Abstract
Entecavir (ETV) exhibits potent antiviral activity in patients chronically infected with wild-type or lamivudine (3TC)-resistant (3TCr) hepatitis B virus (HBV). Among the patients treated in phase II ETV clinical trials, two patients for whom previous therapies had failed exhibited virologic breakthrough while on ETV. Isolates from these patients (arbitrarily designated patients A and B) were analyzed genotypically for emergent substitutions in HBV reverse transcriptase (RT) and phenotypically for reduced susceptibility in cultures and in HBV polymerase assays. After 54 weeks of 3TC therapy, patient A (AI463901-A) received 0.5 mg of ETV for 52 weeks followed by a combination of ETV and 100 mg of 3TC for 89 weeks. Viral rebound occurred at 133 weeks after ETV was started. The 3TCr RT substitutions rtV173L, rtL180M, and rtM204V were present at study entry, and the additional substitutions rtI169T and rtM250V emerged during ETV-3TC combination treatment. Reduced ETV susceptibility in vitro required the rtM250V substitution in addition to the 3TCr substitutions. For liver transplant patient B (AI463015-B), previous famciclovir, ganciclovir, foscarnet, and 3TC therapies had failed, and RT changes rtS78S/T, rtV173L, rtL180M, rtT184S, and rtM204V were present at study entry. Viral rebound occurred after 76 weeks of therapy with ETV at 1.0 mg, with the emergence of rtT184G, rtI169T, and rtS202I substitutions within the preexisting 3TCr background. Reduced susceptibility in vitro was highest when both the rtT184G and the rtS202I changes were combined with the 3TCr substitutions. In summary, infrequent ETV resistance can emerge during prolonged therapy, with selection of additional RT substitutions within a 3TCr HBV background, leading to reduced ETV susceptibility and treatment failure.
Nearly 400 million people are chronically infected with hepatitis B virus (HBV) worldwide (16, 19). After prolonged infections, often lasting decades, patients frequently develop severe liver disease that can lead to cirrhosis and hepatocellular carcinoma. Chronically infected patients also serve as sources of HBV transmission. There are currently three approved therapies for chronic HBV infections: interferon, lamivudine (3TC; β-l-2′,3′-dideoxy-3′-thiacytidine), and adefovir-dipivoxil, the prodrug of adefovir [ADV; 9-(2-phosphonylmethoxyethyl)adenine] (18). Interferon is administered subcutaneously and is associated with numerous adverse events, some of which can be severe, and a sustained antiviral response in only 30 to 40% of treated patients (30). 3TC treatment, administered orally, is effective in reducing viral loads but results in the frequent emergence of drug-resistant HBV due to substitutions at the Tyr-Met-Asp-Asp (YMDD) nucleotide binding site motif of viral DNA polymerase. Data from four large clinical trials revealed 3TC resistance (3TCr) mutations in 24, 42, 53, and 70% of patients after 1, 2, 3, and 4 years of therapy, respectively (15). Treatment with ADV, recently approved by the U.S. Food and Drug Administration, can be associated with dose-limiting nephrotoxicity (6), although the reported frequency of resistance to ADV is considerably lower than that to 3TC (2, 28). Therefore, more potent and safer oral therapies, with less frequent development of viral resistance, are needed for the treatment of chronic HBV infections.
Entecavir (ETV; BMS-200475) is a novel deoxyguanosine analog with potent activity against HBV in vitro, in animal studies, and in patients chronically infected with HBV (7, 11, 14, 17, 21, 23, 29). In enzymatic studies, 3TCr HBV polymerases were shown to have reduced susceptibility to ETV (20). Due to its intrinsic potency and efficient phosphorylation in cells, however, sufficient intracellular concentrations of ETV triphosphate (ETV-TP) can be generated at clinical exposure levels (31) to enable potent inhibition of 3TCr HBV in cell cultures (20). These findings are consistent with recent results from clinical studies demonstrating the efficacy of ETV in subjects with 3TCr viruses (4, 26).
Studies with woodchucks chronically infected with woodchuck hepatitis virus suggest a superior resistance profile for ETV relative to 3TC (7). Treatment with a weekly regimen of ETV for up to 3 years was not associated with viral rebound or emergence of a resistant virus (7). In contrast, 3TC treatment of woodchucks resulted in resistance development as early as within 5 months (22, 32). Nevertheless, the emergence of resistant variants occurs with virtually all antiviral therapies and especially those requiring prolonged administration, as required in the treatment of chronic HBV infections. For this reason, the >500 patients treated in ETV phase II clinical trials and patients in ETV phase III clinical trials have been monitored closely for any evidence of resistance emergence or viral rebound that may be related to resistance. Studies to date suggest that resistance will be infrequent. For example, in one phase II study of patients not responding to 3TC treatment, no evidence of genotypic changes associated with virologic breakthrough among 132 patients completing 1 year of ETV therapy was found (4; unpublished results).
