor cocktail and 100 μM IBMX (3-isobutyl-1-methylxanthine) for 30 min, and cAMP levels were measured using the HitHunter cAMP detection kit for membranes or cells from DiscoveRx (13) or α-screen kit from PerkinElmer (39) as described previously. It should be mentioned that this assay was found to be sensitive to cell type, time of incubation, and presence of protease inhibitor cocktail. For example, GPR83 expressed in some batches of HEK cells did not respond to PEN, whereas GPR83 expressed in CHO cells (transiently or stably) consistently gave dose-dependent inhibition when the assays were carried out for 20 to 30 min (and not longer) and only in the presence of protease inhibitor cocktail.
ERK1/2 phosphorylation
Neuro2A cells alone or transfected with control siRNA or GPR83 siRNA (200 pmol) (Santa Cruz Biotechnology, cat. # sc-75191) were treated without or with 1 μM mPEN in the presence of protease inhibitor cocktail for 5 or 30 min, and ERK1/2 phosphorylation was measured as described (13). Briefly, cells were lysed in 2% SDS in 50 mM tris-Cl (pH 6.8) containing protease and phosphatase inhibitor cocktails (cat. # P0044, Sigma-Aldrich). Samples (~30 μg protein) in 1× Laemmli’s sample buffer containing 10% freshly added β-mercaptoethanol were heated for 30 min at 65°C and subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) followed by transfer to nitrocellulose membranes at 30 V overnight. Membranes were blocked with 10% nonfat dried milk in TBS-T [50 mM tris-Cl (pH 7.4) containing 150 mM NaCl, 1 mM CaCl2, and 0.1% Tween 20]. After four washes (15 min each) with TBS-T, membranes were probed overnight at 4°C with either phospho-ERK1/2 (1:1000), total ERK1/2 (1:1000), or tubulin (1:50,000) antibodies diluted in 30% Odyssey Blocking Buffer (cat. # 927-40000, LI-COR) in TBS-T. After four washes (15 min each) with TBS-T, membranes were incubated with rabbit IRDye 800 and mouse IRDye 680 antibodies (1:10,000 in TBS-T containing 30% Odyssey Blocking Buffer) for 1 hour at 37°C. After four washes (15 min each) with TBS-T, blots were imaged and quantified using the Odyssey Imaging System (LI-COR).
Animals
Animal care and all experimental procedures were in accordance with guidelines from the National Institutes of Health and approved in advance by the University of California, San Francisco, as well as the Icahn School of Medicine at Mount Sinai, New York, Institutional Animal Care and Use Committees.
Slice preparation and electrophysiology
Recordings were made in control male Sprague-Dawley rats (>200 g). Rats were thoroughly anesthetized with isoflurane and then decapitated. Coronal brain slices containing the PVN (200 to 250 μm thick) were prepared using a vibratome (Leica Instruments) in ice-cold Ringer’s solution (119 mM NaCl, 2.5 mM KCl, 1.3 mM MgSO4, 1.0 mM NaH2PO4, 2.5 mM CaCl2, 26.2 mM NaHCO3, and 11 mM glucose) saturated with 95% O2/5% CO2. After recovery at 33°C for at least 1 hour, slices were visualized under a Zeiss Axioskop FS 2 Plus or Axioskop D1 with differential interference contrast optics and infrared illumination using a Zeiss AxioCam MRm and AxioVision 4 (Zeiss) or Microlucida (MBF Biosciences) software. Whole-cell recordings were made at 33°C using 2.5 to 5 megohm pipettes containing 123 mM K-gluconate, 10 mM Hepes, 0.2 mM EGTA, 8 mM NaCl, 2 mM MgATP, 0.3 mM Na3GTP, and 0.1% biocytin (pH 7.2; osmolarity adjusted to 275). Liquid junction potentials were not corrected during recordings. Series resistance and input resistance were sampled throughout the experiment with 4-mV 200-ms hyperpolarizing steps. For all experiments, neurons in which there was change in series resistance of either more than 5 megohm or 15% of baseline were excluded from analysis. Recordings were made using an Axopatch 1-D (Axon Instruments), filtered at 5 kHz, and collected at 20 kHz using IGOR Pro (WaveMetrics). Cells were recorded in voltage clamp mode (V = −70 mV), and glutamate-mediated EPSCs were pharmacologically isolated with either the Cl− channel blocker picrotoxin (100 μM) or the GABAAR antagonist gabazine (10 μM). In a subset of experiments, 6,7-dinitroquinoxaline-2,3(1H,4H)-dione (DNQX; 10 μM) was applied at the end of the experiment to confirm the glutamatergic nature of the recorded synaptic events. Spontaneous events were detected by searching the smoothed first derivative of the data trace for values that exceeded a set threshold, and these events were confirmed visually. For measuring evoked EPSCs, stimulating electrodes were placed 60 to 150 μm lateral to the patched cell, and two pulses (50-ms interval) were delivered once every 10 s. Each evoked EPSC amplitude was calculated by comparing a 2-ms period around the peak to a 2-ms interval just before the stimulation. mPEN effects (in the presence of protease inhibitor cocktail) were statistically evaluated by comparing the last 4 min of baseline data to the last 4 min during drug application using Student’s paired t test.
