Abstract
The emergence of fluoroquinolone resistance in sterile-site isolates of Streptococcus pneumoniae is documented in this study characterizing all invasive levofloxacin-resistant (MIC, ≥8 mg/liter) S. pneumoniae isolates (n = 50) obtained from the Centers for Disease Control and Prevention Active Bacterial Core Surveillance from 1998 to 2002. Resistance among all isolates increased from 0.1% in 1998 to 0.6% in 2001 (P = 0.008) but decreased to 0.4% in 2002, while resistance among vaccine serotypes continued to increase from 0.3% in 1998 to 1.0% in 2002, suggesting that fluoroquinolones continue to exert selective pressure on these vaccine serotypes. Only 22% of resistant isolates were not covered by the conjugate vaccine serogroups. Multilocus sequence typing revealed that 58% of resistant strains were related to five international clones identified by the Pneumococcal Molecular Epidemiology Network, with the Spain23F-1 clone being most frequent (16% of all isolates). Thirty-six percent of the isolates were coresistant to penicillin, 44% were coresistant to macrolides, and 28% were multiresistant to penicillin, macrolides, and fluoroquinolones. Fifty percent of the isolates were resistant to any three drug classes. Ninety-four percent of the isolates had multiple mutations in the quinolone resistance-determining regions of the gyrA, gyrB, parC, and parE genes. In 16% of the isolates, there was evidence of an active efflux mechanism. An unusual isolate was found that showed only a single parE mutation and for which the ciprofloxacin MIC was lower (2 mg/liter) than that of levofloxacin (8 mg/liter). Our results suggest that invasive pneumococcal isolates resistant to levofloxacin in the United States show considerable evidence of multiple resistance and of clonal spread.
Streptococcus pneumoniae is a common cause of invasive bacterial disease worldwide and occurs in all age groups. It is a major pathogen causing community-acquired pneumonia and acute exacerbations of chronic bronchitis, meningitis, sinusitis, and otitis media. Invasive pneumococcal disease (IPD) is defined as the isolation of S. pneumoniae from a normally sterile site, e.g., blood, cerebrospinal fluid, or pleural fluid. Before the introduction of conjugate vaccine, the overall incidence of IPD reported in the United States was 23.2 per 100,000 population (35). Overall, 10% of those with invasive disease die, but the case fatality ratio can be as high as 30.8% among residents of long-term care facilities aged 65 years or older (24). One of the determinants of outcome in IPD is the appropriateness of antimicrobial therapy (12).
Whereas most studies have shown no increase in mortality in patients with invasive disease infected with penicillin-resistant pneumococci (11, 16), fluoroquinolone resistance has been associated with clinical failure (7). Therefore, monitoring of fluoroquinolone resistance in invasive pneumococci is of both clinical and epidemiological importance.
Numerous studies of penicillin-resistant pneumococci in the past 30 years show that the emerging increase in penicillin resistance is mainly driven by the spread of a few successful clones (23) that are also able to switch their serotypes. Twenty-six international multiresistant clones (30; http://www.sph.emory.edu/PMEN) have been defined. The assignment of isolates to one of the international clones is based on multilocus sequence typing (MLST) or pulsed-field gel electrophoresis (PFGE) patterns of restriction enzyme-digested DNA.
In contrast to penicillin resistance, the importance of clonal spread for the distribution of fluoroquinolone resistance is controversial. In some areas, such as Hong Kong, there has been an alarming increase in fluoroquinolone resistance (19), and genetic analysis revealed that the overwhelming majority of strains were identical or closely related to a single clone (18). In contrast, surveillance studies to date from North America have shown only a low prevalence of fluoroquinolone resistance, and genetic analysis of these isolates revealed a wider genetic diversity with only small numbers related to the international clones (5, 9, 20, 42).
Resistance to fluoroquinolones in pneumococci occurs in a stepwise fashion, with mutations observed in either parC or gyrA (depending on the selecting fluoroquinolone) or both, leading to decreased fluoroquinolone susceptibility (33). Strains usually become fully fluoroquinolone resistant with the addition of a mutation in the other target gene (either gyrA or parC) (33). Mutations in parE and gyrB may contribute to resistance, and efflux also contributes to ciprofloxacin resistance but influences resistance to other more active fluoroquinolones, usually to a lesser extent (15, 21, 33, 35, 38). The objectives of this study were to determine whether invasive levofloxacin-resistant (levofloxacin MICs, ≥8 mg/liter) clinical isolates of S. pneumoniae collected from 1998 to 2002 in the Active Bacterial Core Surveillance (ABCS) sites were cross-resistant to other fluoroquinolones and to classes of other antibiotics and whether these isolates were genetically related to each other or to the major pandemic clones and to explore the mechanisms of fluoroquinolone resistance in these isolates.
MATERIALS AND METHODS
Isolates.