In this study, we have analyzed two patients (arbitrarily designated patients A and B) who experienced virologic breakthrough after more than 1 year of ETV therapy in phase II trials. Both patients had experienced failed previous treatment(s) and harbored resistant HBV at study entry. Breakthrough HBV isolated from both contained the preexisting, signature 3TCr substitutions (rtL180M and rtM204V) in addition to unique substitutions that emerged during ETV therapy. In vitro studies with recombinant HBV were used to show that patient isolates with emergent substitutions displayed reduced susceptibility to ETV in cell cultures and to ETV-TP in in vitro endogenous polymerase assays. Experiments with recombinant HBV clones were also used to show that complex changes, requiring novel reverse transcriptase (RT) substitutions in combination with those conferring 3TCr, were required for reduced susceptibility to ETV. Therefore, while viral resistance to 3TC (1) or ADV (2, 28) can result from single amino acid substitutions, at least three different concomitant amino acid substitutions in HBV RT were required to achieve clinically relevant levels of viral resistance to ETV in these patients.
MATERIALS AND METHODS
Cells, viruses, and antiviral compounds.
HepG2 human hepatoma cells were maintained in RPMI 1640 (Invitrogen, Carlsbad, Calif.) supplemented with 10% heat-inactivated fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 2 mM l-glutamine. These cells were grown on plastic culture surfaces previously coated with type I collagen (BD Biosciences, Bedford, Mass.). The laboratory clone of HBV was the genotype D serotype ayw clone in plasmid pCMV-HBV; it was kindly provided by Stephen Goff (12). ETV, 3TC, and ADV were synthesized at Bristol-Myers Squibb Pharmaceutical Research Institute (BMS). l-Deoxyribosylthymine (l-dT) was purchased from Moravek Biochemicals, Brea, Calif.
HBV polymerase genotyping.
HBV DNA was extracted from serum samples by using either a QIAamp viral RNA minikit or an UltraSens virus kit according to the manufacturer's instructions (Qiagen, Valencia, Calif.). The HBV polymerase domain, amino acids 1 to 344 (25), was PCR amplified by using primers with the sequences 5′-CCT CAG GCC ATG CAG TGG AA-3′ and 5′-CCT GCT GCG CGC AAA ACA AGC GGC TAG GAG TTC CGC AGT ATG GA-3′ and high-fidelity Platinum Taq DNA polymerase (Life Technologies, Gaithersburg, Md.) according to the manufacturer's instructions. PCR amplification consisted of 40 cycles of 94°C for 15 s, 55°C for 15 s, and 68°C for 3 min. PCR products were analyzed by agarose gel electrophoresis, purified by using a Qiagen PCR purification kit, and sequenced directly by using primers HBVFS4 (5′-TGT ATT CCC ATC CCA TC-3′), HBVRS4 (5′-GAC ATA CTT TCC AAT CAA TAG G-3′), HBVRS3 (5′-AGA ACC AAC AAG AAG ATG AGG-3′), and HBV4 (5′-GCT AGG AGT TCC GCA GTA TGG A-3′) and an ABI Prism Big Dye terminator cycle sequencing ready reaction kit. The sequencing reactions were run on an ABI 3100 automated sequencer (PE Applied Biosystems), and the data were analyzed by using Sequencher software (Gene Codes Corp., Ann Arbor, Mich.). Amino acid substitutions that emerged during ETV treatment, other than naturally occurring polymorphic changes, were revealed by comparison to 250 wild-type (wt) HBV genomes (GenBank) and are presented in Tables 1 and 2. In addition, other amino acid substitutions reported to occur in response to antiviral therapy for HBV (25) are also shown.
TABLE 1.