Neurite outgrowth assays
Neuro2A cells alone or cells expressing GPR83 siRNA (200 pmol) were plated into poly-L-lysine–coated 12-well plates (20,000 cells per well) in growth media containing 10% FBS. On the following day, the media were substituted with growth media lacking FBS, and cells were treated with either mPEN, mbigLEN, or rSAAS (1 μM final concentration) in the presence of protease inhibitor cocktail for 16 to 20 hours at 37°C. Vehicle-treated cells were used as controls. Cells were imaged using a Nikon microscope at 40× objective. To avoid sampling bias sequential images were taken to cover ~300 to 500 cells. In addition, experiments were repeated on different days. ImageJ software was used to determine the cell diameter as well as the length of the neurites. Cells were scored (by an experimenter blinded to treatment conditions) as positive for neurite outgrowth when the length of the neurite was more than two times that of the cell diameter.
Measurement of intracellular Ca2+ signals
These assays were carried out as described previously (13). Briefly, CHO cells expressing either myc-tagged mGPR19, mGPR108, mGPR165, mGPR171, or untagged hGPR83 along with a chimeric hGα16/i3 protein were plated into black poly-L-lysine–coated 96-well clear-bottom plates (40,000 cells per well). On the next day, the growth medium was removed, and cells were washed twice in HBSS containing 20 mM Hepes buffer. Cells were incubated with Fluo-4 NW calcium dye (3 μM in 100-μl HBSS buffer containing protease inhibitor cocktail) for 1 hour at 37°C and treated with buffer or with 1 μM of scrambled peptide, hPEN, rlittleLEN, mbigLEN, or ATP. Increases in intracellular Ca2+ levels were measured for ~210 s at excitation 494 nm and emission 516 nm. In another set of studies, Neuro2A cells or CHO cells expressing hGPR83 along with a chimeric hGα16/i3 protein were treated with mPEN, hPEN (0 to 1 μM), or ATP (1 μM) in HBSS buffer containing protease inhibitor cocktail, and increases in intracellular Ca2+ levels were measured.
Western blot analysis
For experiments using GPR83 siRNA, lysates (~30 μg of protein) from Neuro2A cells expressing either control siRNA or GPR83 siRNA (50, 100, or 200 pmol) were subjected to Western blot analysis essentially as described above for ERK1/2 phosphorylation except that blots were blocked in Odyssey Blocking Buffer, and 0.1% Triton X-100 was added to the TBS-T wash buffer. GPR83 antibody raised in rabbit (1:1000) was used as the primary antibody. Tubulin antibodies raised in mouse (1:50,000) were used as loading control. Secondary antibodies used were rabbit IRDye 800 (1:10,000) and mouse IRDye 680 secondary antibodies (1:10,000). For experiments examining the specificity of GPR83 antibody raised in rabbit and GPR171 antibody raised in rat, membranes from CHO cells alone or expressing HA-tagged mGPR83 or myc-tagged mGPR171, from Neuro2A cells alone or expressing either myc-tagged mGPR171, GPR83 shRNA, or GPR171 shRNA, and from the hypothalamus of individual wild-type and GPR83 knockout mouse (~20 to 35 μg of protein) were used. Membranes were solubilized in 2% SDS in 50 mM tris-Cl (pH 6.8) containing protease and phosphatase inhibitor cocktails. Samples (~20 to 30 μg of protein) in 1× Laemmli’s sample buffer containing 10% freshly added β-mercaptoethanol and 8 M urea were heated for 30 min at 65°C and subjected to SDS-PAGE and Western blot analysis as described above. The GPR83 antibody raised in rabbit (1:1000) and GPR171 antibody raised in rat (1:5000) were used as the primary antibodies. Tubulin antibodies raised in mouse (1:50,000) were used as loading control. Secondary antibodies used recognized rabbit IRDye 800 (1:10,000), rat IRDye 800 (1:10,000), or mouse IRDye 680 (1:10,000).