All sterile-site isolates for which the levofloxacin MIC was ≥8 mg/liter that were collected from 1998 to 2002 by the Centers for Disease Control and Prevention (CDC) as part of the ABCS were studied. The methods for case identification and isolate collection have been previously described (42). The isolates were recovered from California (San Francisco County), Connecticut, Georgia (20-county Atlanta area), Maryland (6-county Baltimore area), New York (7-county Rochester and 8-county Albany areas), Oregon (3-county Portland area), and Tennessee (11 urban counties). The surveillance areas comprised ∼18.5 million persons at the beginning of 1999, and this increased to ∼19.8 million by mid-1999 with the addition of six counties in Tennessee.
Serotyping.
Serotyping of strains was performed at the CDC by the standard Quellung method, as described previously (14).
Antimicrobial susceptibility testing and efflux pump testing.
Susceptibility to fluoroquinolones (levofloxacin, ciprofloxacin, moxifloxacin, gatifloxacin, gemifloxacin, and garenoxacin) was determined by the agar dilution method in O2 according to standard procedures (31, 39). Fluoroquinolones were provided by the manufacturers.
The presence of an efflux pump was investigated by determination of the MICs of ciprofloxacin by the agar dilution method in the presence of reserpine (10 mg/liter). A fourfold decrease in the MIC in the presence of reserpine (two dilution steps) was considered evidence for the presence of an efflux mechanism. Data on susceptibility to other antibiotics were obtained from the CDC ABCS database. The MICs in this database were determined by broth microdilution according to NCCLS guidelines (31) at the CDC and the University of Texas Health Science Center at San Antonio.
Levofloxacin MICs were tested by both broth dilution and agar dilution and revealed similar results (all MICs are ±1 dilution).
PFGE.
Genomic DNA was prepared in situ in agarose blocks as described previously (25, 28) and was digested with SmaI (Life Technologies, Gaithersburg, Md.). The fragments were resolved by PFGE in 1% agarose (SeaKem GTG agarose; BioWhittaker Molecular Applications, Rockland, Maine) in 0.5× Tris-borate-EDTA buffer at 14°C and 6 V/cm in a CHEF-DR III system (Bio-Rad Laboratories, Hercules, Calif.). The parameters for block 1 were an initial pulse time of 1 s increased to 30 s over 17 h; those for block 2 were 5 s increased to 9 s over 6 h. Bionumerics software (Applied-Maths, Kortrijk, Belgium) was used to construct unweighted pair group method with arithmetic means dendrograms of fragment patterns with the Dice coefficient. The Dice similarity coefficient was used with optimization and position tolerance settings of 1.0 and 1.5%, respectively. PFGE-based clusters were defined as isolates with ≥80% genetic relatedness on the dendrogram.
MLST.
MLST was performed as previously described (10) using the modified primers designed by Gertz et al. (13). Sequencing was performed with an ABI 3100 apparatus (Applied Biosystems, Foster City, Calif.). For analysis, MLST alleles were downloaded from http://www.mlst.net for convenient screening with the Wisconsin version 10.2 software package. Sequence types (STs) were assigned by comparing the allelic profile to the reference ST set at http://www.mlst.net. Within the analyzed isolates, three new STs were discovered and submitted to http://www.mlst.net.
PCR and DNA sequencing of the QRDR.
The quinolone resistance-determining regions (QRDRs) of the topoisomerase type II genes parC, parE, gyrA, and gyrB were amplified from extracted chromosomal DNA by PCR using the primers and cycling conditions described by Pan et al. (33). The amplification products were purified with ExoSAP-IT (USB Corp., Cleveland, Ohio). DNA sequencing was performed using BigDye Terminator 1.1 Cycle (Applied Biosystems) with the ABI 3100 automated sequencer.
Protein sequence accession numbers.
The National Center for Biotechnology Information Blastx program (http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/bl2.html) was used for comparison of the QRDR nucleotide sequences with the protein sequences of wild-type S. pneumoniae (R6). The following protein sequence accession numbers were used as references: gyrA, gi1503142; gyrB, gi 15902759; parC, gi 15902801; and parE, gi 15902800.
RESULTS
Demographics and trends.
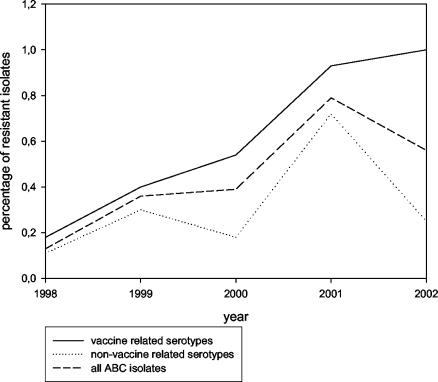
Fifty levofloxacin-resistant isolates (MICs, ≥8 mg/liter) out of 15,432 ABC isolates tested (overall prevalence, 0.32%) were identified in eight states from 1998 to 2002 (Table 1). The absolute number of strains detected increased annually from 1998 to 2001 and decreased in 2002. The proportion of resistant strains increased from 0.1% in 1998 to 0.6% in 2001 (P = 0.008; Yates corrected chi-square test) but decreased to 0.4% in 2002. Resistance among conjugate vaccine serotypes continuously increased from 0.3% in 1998 to 1.0% in 2002 (Table 1 and Fig. 1).