Genotypes of patient A HBV isolates
| Treatmenta | Total no. of wks of treatment | Viral load (log10) copies/mlb | Patient A RT substitution(s) in wt residuec:
|
|||||
|---|---|---|---|---|---|---|---|---|
| L80 | I169 | V173 | L180 | M204 | M250 | |||
| ETV | 0 | 9.53 | L/I | — | V/L | L/M | I/V | — |
| 24 | 8.26 | — | — | V/L | M | V | — | |
| 48 | 7.61 | — | — | V/L | M | V | — | |
| ETV + 3TC | 56 | 6.35 | — | — | V/L | M | V | — |
| 68 | 7.26 | — | I/T | V/L | M | V | M/V | |
| 72 | 8.08 | — | I/T | V/L | M | V | M/V | |
| 76 | 7.02 | — | I/T | V/L | M | V | M/V | |
| 80 | 7.28 | — | I/T | V/L | M | V | M/V | |
| 106 | 6.34 | — | T | V/L | M | V | M/V | |
| 108 | 6.68 | — | T | V/L | M | V | M/V | |
| 133 | 7.89 | — | T | V/L | M | V | M/V | |
Once-daily treatments were 0.5 mg of ETV for 52 weeks, 0.5 mg of ETV and 100 mg of 3TC for 61 weeks (to week 117), and 1.0 mg of ETV and 100 mg of 3TC.
Determined by the Roche Amplicor method.
Emergent substitutions on therapy in conserved residues or in positions previously noted to change in response to antiviral therapy (25). —, wt residue; multiple residues indicate mixtures.
TABLE 2.
Genotypes of patient B HBV isolates
| Total no. of wks of treatmenta | Viral load (log10 copies/ml)b | Patient B RT substitution(s) in wt residuec:
|
||||||
|---|---|---|---|---|---|---|---|---|
| S78 | I169 | V173 | L180 | T184 | S202 | M204 | ||
| 0 | 8.34 | S/T | — | V/L | M | T/S | — | V |
| 4 | 6.81 | S/T | — | V/L | M | T/S | — | V |
| 12 | 6.46 | S/T | — | V/L | M | T/S | — | V |
| 24 | 5.99 | S/T | — | V/L | M | T/S | — | V |
| 37 | 6.08 | T | — | V/L | M | T/S | — | V |
| 48 | 6.18 | T | — | V/L | M | T/S | — | V |
| 60 | 5.90 | T | — | — | M | S | — | V |
| 68 | ND | — | — | — | M | G | I | V |
| 76 | 9.52 | S/T | — | — | M | G | I | V |
| 84 | ND | S/T | I/T | — | M | G | I | V |
| 92 | 9.70 | S/T | I/T | — | M | G | I | V |
Treatment was ETV at 1.0 mg once daily.
Determined by the Roche Amplicor method. ND, not determined.
Emergent substitutions on therapy in conserved residues or in positions previously noted to change in response to antiviral therapy (25). —, wt residue; multiple residues indicate mixtures. Emergent changes in naturally polymorphic residues were A38E, Y/H53H, N/D76D, L/I91I, N/T128N, R/Q153R, and N238N/D.
Phenotypic analysis of cell cultures.
The susceptibility of recombinant HBV clones was determined by quantitating the level of extracellular HBV DNA after expression in HepG2 cells cultured in the presence of antiviral compounds. HBV was expressed by transient transfection of full-length HBV genomes in plasmid pCMV-HBV 180B3 (formerly pBAC-HBV) (20), expressing the HBV genotype D pregenomic RNA under the transcriptional control of the cytomegalovirus immediate-early promoter. The AvrII-NsiI fragment (RT amino acids 17 to 311) from patient isolates was used to replace the corresponding sequence in pCMV-HBV 180B3 for patient RT background clones. Alternatively, for clones with the laboratory RT background, site-directed mutagenesis was performed by the quick-change method as described by the manufacturer (Stratagene, La Jolla, Calif.). HepG2 cells in growth medium were used to seed six-well plates at 1.5 × 106 cells/well. On the following day, wells were transfected for 5 h with various pCMV-HBV constructs (4 μg/well) by using Lipofectamine 2000 as described by the manufacturer (Invitrogen). Following transfection, cells were trypsinized, used to seed 96-well plates at ∼5 × 104 cells/well for susceptibility assays, and fed with assay medium (same as growth medium but with 5% heat-inactivated fetal bovine serum) containing 0.125% dimethyl sulfoxide and containing or not containing dilutions of antiviral compounds for 5 days.