Reverse transcription PCR
Total RNA was isolated from Neuro2A cells, and cDNA was prepared as described (13). Briefly, 3 × 108 cells were homogenized with TRIzol (Invitrogen). Total RNA was isolated using RNeasy Mini Kit (cat. # 74104, Qiagen) as per the manufacturer’s protocol. Real-time PCRs were performed using ABI PRISM 7900HT sequence detection system, and amplifications were done using the SYBR Green PCR Master Mix (Applied Biosystems). Quantitative reverse transcription PCR (RT-PCR) was performed using sense (5′-CTGGCGGTGTCTAATTTGTG-3′) and antisense (5′-TTTCTTCCAGAGGCTTGCTC-3′) primers for mGPR83 and sense (5′-TGAAGGTCGGTGTGAACG-3′) and antisense (5′-CAATCTCCACTTTGCCACTG-3′) primers for mouse glyceraldehyde-3-phosphate dehydrogenase (Sigma-Aldrich). Quantitative analysis was performed using the ΔΔCt method and REST software (www.gene-quantification.de) (51).
Immunohistochemistry
Mice were deeply anesthetized with ketamine (100 mg/kg, intraperitoneally) and transcardially perfused through the ascending aorta with 4% paraformaldehyde (200 ml). Brains were postfixed with 4% paraformaldehyde for 1 hour and stored in 1× phosphate-buffered saline (PBS). Immunohistochemistry was performed on free-floating coronal brain slices (40 μm) containing hypothalamus. Sections were incubated in 1% sodium borohydride in PBS for 30 min followed by blocking buffer (5% normal goat serum and 0.3% Triton X-100 in PBS) for 1 hour. Tissue sections were incubated overnight at 4°C in primary antibodies against GPR83 (rabbit, 1:250; Abcam) and GPR171 (rat, 1:100) in 1% bovine serum albumin and 0.1% Triton X-100. Specificity of GPR83 and GPR171 antibodies was tested with tissues of animals lacking the receptors, with CHO cells alone or expressing either myc-tagged mGPR171 or HA-tagged mGPR83, and with Neuro2A cells alone or expressing myc-tagged mGPR171 and shRNA to mGPR171 or to mGPR83 (fig. S8). Antibodies were visualized with donkey anti-rat 594 and goat anti-rabbit 488 (Life Technologies). After 5 min of incubation with DAPI (4′,6-diamidino-2-phenylindole) (100 ng/ml), sections were mounted with ProLong Gold Antifade (Molecular Probes). Images were taken with a Leica DM6000 microscope at the Microscopy CORE at the Icahn School of Medicine at Mount Sinai.
Receptor internalization assay
Receptor internalization assays were carried out using previously described protocols (52, 53). Briefly, HEK-293 cells expressing HA-tagged mGPR83 were seeded 24 hours after transfection into 24-well plates (2 × 105 cells per well). Cells were labeled with GPR83 antibodies raised in rabbit (1:1000 in PBS containing 1% FBS) for 1 hour at 4°C, washed three times with growth media, followed by treatment with 100 nM of either mPEN or hPEN in media containing protease inhibitor cocktail for different time periods (0 to 120 min) at 37°C. Cells were briefly fixed (3 min) with 4% paraformaldehyde followed by three washes (5 min each) with PBS followed by incubation with anti-rabbit antibody coupled to horse-radish peroxidase (1:2000 in PBS containing 1% FBS) for 1 hour at 37°C. Cells were washed three times with 1% FBS in PBS (5 min each wash), and color was developed by the addition of the substrate o-phenylenediamine [5 mg/10 ml in 0.15-m citrate buffer (pH 5) containing 15 μl of H2O2]. Absorbance at 490 nm was measured with a Bio-Rad ELISA reader. In a separate set of experiments, CHO cells expressing untagged hGPR83 (2 × 105 cells) without or with myc-tagged mGPR171 were labeled with GPR83 antibodies raised in rabbit and treated with 100 nM of mPEN in media containing protease inhibitor cocktail for different time periods (0 to 30 min), and cell surface levels of hGPR83 were measured using anti-rabbit antibody coupled to horseradish peroxidase as described above. In a parallel set of studies, CHO cells expressing myc-tagged mGPR171 (2 × 105 cells) without or with untagged hGPR83 were labeled with myc antibodies raised in rabbit (1:1000 in PBS containing 1% FBS) and treated with 100 nM of bigLEN for different time periods (0 to 30 min) in media containing protease inhibitor cocktail, and cell surface levels of mGPR171 were measured as described above.