TABLE 1.
Proportions of levofloxacin-resistant isolates per year among sterile-site pneumococci collected through ABCs
| Parameter | Result in:
|
||||
|---|---|---|---|---|---|
| 1998 | 1999 | 2000 | 2001 | 2002 | |
| Total no. of isolates | 2,941 | 3,403 | 3,469 | 3,092 | 2,553 |
| No. of vaccine serotypesa | 1,099 | 1,265 | 1,292 | 1,178 | 800 |
| Total no. of LFX-resistant isolates | 4 | 8 | 9 | 18 | 11 |
| No. of LFX-resistant isolates vaccine serotypesa | 3 | 4 | 7 | 11 | 8 |
| Proportion of LFX resistance among all isolates (%) | 0.1 | 0.2 | 0.3 | 0.6 | 0.4 |
| Proportion of LFX resistance among vaccine serotypes (%)a | 0.3 | 0.3 | 0.5 | 0.9 | 1.0 |
Isolates of serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F recovered from patients >35 years of age.
FIG. 1.
Proportion of levofloxacin-resistant invasive isolates per year among vaccine serotypes and nonvaccine serotypes recovered from patients ≥35 years old (ABCS, 1998 to 2002).
Most isolates came from residents of Connecticut (n = 18), Maryland (n = 10), and Georgia (n = 9). The majority of strains were recovered from blood (n = 44; 88%), three strains were isolated from pleural fluid, two from synovial fluid, and one from peritoneal fluid. The most common diagnosis was pneumonia (n = 40; 80%). In five cases of bacteremia, no focus could be identified. Three patients presented with cellulitis, and one each presented with septic arthritis and peritonitis. No isolates from children were identified. The median age was 77 years (range, 36 to 98 years), with 50% of the patients between 61 and 85 years.
Serotypes and antimicrobial susceptibilities.
Seventeen different serotypes were represented. The majority of the strains belonged to serotypes 6B (n = 9; 18%), 23F (n = 6; 12%), and 14 (n = 6; 12%). Six percent (n = 3) of the strains were of serotype 35B, a type not found in the 23-valent vaccine.
The MICs for six fluoroquinolones and other antibiotics tested are shown in Table 2. Eighteen isolates (36%) were resistant to penicillin, 22 (44%) were resistant to macrolides, and 14 (28%) exhibited resistance to both. Twenty-five of the isolates (50%) were multiresistant (i.e., resistant to ≥3 different classes of antibiotics). Strains of serotypes 14 and 23F were all multiresistant. Isolates of serotypes 3 (n = 3), 18C (n = 3), and 22F (n = 2) were resistant only to fluoroquinolones and susceptible to all other tested antibiotics.
TABLE 2.
Antibiotic susceptibilities of 50 investigated levofloxacin-resistant isolates
| Drug | MIC50 (μg/ml) | MIC90 (μg/ml) | Range (μg/ml) | % Nonsusceptible | % Resistant |
|---|---|---|---|---|---|
| Levofloxacin | 16 | 16 | 8-32 | 100 | 100 |
| Ciprofloxacin | 32 | 64 | 2-64 | 100 | 98 |
| Garenoxacin | 0.5 | 1 | 0.125-2 | NAa | NAa |
| Gatifloxacin | 4 | 8 | 0.25-8 | 92 | 84 |
| Moxifloxacin | 2 | 4 | 0.125-4 | 94 | 30 |
| Gemifloxacin | 0.25 | 0.5 | 0.125-1 | 44 | 44 |
| Penicillin | 0.06 | 4 | 0.03-8 | 42 | 36 |
| Amoxicillin | 0.06 | 2 | 0.03-8 | 28 | 18 |
| Cefuroxime | 0.25 | 8 | 0.12-8 | 42 | 42 |
| Cefotaxime | 0.06 | 2 | 0.06-8 | 32 | 14 |
| Meropenem | 0.12 | 1 | 0.06-2 | 38 | 20 |
| Erythromycin | 0.12 | 32 | 0.03-64 | 44 | 44 |
| Clindamycin | 0.06 | 2 | 0.03-4 | 14 | 14 |
| Chloramphenicol | 4 | 16 | 2-16 | 18 | 18 |
| Rifampin | 2 | 4 | 1-4 | 2 | 0 |
| Tetracycline | 2 | 8 | 1-8 | 20 | 20 |
| Co-trimoxazole | 2 | 8 | 0.12-8 | 50 | 46 |
| Vancomycin | 0.25 | 0.5 | 0.12-0.5 | 0 | 0 |
| Quinupristin-dalfopristin | 1 | 4 | 1-4 | 26 | 24 |
NA, not applicable, since there are no breakpoints defined yet.