Extracellular HBV was quantitated by DNA dot blot hybridization on Nytran supercharged membranes (Schleicher & Schuell) with a chemiluminescence-labeled 3.2-kbp HBV ayw genome fragment probe according to the manufacturer's instructions (Amersham Gene Images random prime labeling module) and as described previously (20). Supernatant HBV was quantitated directly or after immunocapture of HBV virions. Both methods yielded equivalent 50% effective concentrations (EC50s). HBV virions were immunocaptured after adjustment of supernatants to 0.8% (vol/vol) NP-40 on plates coated with protein A and anti-HBV core protein polyclonal antibody (Dako, Carpinteria, Calif.). Captured nucleocapsids were washed thoroughly with Tris-buffered saline (50 mM Tris-HCl [pH 8.0], 150 mM NaCl) before release and denaturation of HBV DNA with 1 N NaOH-1.5 M NaCl-0.15 M sodium citrate and then dot blot hybridization. Densitometric scans of Kodak BioMax MR film exposures of the blots were used to determine HBV DNA levels. Parallel hybridization to dilutions of the HBV genotype D genome fragment was used for quantitation and to ensure that exposures were within the linear range of the film. Data were plotted as percent inhibition, and the EC50 was calculated as the drug concentration at which 50% inhibition of extracellular HBV occurred by using Microsoft Excel XLfit equation 205: y = A + [(B − A)]/{1 + [(C/x)D]}, where A is the percent inhibition at the lowest inhibitor concentration, B is the percent inhibition at the highest inhibitor concentration, C is the inhibitor concentration at the middle of the curve, and D is the slope of the curve.
Replicative capacity.
Virus replication rates were measured by cotransfecting recombinant HBV clones with a Renilla luciferase reporter plasmid (phRL-CMV; Promega) to normalize transfection efficiency. HepG2 cells were transfected in six-well plates as described above. The luciferase activity of phRL-CMV was measured at 16 h posttransfection according to the manufacturer's instructions. Cell culture medium was replaced daily, and extracellular HBV was quantitated by dot blot hybridization as described above. Intracellular replicative intermediates within HBV nucleocapsids were also quantitated from cell lysates. Briefly, cell monolayers were washed with phosphate-buffered saline and lysed with 50 mM Tris-Cl (pH 8.0)-150 mM NaCl-5 mM EDTA-0.8% NP-40. Lysates were clarified by centrifugation at 4,000 × g for 5 min and then added to plates coated with protein A and anti-HBV core protein polyclonal antibody. The plates were washed thoroughly with Tris-buffered saline before release and denaturation of DNA from nucleocapsid-antibody complexes with 1 N NaOH-1.5 M NaCl-0.15 M sodium citrate. HBV DNA was quantitated by dot blot hybridization as described above. Virus replication curves were plotted as picograms of HBV DNA per relative light unit.
Intracellular HBV capsid production and isolation.
Intracellular HBV nucleocapsids were isolated from T-175 flasks of transfected HepG2 cells. Cells at 80 to 90% confluence were transfected with 36 μg of plasmid DNA by using Lipofectamine 2000 according to the manufacturer's instructions, except that the DNA-liposome complexes were replaced after 4 h with fresh growth medium containing 1 mM phosphonoformic acid (PFA; Sigma). PFA was added to enable the isolation of capsids that did not progress beyond the initial stages of DNA elongation as described previously (24). Cells were harvested by scraping at 2 days posttransfection, and the cell pellets were stored at −80°C until further processing.
Nucleocapsids were isolated as described previously (24) with modifications. Thawed cell pellets from one to three flasks were resuspended in 20 ml of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 10 mM EDTA, 0.7% NP-40, 3 mM dithiothreitol) for 15 min. Crude lysates were clarified by centrifugation at 2,000 × g for 15 min at 4°C, and supernatants were partially digested with 1 mg of proteinase K (Invitrogen) and 90 U of micrococcal nuclease (U.S. Biochemical Corp., Cleveland, Ohio) plus 15 mM CaCl2 for 15 min at 37°C. Digests were clarified by centrifugation at 10,000 × g for 20 min at 4°C. Supernatants were layered onto a 15-ml 25% sucrose cushion prepared in 10 mM Tris-HCl (pH 7.4)-150 mM NaCl-1 mM EDTA-0.75% NP-40 and centrifuged at 140,000 × g for >15 h. Nucleocapsid pellets were resuspended in 100 μl of 10 mM Tris-HCl (pH 7.4)-150 mM NaCl-1 mM EDTA per original T-175 flask and stored at −80°C until use. Nucleocapsids were quantified on the basis of their content of HBV DNA template by using quantitative PCR as described previously (20), except that primers and probes were used at 0.4 and 0.13 μM, respectively.
In vitro endogenous polymerase assays.