Proximity ligation assay
Mice were deeply anesthetized with ketamine (100 mg/kg, intraperitoneally) and transcardially perfused through the ascending aorta with 4% paraformaldehyde (200 ml). Brains were postfixed for 1 hour with 4% paraformaldehyde and stored in 1× PBS. PLA was performed on coronal brain slices (40 μm) containing hypothalamus. The presence/absence of receptor-receptor molecular proximity in these samples was detected using the Duolink II in situ PLA detection kit (Sigma-Aldrich). For cells, CHO cells expressing myc-tagged mGPR171, mGPR83, or both receptors were fixed in 4% paraformaldehyde for 15 min and washed with PBS containing 20 mM glycine, and PLA was carried out as described below. Briefly, the GPR171 antibodies raised in rabbit [cat. # GTX108131, GeneTex; for receptor specificity of this antibody, see (13)] were linked to a plus PLA probe, and the GPR83 antibody raised in rabbit (Abcam; for receptor specificity of this antibody, see fig. S8) was linked to a minus PLA probe following the manufacturer’s instructions. After incubation for 1 hour at 37°C with the blocking solution in a preheated humidity chamber, cells were incubated overnight with the PLA probe-linked antibodies (final concentration of 132 μg/ml) at 4°C. The cells were washed with buffer A (0.01 M tris-Cl pH 7.4 containing 0.15 M NaCl and 0.05% Tween 20) at room temperature and then incubated with the ligation solution for 1 hour at 37°C in a humidity chamber. After washes with buffer A, samples were incubated with the amplification solution for 100 min at 37°C in a humidity chamber and then washed with buffer B [0.2 M tris-Cl (pH 7.5) containing 0.1 M NaCl] followed by another wash with 100× diluted buffer B. Samples were mounted using an aqueous mounting medium with DAPI (nuclear staining) and visualized using Leica DM6000 microscope at the Microscopy CORE at the Icahn School of Medicine at Mount Sinai.
Data and statistical analyses
Results are expressed as means ± SE. Data with one variable were analyzed using one-way ANOVAs, whereas data with two variables were analyzed with two-way ANOVAs followed by Tukey’s multiple comparisons or Dunnett’s post hoc comparisons. Data comparing two groups were analyzed using two-tailed unpaired Student’s t tests. Significance was set at P < 0.05. Statistical analyses of data were generated by using Prism software (version 6.0; GraphPad Software). For details for each analysis, see table S1.
Supplementary Material
Acknowledgments
We thank C. Moraje and K. Gagnidze for technical support and members of the Devi Lab for active scientific discussions.
Funding: This work was supported by NIH awards DA008863 and NS026880 to L.A.D., DA030529 to E.B.M., and DA004494 to L.D.F. E.N.B. is supported by T32 DA007135. S.S. is supported by a grant from Alfonso Martin Escudero Foundation. This work was also supported by the Deutsche Forschungsgemeinschaft (DFG; TS226/1-1) and Else Kröner-Fresenius-Stiftung (EKFS) project 2014_A114.