Mechanisms of fluoroquinolone resistance.
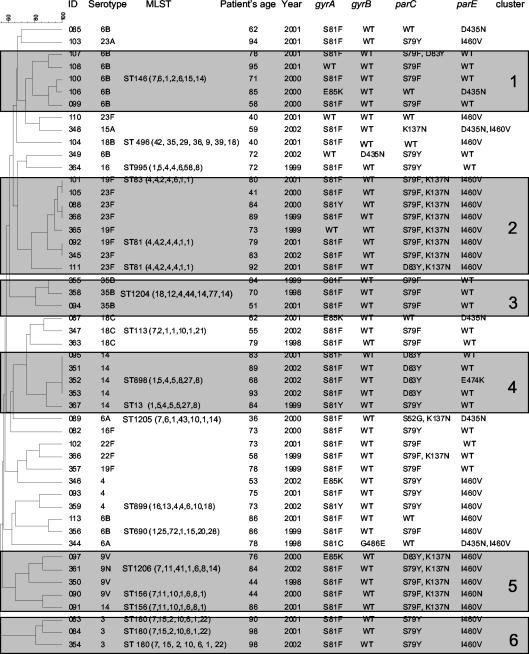
Sequencing of the QRDRs of gyrA, gyrB, parC, and parE showed that most of the isolates had mutations in both target enzymes (n = 47; 94%) (Fig. 2). In 13 strains, as many as four mutations could be detected. Mutations were found most frequently in the products of gyrA (n = 43) and parC (n = 38). The majority of the isolates also displayed mutations in the product of parE (n = 30), but only two strains with mutations in the product of gyrB were found. In the gyrA product, only single mutations were found, whereas 13 strains had double mutations in the product of parC and four had double or triple mutations in the product of parE. Isolate 110 displayed only the I460V mutation in the product of parE. In this isolate, no evidence for an efflux pump was obtained, suggesting the existence of a novel mechanism of resistance that may include the presence of mutations outside the QRDR. Unusually, the ciprofloxacin MIC for this isolate (2 mg/liter) was fourfold lower than the levofloxacin MIC for it.
FIG. 2.
Genetic relatedness of levofloxacin-resistant invasive isolates in the United States. ID, identification number; Year, year collected; WT, wild type.
Thirteen different mutations could be detected: four in the product of gyrA (at S81 and E85), two in the product of gyrB (at G486 and D435), four in the product of parC (at S79, D83, and K137), and three in the product of parE (at I460, D435, and E474). The presence or absence of parE and gyrB gene product mutations had no discernible influence on the MICs.
In 8 out of the 50 isolates (16%), there was evidence for the presence of an active efflux mechanism. Isolates without mutations in the parC product (n = 7) exhibited significantly more frequent efflux (43%) than isolates with mutations in the parC product (n = 43; 11% with efflux; P = 0.04; Wilcoxon signed rank test). The ciprofloxacin MICs were most affected by the presence of an efflux pump. The pump had less effect on levofloxacin MICs and had no effect on moxifloxacin MICs (data not shown).
PFGE, MLST, and comparison with international clones.
The PFGE analysis revealed that the majority of the isolates (n = 41; 82%) could be grouped into eight smaller clusters (defined as a group of strains with n of ≥2) and four large clusters (n ≥ 5). Interestingly, the majority of the strains within one cluster (cluster 2), related to the international clone Spain23F-1, displayed identical or very similar QRDR patterns (Fig. 3). These strains were isolated from four states across the United States. Cluster 2, the largest identified, included eight isolates with serotypes 23F (n = 5) and 19F (n = 3). ST 81, which is the ST of the Spain23F-1 clone, was identified in two isolates, and ST 83 was identified in another isolate (a single-locus variant of ST 81).
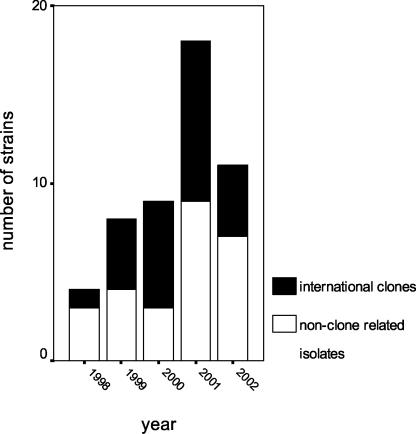
FIG. 3.
Distribution of international clones among levofloxacin-resistant invasive pneumococci by year (ABCS, 1998 to 2002).
Cluster 1 had five isolates of serotype 6B; MLST of one randomly chosen isolate identified it as ST 146. ST 146 is a double-locus variant of the international Greece6B-22 clone.
There was one cluster (cluster 3; n = 3) of the non-vaccine-related serotype 35B. The isolates were recovered from three different states, but all displayed the same mutation pattern. One of the isolates was analyzed by MLST and assigned to a new ST that is a single-locus variant of the Utah35B-24 clone. Two of these isolates were resistant to penicillin (MIC, 2 mg/liter), and one was nonsusceptible (MIC, 1 mg/liter).