Footnotes
Author contributions: L.A.D., I.G., E.B.M., and L.D.F. designed the research; I.G., E.N.B., A.G., S.S., E.B.M., W.F., A.M., and A.K.F. performed the research; I.G., L.A.D., S.S., E.B.M., A.K.F., and A.G. analyzed the data; T.D.M., M.H.T., and G.K. supplied reagents and helped design some of the studies; L.A.D., I.G., A.K.F., and L.D.F. wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The GPR83 knockout mice can be commercially obtained from Jackson Laboratories.
www.sciencesignaling.org/cgi/content/full/9/425/ra43/DC1
Fig. S1. Variable results with studies examining the effect of mPEN on adenylyl cyclase activity in hypothalamic membranes.
Fig. S2. The binding of mPEN to hypothalamic membranes from individual male mice.
Fig. S3. Variable results with studies examining the effect of mPEN on PLC activity in hippocampal membranes.
Fig. S4. Evoked EPSC amplitude after sequential application of increasing concentrations of mPEN and washout.
Fig. S5. mPEN-stimulated neurite outgrowth in Neuro2A cells.
Fig. S6. Expression of GPR83 in heterologous cells confers PEN signaling and receptor endocytosis.
Fig. S7. Quantitative RT-PCR to confirm the presence of GPR83 mRNA in Neuro2A cells.
Fig. S8. Specificity of the GPR83 and GPR171 antibodies.
Fig. S9. Confirmation of antibody specificity and accuracy of colocalization by analysis of the lateral septum.
Table S1. Description of statistical analysis for different figures.
REFERENCES AND NOTES
- 1.Fricker LD, McKinzie AA, Sun J, Curran E, Qian Y, Yan L, Patterson SD, Courchesne PL, Richards B, Levin N, Mzhavia N, Devi LA, Douglass J. Identification and characterization of proSAAS, a granin-like neuroendocrine peptide precursor that inhibits prohormone processing. J Neurosci. 2000;20:639–648. doi: 10.1523/JNEUROSCI.20-02-00639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fricker LD. Analysis of mouse brain peptides using mass spectrometry-based peptidomics: Implications for novel functions ranging from non-classical neuropeptides to microproteins. Mol BioSyst. 2010;6:1355–1365. doi: 10.1039/c003317k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wardman JH, Zhang X, Gagnon S, Castro LM, Zhu X, Steiner DF, Day R, Fricker LD. Analysis of peptides in prohormone convertase 1/3 null mouse brain using quantitative peptidomics. J Neurochem. 2010;114:215–225. doi: 10.1111/j.1471-4159.2010.06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Pan H, Peng B, Steiner DF, Pintar JE, Fricker LD. Neuropeptidomic analysis establishes a major role for prohormone convertase-2 in neuropeptide biosynthesis. J Neurochem. 2010;112:1168–1179. doi: 10.1111/j.1471-4159.2009.06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Che F-Y, Berezniuk I, Sonmez K, Toll L, Fricker LD. Peptidomics of Cpefat/fat mouse brain regions: Implications for neuropeptide processing. J Neurochem. 2008;107:1596–1613. doi: 10.1111/j.1471-4159.2008.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayah M, Fortenberry Y, Cameron A, Lindberg I. Tissue distribution and processing of proSAAS by proprotein convertases. J Neurochem. 2001;76:1833–1841. doi: 10.1046/j.1471-4159.2001.00165.x. [DOI] [PubMed] [Google Scholar]
- 7.Fricker LD. Neuropeptides and Other Bioactive Peptides: From Discovery to Function. Morgan & Claypool Life Sciences; Princeton, NJ: 2012. pp. 1–109. [Google Scholar]
- 8.Wardman JH, Berezniuk I, Di S, Tasker JG, Fricker LD. ProSAAS-derived peptides are colocalized with neuropeptide Y and function as neuropeptides in the regulation of food intake. PLOS One. 2011;6:e28152. doi: 10.1371/journal.pone.0028152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luquet S, Perez FA, Hnasko TS, Palmiter RD. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310:683–685. doi: 10.1126/science.1115524. [DOI] [PubMed] [Google Scholar]
- 10.Krashes MJ, Koda S, Ye C, Rogan SC, Adams AC, Cusher DS, Maratos-Flier E, Roth BL, Lowell BB. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei S, Feng Y, Che F-Y, Pan H, Mzhavia N, Devi LA, McKinzie AA, Levin N, Richards WG, Fricker LD. Obesity and diabetes in transgenic mice expressing proSAAS. J Endocrinol. 2004;180:357–368. doi: 10.1677/joe.0.1800357. [DOI] [PubMed] [Google Scholar]
- 12.Morgan DJ, Wei S, Gomes I, Czyzyk T, Mzhavia N, Pan H, Devi LA, Fricker LD, Pintar JE. The propeptide precursor proSAAS is involved in fetal neuropeptide processing and body weight regulation. J Neurochem. 2010;113:1275–1284. doi: 10.1111/j.1471-4159.2010.06706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes I, Aryal DK, Wardman JH, Gupta A, Gagnidze K, Rodriguiz RM, Kumar S, Wetsel WC, Pintar JE, Fricker LD, Devi LA. GPR171 is a hypothalamic G protein-coupled receptor for BigLEN, a neuropeptide involved in feeding. Proc Natl Acad Sci USA. 2013;110:16211–16216. doi: 10.1073/pnas.1312938110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan DJ, Mzhavia N, Peng B, Pan H, Devi LA, Pintar JE. Embryonic gene expression and pro-protein processing of proSAAS during rodent development. J Neurochem. 2005;93:1454–1462. doi: 10.1111/j.1471-4159.2005.03138.x. [DOI] [PubMed] [Google Scholar]
- 15.Dubois CJ, Ramamoorthy P, Whim MD, Liu SJ. Activation of NPY type 5 receptors induces a long-lasting increase in spontaneous GABA release from cerebellar inhibitory interneurons. J Neurophysiol. 2012;107:1655–1665. doi: 10.1152/jn.00755.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomes I, Grushko JS, Golebiewska U, Hoogendoorn S, Gupta A, Heimann AS, Ferro ES, Scarlata S, Fricker LD, Devi LA. Novel endogenous peptide agonists of cannabinoid receptors. FASEB J. 2009;23:3020–3029. doi: 10.1096/fj.09-132142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fricker AD, Rios C, Devi LA, Gomes I. Serotonin receptor activation leads to neurite outgrowth and neuronal survival. Brain Res Mol Brain Res. 2005;138:228–235. doi: 10.1016/j.molbrainres.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 18.Rozenfeld R, Gupta A, Gagnidze K, Lim MP, Gomes I, Lee-Ramos D, Nieto N, Devi LA. AT1R-CB1R heteromerization reveals a new mechanism for the pathogenic properties of angiotensin II. EMBO J. 2011;30:2350–2363. doi: 10.1038/emboj.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bromberg KD, Ma’ayan A, Neves SR, Iyengar R. Design logic of a cannabinoid receptor signaling network that triggers neurite outgrowth. Science. 2008;320:903–909. doi: 10.1126/science.1152662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regard JB, Sato IT, Coughlin SR. Anatomical profiling of G protein-coupled receptor expression. Cell. 2008;135:561–571. doi: 10.1016/j.cell.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen T-M, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong H-W, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf K-R, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 22.Gomes I, Ayoub MA, Fujita W, Jaeger WC, Pfleger KDG, Devi LA. G protein–coupled receptor heteromers. Annu Rev Pharmacol Toxicol. 2016;56:403–425. doi: 10.1146/annurev-pharmtox-011613-135952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sah R, Parker SL, Sheriff S, Eaton K, Balasubramaniam A, Sallee FR. Interaction of NPY compounds with the rat glucocorticoid-induced receptor (GIR) reveals similarity to the NPY-Y2 receptor. Peptides. 2007;28:302–309. doi: 10.1016/j.peptides.2006.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Moerlooze L, Williamson J, Liners F, Perret J, Parmentier M. Cloning and chromosomal mapping of the mouse and human genes encoding the orphan glucocorticoid-induced receptor (GPR83) Cytogenet Cell Genet. 2000;90:146–150. doi: 10.1159/000015650. [DOI] [PubMed] [Google Scholar]