Cluster 4 included four isolates of serotype 14 from Connecticut and one from Tennessee. The MLST result for one isolate of this group was ST 898, a double-locus variant of the England14-9 clone; the Tennessee isolate exhibited ST 13 (a single-locus variant of the England14-9 clone). Also in this cluster, three strains with the same mutation pattern were found; the fourth isolate had an additional E474K mutation in the product of parE. MLST analysis of two randomly chosen isolates from a cluster of five (cluster 5) assigned them to ST 156, which is the ST of the Spain9V-3 clone.
MLST of the three serotype 3 isolates from Georgia assigned them all to ST 180 (cluster 6). All three isolates displayed exactly the same mutation pattern and nearly identical MICs (±1 dilution) for all 19 tested antibiotics.
Thus, five of the international clones accounted for 26 (52%) of the isolates. The most common was Spain23F-1 (n = 8; 16%).
Figure 3 shows the numbers of the clone-related and the non-clone-related levofloxacin-resistant isolates per year.
DISCUSSION
Fluoroquinolone-resistant pneumococci are currently rare in the United States (1). To date, the highest prevalence described has been <1.3% in adults (22). Despite an increase in the prevalence of levofloxacin-resistant isolates during the surveillance period of our study (1998 to 2002), the overall proportion of levofloxacin-resistant strains has remained <1%. However, the experience with exponentially increasing resistance to penicillins and macrolides around the globe shows that clonal spread is one of the main drivers of antibiotic resistance in the pneumococcus (23).
This paper analyzes only levofloxacin-resistant strains based on the NCCLS breakpoint of 8 mg/liter. There is concern that there may be a high prevalence of single mutations in topoisomerase type II genes among levofloxacin-susceptible (MIC, ≤2 mg/liter) isolates (8, 26).
Fluoroquinolones have been the antibiotics of choice for infections caused by pneumococci resistant to penicillin and macrolides in adults or for adult patients with allergies to beta-lactams. They have recently been recommended in several guidelines for empirical therapy of community-acquired pneumonia (27, 37). The possibility of clonal spread of fluoroquinolone resistance in pneumococci, as has been seen with other antibiotics, is an issue of major concern. Whereas clonal outbreaks of fluoroquinolone-resistant strains have already been described (18, 39), large surveillance studies have shown a far wider clonal diversity among the fluoroquinolone-resistant pneumococci than among penicillin-resistant pneumococci (9, 42). Therefore, many have believed that fluoroquinolone resistance is largely associated with spontaneous mutations in unrelated susceptible pneumococci exposed to these drugs.
However, an observation supporting clonal spread as a mechanism for increasing fluoroquinolone resistance is the detection of fluoroquinolone-resistant international clones (30). The presence of fluoroquinolone resistance within the international clones has been reported for Spain23F-1 (18), Spain9V-3 (29), and recently, Tennessee23F-4 (4), Spain14-5 (34), England14-9, Taiwan19F-14, Taiwan23F-15, and Maryland6B-17 (4). As in other North American surveillance studies, we also found several isolates which were identical to the Spain23F-1 (6, 9, 20) and the Spain9V-3 (6, 20) international clones, and in addition, strains closely related to the Greece6B-22, Utah35B-24, and England14-5 clones. Altogether, 25 isolates (50%) were identical or closely related to international clones. These results show a higher degree of clonality than that found among U.S. levofloxacin-resistant (MIC, ≥8 mg/liter) isolates from the respiratory tract reported in the TRUST (Tracking Resistance in the United States Today) surveillance study (25%) (9).
The absolute number and the prevalence of levofloxacin-resistant isolates increased until 2001 and appeared to decline in 2002. Likewise, the proportion of clones among the levofloxacin-resistant isolates increased, reaching 75% in 2000, and decreased in 2002. The decrease may be related to the introduction of the pneumococcal conjugate vaccine in 2000, since the serotypes of the great majority of the clones are covered by the vaccine. However, a separate analysis of conjugate vaccine serotypes (Fig. 2) showed that (i) the proportion of levofloxacin-resistant strains in this group is higher than that in nonvaccine serotypes and that in the overall pneumococcal population and (ii) this proportion continuously increased during the surveillance period. Therefore, the introduction of the vaccine may have changed a continuous upward trend in levofloxacin-resistant strains by reducing the absolute numbers of levofloxacin-resistant strains within vaccine serotypes, but selection for fluoroquinolone resistance among the diminishing numbers of these vaccine serotypes continues to occur.
An observation in our study is the existence of fluoroquinolone-resistant serotype 3 isolates. The serotype 3 isolates in our study all displayed the same ST 180 and exhibited identical QRDR mutations and MICs. They were all recovered from patients in the state of Georgia. Serotype 3 is considered the most virulent of S. pneumoniae serotypes, and it is commonly associated with invasive disease in adults. In addition, pneumococci with this serotype and ST 180 tend to spread clonally (13, 17). However, serotype 3 isolates have not yet been defined as one of the major antibiotic-resistant international clones, since most serotype 3 isolates are broadly susceptible to antibiotics (32, 36).