- 25.Hansen W, Westendorf AM, Toepfer T, Mauel S, Geffers R, Gruber AD, Buer J. Inflammation in vivo is modulated by GPR83 isoform-4 but not GPR83 isoform-1 expression in regulatory T cells. Genes Immun. 2010;11:357–361. doi: 10.1038/gene.2010.5. [DOI] [PubMed] [Google Scholar]
- 26.Toms C, Jessup H, Thompson C, Baban D, Davies K, Powrie F. Gpr83 expression is not required for the maintenance of intestinal immune homeostasis and regulation of T-cell-dependent colitis. Immunology. 2008;125:302–312. doi: 10.1111/j.1365-2567.2008.02857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller TD, Müller A, Yi C-X, Habegger KM, Meyer CW, Gaylinn BD, Finan B, Heppner K, Trivedi C, Bielohuby M, Abplanalp W, Meyer F, Piechowski CL, Pratzka J, Stemmer K, Holland J, Hembree J, Bhardwaj N, Raver C, Ottaway N, Krishna R, Sah R, Sallee FR, Woods SC, Perez-Tilve D, Bidlingmaier M, Thorner MO, Krude H, Smiley D, DiMarchi R, Hofmann S, Pfluger PT, Kleinau G, Biebermann H, Tschöp MH. The orphan receptor Gpr83 regulates systemic energy metabolism via ghrelin-dependent and ghrelin-independent mechanisms. Nat Commun. 2013;4:1968. doi: 10.1038/ncomms2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wardman JH, Fricker LD. ProSAAS-derived peptides are differentially processed and sorted in mouse brain and AtT-20 cells. PLOS One. 2014;9:e104232. doi: 10.1371/journal.pone.0104232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatcher NG, Atkins N, Jr, Annangudi SP, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. Mass spectrometry-based discovery of circadian peptides. Proc Natl Acad Sci USA. 2008;105:12527–12532. doi: 10.1073/pnas.0804340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pesini P, Detheux M, Parmentier M, Hökfelt T. Distribution of a glucocorticoid-induced orphan receptor (JP05) mRNA in the central nervous system of the mouse. Brain Res Mol Brain Res. 1998;57:281–300. doi: 10.1016/s0169-328x(98)00099-0. [DOI] [PubMed] [Google Scholar]
- 31.Brézillon S, Detheux M, Parmentier M, Hökfelt T, Hurd YL. Distribution of an orphan G-protein coupled receptor (JP05) mRNA in the human brain. Brain Res. 2001;921:21–30. doi: 10.1016/s0006-8993(01)03068-2. [DOI] [PubMed] [Google Scholar]
- 32.Sah R, Pritchard LM, Richtand NM, Ahlbrand R, Eaton K, Sallee FR, Herman JP. Expression of the glucocorticoid-induced receptor mRNA in rat brain. Neuroscience. 2005;133:281–292. doi: 10.1016/j.neuroscience.2005.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baughman G, Harrigan MT, Campbell NF, Nurrish SJ, Bourgeois S. Genes newly identified as regulated by glucocorticoids in murine thymocytes. Mol Endocrinol. 1991;5:637–644. doi: 10.1210/mend-5-5-637. [DOI] [PubMed] [Google Scholar]
- 34.Harrigan MT, Campbell NF, Bourgeois S. Identification of a gene induced by glucocorticoids in murine T-cells: A potential G protein-coupled receptor. Mol Endocrinol. 1991;5:1331–1338. doi: 10.1210/mend-5-9-1331. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Herman JP, Pritchard LM, Spitzer RH, Ahlbrand RL, Kramer GL, Petty F, Sallee FR, Richtand NM. Cloning, expression, and regulation of a glucocorticoid-induced receptor in rat brain: Effect of repetitive amphetamine. J Neurosci. 2001;21:9027–9035. doi: 10.1523/JNEUROSCI.21-22-09027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams F, Grassie M, Shahid M, Hill DR, Henry B. Acute oral dexamethasone administration reduces levels of orphan GPCR glucocorticoid-induced receptor (GIR) mRNA in rodent brain: Potential role in HPA-axis function. Brain Res Mol Brain Res. 2003;117:39–46. doi: 10.1016/s0169-328x(03)00280-8. [DOI] [PubMed] [Google Scholar]