Also noteworthy is the description of a cluster of fluoroquinolone-resistant serotype 35B isolates, a serotype not in currently available vaccines. The emergence of this clone of penicillin-resistant 35B pneumococci in the United States has recently been described (2). All three isolates in our study were nonsusceptible to both penicillin and levofloxacin.
The most active of the licensed fluoroquinolones in our study were gemifloxacin, with a MIC at which 90% of isolates were inhibited (MIC90) of 0.5 mg/liter, and moxifloxacin, with a MIC90 of 4 mg/liter. Twenty-two out of the 50 isolates, however, remained resistant to gemifloxacin, while 15 isolates remained resistant to moxifloxacin. The clinical relevance of these MICs depends on the pharmacodynamic properties of the drugs.
In conclusion, our study found a higher proportion of fluoroquinolone-resistant isolates related to the international clones than has been described in other studies (3, 9, 20). The introduction of the conjugate pneumococcal vaccine in 2000 and the demonstrated impact on the pneumococcal population and distribution of serotypes in adults (40) may have changed the course of an upward trend in fluoroquinolone resistance among invasive isolates, leading to the observed decrease in resistance in 2002, by reducing the absolute numbers of levofloxacin-resistant isolates within vaccine serotypes. Among vaccine serotypes, however, we found a continuously increasing resistance rate reaching 1.0% in 2002. Ongoing surveillance is needed to closely monitor the further evolution of fluoroquinolone resistance in this era of conjugate pneumococcal vaccine use.
Acknowledgments
The study was supported by a research grant from Bayer. Mathias W. Pletz was supported by a scholarship from the German Research Foundation (Deutsche Forschungsgemeinschaft).
We thank the clinical laboratory and surveillance personnel participating in ABCS and acknowledge the CDC National Vaccine Program Office and Opportunistic Infections Working Group for their contributions to this work, and specifically, we thank Zhongya Li for assistance with automated sequencing. We thank those members of the Active Bacterial Core Surveillance Team who contributed to the present study. We also thank Bayer, Bristol-Myers Squibb, Johnson Pharmaceuticals, LG Life Sciences, Ltd., and Ortho McNeil for providing the tested fluoroquinolones.
Contributing members of the ABCS team include the following: Tamar Pilishvili, Tami Slcoff, Carolyn Wright, Chris van Beneden, and Delois Jackson (Centers for Disease Control and Prevention); Arthur Reingold (University of California at Berkeley School of Public Health); James Hadler (Connecticut Department of Public Health); Monica M. Farley (Emory University); Lee H. Harrison (University of Pittsburgh); Nancy M. Bennett (Monroe County Department of Health); Paul R. Cieslak (Oregon Department of Human Services); and William Schaffner (Vanderbilt Medical Center).
REFERENCES
- 1.Anonymous. 2001. Resistance of Streptococcus pneumoniae to fluoroquinolones—United States, 1995-1999. Morb. Mortal. Wkly. Rep. 50:800-804. [PubMed] [Google Scholar]
- 2.Beall, B., M. C. McEllistrem, R. E. Gertz, Jr., D. J. Boxrud, J. M. Besser, L. H. Harrison, J. H. Jorgensen, and C. G. Whitney. 2002. Emergence of a novel penicillin-nonsusceptible, invasive serotype 35B clone of Streptococcus pneumoniae within the United States. J. Infect. Dis. 186:118-122. [DOI] [PubMed] [Google Scholar]
- 3.Brueggemann, A. B., S. L. Coffman, P. Rhomberg, H. Huynh, L. Almer, A. Nilius, R. Flamm, and G. V. Doern. 2002. Fluoroquinolone resistance in Streptococcus pneumoniae in United States since 1994-1995. Antimicrob. Agents Chemother. 46:680-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canton, R., M. Morosini, M. C. Enright, and I. Morrissey. 2003. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the global PROTEKT surveillance programme. J. Antimicrob. Chemother. 52:944-952. [DOI] [PubMed] [Google Scholar]
- 5.Chen, D. K., A. McGeer, J. C. de Azavedo, D. E. Low, and The Canadian Bacterial Surveillance Network. 1999. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N. Engl. J. Med. 341:233-239. [DOI] [PubMed] [Google Scholar]
- 6.Corso, A., E. P. Severina, V. F. Petruk, Y. R. Mauriz, and A. Tomasz. 1998. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb. Drug Resist. 4:325-337. [DOI] [PubMed] [Google Scholar]
- 7.Davidson, R., R. Cavalcanti, J. L. Brunton, D. J. Bast, J. C. de Azavedo, P. Kibsey, C. Fleming, and D. E. Low. 2002. Resistance to levofloxacin and failure of treatment of pneumococcal pneumonia. N. Engl. J. Med. 346:747-750. [DOI] [PubMed] [Google Scholar]
- 8.Davies, T. A., A. Evangelista, S. Pfleger, K. Bush, D. F. Sahm, and R. Goldschmidt. 2002. Prevalence of single mutations in topoisomerase type II genes among levofloxacin-susceptible clinical strains of Streptococcus pneumoniae isolated in the United States in 1992 to 1996 and 1999 to 2000. Antimicrob. Agents Chemother. 46:119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, T. A., R. Goldschmidt, S. Pfleger, M. Loeloff, K. Bush, D. F. Sahm, and A. Evangelista. 2003. Cross-resistance, relatedness and allele analysis of fluoroquinolone-resistant US clinical isolates of Streptococcus pneumoniae (1998-2000). J. Antimicrob. Chemother. 52:168-175. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 11.Ewig, S., M. Ruiz, A. Torres, F. Marco, J. A. Martinez, M. Sanchez, and J. Mensa. 1999. Pneumonia acquired in the community through drug-resistant Streptococcus pneumoniae. Am. J. Respir. Crit. Care Med. 159:1835-1842. [DOI] [PubMed] [Google Scholar]
- 12.Georges, H., O. Leroy, C. Vandenbussche, B. Guery, S. Alfandari, L. Tronchon, and G. Beaucaire. 1999. Epidemiological features and prognosis of severe community-acquired pneumococcal pneumonia. Intensive Care Med. 25:198-206. [DOI] [PubMed] [Google Scholar]
- 13.Gertz, R. E., Jr., M. C. McEllistrem, D. J. Boxrud, Z. Li, V. Sakota, T. A. Thompson, R. R. Facklam, J. M. Besser, L. H. Harrison, C. G. Whitney, and B. Beall. 2003. Clonal distribution of invasive pneumococcal isolates from children and selected adults in the United States prior to 7-valent conjugate vaccine introduction. J. Clin. Microbiol. 41:4194-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gherardi, G., C. G. Whitney, R. R. Facklam, and B. Beall. 2000. Major related sets of antibiotic-resistant pneumococci in the United States as determined by pulsed-field gel electrophoresis and pbp1a-pbp2b-pbp2x-dhf restriction profiles. J. Infect. Dis. 181:216-229. [DOI] [PubMed] [Google Scholar]
- 15.Gill, M., N. P. Brenwald, and R. Wise. 1999. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:187-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein, E. J., and S. M. Garabedian-Ruffalo. 2002. Widespread use of fluoroquinolones versus emerging resistance in pneumococci. Clin. Infect. Dis. 35:1505-1511. [DOI] [PubMed] [Google Scholar]
- 17.Henriques, B., M. Kalin, A. Ortqvist, B. Olsson Liljequist, M. Almela, T. J. Marrie, M. A. Mufson, A. Torres, M. A. Woodhead, S. B. Svenson, and G. Kallenius. 2000. Molecular epidemiology of Streptococcus pneumoniae causing invasive disease in 5 countries. J. Infect. Dis. 182:833-839. [DOI] [PubMed] [Google Scholar]
- 18.Ho, P. L., W. C. Yam, T. K. Cheung, W. W. Ng, T. L. Que, D. N. Tsang, T. K. Ng, and W. H. Seto. 2001. Fluoroquinolone resistance among Streptococcus pneumoniae in Hong Kong linked to the Spanish 23F clone. Emerg. Infect. Dis. 7:906-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho, P. L., R. W. Yung, D. N. Tsang, T. L. Que, M. Ho, W. H. Seto, T. K. Ng, W. C. Yam, and W. W. Ng. 2001. Increasing resistance of Streptococcus pneumoniae to fluoroquinolones: results of a Hong Kong multicentre study in 2000. J. Antimicrob. Chemother. 48:659-665. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, C. N., W. H. Benjamin, Jr., S. A. Moser, S. K. Hollingshead, X. Zheng, M. J. Crain, M. H. Nahm, and K. B. Waites. 2003. Genetic relatedness of levofloxacin-nonsusceptible Streptococcus pneumoniae isolates from North America. J. Clin. Microbiol. 41:2458-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jorgensen, J. H., L. M. Weigel, M. J. Ferraro, J. M. Swenson, and F. C. Tenover. 1999. Activities of newer fluoroquinolones against Streptococcus pneumoniae clinical isolates including those with mutations in the gyrA, parC, and parE loci. Antimicrob. Agents Chemother. 43:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlowsky, J. A., C. Thornsberry, I. A. Critchley, M. E. Jones, A. T. Evangelista, G. J. Noel, and D. F. Sahm. 2003. Susceptibilities to levofloxacin in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis clinical isolates from children: results from 2000-2001 and 2001-2002 TRUST studies in the United States. Antimicrob. Agents Chemother. 47:1790-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klugman, K. P. 2002. The successful clone: the vector of dissemination of resistance in Streptococcus pneumoniae. J. Antimicrob. Chemother. 50(Suppl. 2):1-5. [DOI] [PubMed] [Google Scholar]