- 37.Vollmer LE, Ghosal S, Rush JA, Sallee FR, Herman JP, Weinert M, Sah R. Attenuated stress-evoked anxiety, increased sucrose preference and delayed spatial learning in glucocorticoid-induced receptor-deficient mice. Genes Brain Behav. 2013;12:241–249. doi: 10.1111/j.1601-183X.2012.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller A, Kleinau G, Piechowski CL, Müller TD, Finan B, Pratzka J, Grüters A, Krude H, Tschöp M, Biebermann H. G-protein coupled receptor 83 (GPR83) signaling determined by constitutive and zinc(II)-induced activity. PLOS One. 2013;8:e53347. doi: 10.1371/journal.pone.0053347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller A, Leinweber B, Fischer J, Müller TD, Grüters A, Tschöp MH, Knäuper V, Biebermann H, Kleinau G. The extracellular N-terminal domain of G-protein coupled receptor 83 regulates signaling properties and is an intramolecular inverse agonist. BMC Res Notes. 2014;7:913. doi: 10.1186/1756-0500-7-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin AL, Steurer MA, Aronstam RS. Constitutive activity among orphan class-A G protein coupled receptors. PLOS One. 2015;10:e0138463. doi: 10.1371/journal.pone.0138463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen W, Loser K, Westendorf AM, Bruder D, Pfoertner S, Siewert C, Huehn J, Beissert S, Buer J. G protein-coupled receptor 83 overexpression in naive CD4+CD25−T cells leads to the induction of Foxp3+ regulatory T cells in vivo. J Immunol. 2006;177:209–215. doi: 10.4049/jimmunol.177.1.209. [DOI] [PubMed] [Google Scholar]
- 42.Hansen W, Westendorf AM, Buer J. Regulatory T cells as targets for immunotherapy of autoimmunity and inflammation. Inflamm Allergy Drug Targets. 2008;7:217–223. doi: 10.2174/187152808786848360. [DOI] [PubMed] [Google Scholar]
- 43.Lu L-F, Gavin MA, Rasmussen JP, Rudensky AY. G protein-coupled receptor 83 is dispensable for the development and function of regulatory T cells. Mol Cell Biol. 2007;27:8065–8072. doi: 10.1128/MCB.01075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakraborty TR, Tkalych O, Nanno D, Garcia AL, Devi LA, Salton SRJ. Quantification of VGF- and pro-SAAS-derived peptides in endocrine tissues and the brain, and their regulation by diet and cold stress. Brain Res. 2006;1089:21–32. doi: 10.1016/j.brainres.2006.02.124. [DOI] [PubMed] [Google Scholar]
- 45.Mzhavia N, Berman Y, Che F-Y, Fricker LD, Devi LA. ProSAAS processing in mouse brain and pituitary. J Biol Chem. 2001;276:6207–6213. doi: 10.1074/jbc.M009067200. [DOI] [PubMed] [Google Scholar]
- 46.Lindberg I, Yang H-YT. Distribution of met5-enkephalin-arg6-gly7-leu8-immunoreactive peptides in rat brain: Presence of multiple molecular forms. Brain Res. 1984;299:73–78. doi: 10.1016/0006-8993(84)90789-3. [DOI] [PubMed] [Google Scholar]
- 47.Gupta A, Décaillot FM, Gomes I, Tkalych O, Heimann AS, Ferro ES, Devi LA. Conformation state-sensitive antibodies to G-protein-coupled receptors. J Biol Chem. 2007;282:5116–5124. doi: 10.1074/jbc.M609254200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomes I, Ijzerman AP, Ye K, Maillet EL, Devi LA. G protein-coupled receptor heteromerization: A role in allosteric modulation of ligand binding. Mol Pharmacol. 2011;79:1044–1052. doi: 10.1124/mol.110.070847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta A, Mulder J, Gomes I, Rozenfeld R, Bushlin I, Ong E, Lim M, Maillet E, Junek M, Cahill CM, Harkany T, Devi LA. Increased abundance of opioid receptor heteromers after chronic morphine administration. Sci Signaling. 2010;3:ra54. doi: 10.1126/scisignal.2000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Silva NS, Quinn PA. Rapid screening assay for phospholipase C activity in mycoplasmas. J Clin Microbiol. 1987;25:729–731. doi: 10.1128/jcm.25.4.729-731.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta A, Gomes I, Wardman J, Devi LA. Opioid receptor function is regulated by post-endocytic peptide processing. J Biol Chem. 2014;289:19613–19626. doi: 10.1074/jbc.M113.537704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta A, Fujita W, Gomes I, Bobeck E, Devi LA. Endothelin-converting enzyme 2 differentially regulates opioid receptor activity. Br J Pharmacol. 2015;172:704–719. doi: 10.1111/bph.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.