- 24.Kupronis, B. A., C. L. Richards, and C. G. Whitney. 2003. Invasive pneumococcal disease in older adults residing in long-term care facilities and in the community. J. Am. Geriatr. Soc. 51:1520-1525. [DOI] [PubMed] [Google Scholar]
- 25.Lefevre, J. C., G. Faucon, A. M. Sicard, and A. M. Gasc. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 31:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim, S., D. Bast, A. McGeer, J. de Azavedo, and D. E. Low. 2003. Antimicrobial susceptibility breakpoints and first-step parC mutations in Streptococcus pneumoniae: redefining fluoroquinolone resistance. Emerg. Infect. Dis. 9:833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandell, L. A., J. G. Bartlett, S. F. Dowell, T. M. File, Jr., D. M. Musher, and C. Whitney. 2003. Update of practice guidelines for the management of community-acquired pneumonia in immunocompetent adults. Clin. Infect. Dis. 37:1405-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDougal, L., J. Rasheed, J. Biddle, and F. Tenover. 1995. Identification of multiple clones of extended-spectrum cephalosporin-resistant Streptococcus pneumoniae isolates in the United States. Antimicrob. Agents Chemother. 39:2282-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGee, L., C. E. Goldsmith, and K. P. Klugman. 2002. Fluoroquinolone resistance among clinical isolates of Streptococcus pneumoniae belonging to international multiresistant clones. J. Antimicrob. Chemother. 49:173-176. [DOI] [PubMed] [Google Scholar]
- 30.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically—approved standard M7-A6, 6th ed. NCCLS, Wayne, Pa.
- 32.Overweg, K., D. Bogaert, M. Sluijter, J. Yother, J. Dankert, R. de Groot, and P. W. Hermans. 2000. Genetic relatedness within serotypes of penicillin-susceptible Streptococcus pneumoniae isolates. J. Clin. Microbiol. 38:4548-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan, X. S., J. Ambler, S. Mehtar, and L. M. Fisher. 1996. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Trallero, E., J. M. Marimon, A. Gonzalez, and L. Iglesias. 2003. Spain14-5 international multiresistant Streptococcus pneumoniae clone resistant to fluoroquinolones and other families of antibiotics. J. Antimicrob. Chemother. 51:715-719. [DOI] [PubMed] [Google Scholar]
- 35.Robinson, K. A., W. Baughman, G. Rothrock, N. L. Barrett, M. Pass, C. Lexau, B. Damaske, K. Stefonek, B. Barnes, J. Patterson, E. R. Zell, A. Schuchat, and C. G. Whitney. 2001. Epidemiology of invasive Streptococcus pneumoniae infections in the United States, 1995-1998: opportunities for prevention in the conjugate vaccine era. JAMA 285:1729-1735. [DOI] [PubMed] [Google Scholar]
- 36.Sahloul, R. T., R. J. Stanek, and M. A. Mufson. 2000. Surveillance of penicillin-resistant Streptococcus pneumoniae in one American metropolitan area, 1989-1998. Eur. J. Clin. Microbiol. Infect. Dis. 19:704-707. [DOI] [PubMed] [Google Scholar]
- 37.Vegelin, A. L., P. Bissumbhar, J. C. Joore, J. W. Lammers, and I. M. Hoepelman. 1999. Guidelines for severe community-acquired pneumonia in the western world. Neth. J. Med. 55:110-117. [DOI] [PubMed] [Google Scholar]
- 38.Weigel, L. M., G. J. Anderson, R. R. Facklam, and F. C. Tenover. 2001. Genetic analyses of mutations contributing to fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:3517-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss, K., C. Restieri, R. Gauthier, M. Laverdiere, A. McGeer, R. J. Davidson, L. Kilburn, D. J. Bast, J. de Azavedo, and D. E. Low. 2001. A nosocomial outbreak of fluoroquinolone-resistant Streptococcus pneumoniae. Clin. Infect. Dis. 33:517-522. [DOI] [PubMed] [Google Scholar]
- 40.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, N. M. Bennett, R. Lynfield, A. Reingold, P. R. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, and A. Schuchat. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737-1746. [DOI] [PubMed] [Google Scholar]
- 41.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, C. Lexau, A. Reingold, L. Lefkowitz, P. R. Cieslak, M. Cetron, E. R. Zell, J. H. Jorgensen, and A. Schuchat. 2000. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States. N. Engl. J. Med. 343:1917-1924. [DOI] [PubMed] [Google Scholar]
- 42.Zhanel, G. G., A. Walkty, K. Nichol, H. Smith, A. Noreddin, and D. J. Hoban. 2003. Molecular characterization of fluoroquinolone resistant Streptococcus pneumoniae clinical isolates obtained from across Canada. Diagn. Microbiol. Infect. Dis. 45:63-67. [DOI] [PubMed] [Google Scholar]