Isolated HBV nucleocapsids containing HBV polymerase and the viral DNA template were incubated at 37°C for 1 h in 20 μl of endogenous polymerase assay buffer (50 mM Tris-HCl [pH 7.4], 75 mM NH4Cl, 20 mM MgCl2, 0.1 mM β-mercaptoethanol, 0.1% NP-40) plus 1 μM each dATP, dCTP, and dTTP and 2.2 nM [α-33P]dGTP (3,000 Ci/mmol; Perkin-Elmer, Boston, Mass.). For 50% inhibitory concentration (IC50) determinations, ETV-TP was serially diluted from 100 nM for wt polymerases and from either 1,000 or 5,000 nM for mutant enzymes. Nucleocapsids containing approximately 18 to 72 pg of HBV DNA were used per reaction to yield a 10-fold signal over the background in 1 h (the background was determined by complete inhibition with 10 mM PFA). Reactions were stopped by the addition of an equal volume of 20% cold trichloroacetic acid (TCA). Acid-insoluble material was collected by vacuum filtration on GF/B (Perkin-Elmer) 96-well plates, washed with cold 10% TCA, and dried, and counts were determined by liquid scintillation. Other reactions contained 3TC triphosphate (Moravek) or ADV diphosphate (synthesized at BMS). The IC50 was calculated as the drug concentration at which 50% inhibition of TCA-precipitable counts per minute occurred by using Microsoft Excel XLfit equation 205 (see above).
RESULTS
Patient A (AI463901-A) profile.
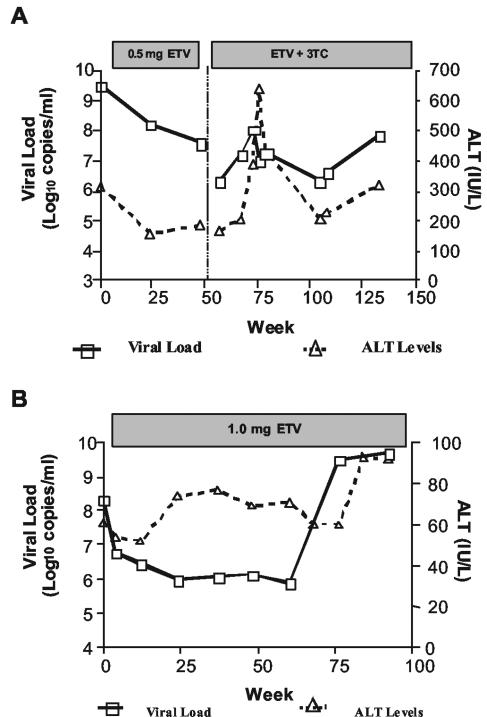
Patient A is a 44-year-old male who had an anti-HBe-positive chronic HBV infection and who started 3TC therapy with serum HBV DNA levels of >8.76 log10 copies/ml. The viral load was suppressed to 6.36 log10 copies/ml following 8 weeks of 3TC treatment but rebounded to 8.38 log10 copies/ml at week 25. 3TC signature mutations, resulting in the amino acid substitutions rtL180M and rtM204V, were detected by DNA sequencing at week 28, and serum alanine transaminase (ALT) levels peaked at 741 IU/liter by week 34. The patient began therapy with 0.5 mg of ETV daily after week 54 of 3TC treatment with a viral load of 9.53 log10 copies/ml (Fig. 1A). After 48 weeks of ETV therapy, the viral DNA and ALT levels decreased to 7.61 log10 copies/ml and 185 IU/liter, respectively. Following 4 more weeks of ETV therapy and 4 weeks without any antiviral therapy, the patient was treated for 61 weeks with 0.5 mg of ETV plus 100 mg of 3TC. However, after both HBV DNA and ALT levels were poorly responsive and showed only transient fluctuations, the patient was switched to 1.0 mg of ETV plus 100 mg of 3TC for 28 weeks. At 133 weeks from the start of ETV therapy, there was an increase in the viral load of >1.5 log10 copies/ml from the nadir, prompting further analysis. At week 145, ETV was discontinued and 10 mg (daily) of adefovir was added while 100 mg (daily) of 3TC was continued. By week 157, HBV DNA was reduced from 7.5 to 5.2 log10 copies/ml.
FIG. 1.
Patient profiles and outcomes with ETV therapy. Serum HBV DNA loads and ALT levels for patient A (A) and patient B (B) at various times of ETV therapy are shown. The duration of therapy is indicated by a bar above the graphs. For patient A, treatment included 0.5 mg of ETV for weeks 0 through 52, 0.5 mg of ETV with 100 mg of 3TC for weeks 56 through 117, and 1.0 mg of ETV with 100 mg of 3TC until week 145.
When ETV therapy was initiated, patient A was infected with HBV of genotype D and serotype ayw and had the 3TCr substitutions rtL180M, rtM204V, and rtV173L (10). Virus isolated at week 133, when increases in the viral load and ALT levels were observed, had the unique substitutions rtI169T and rtM250V in mixtures with wt residues in addition to the 3TCr substitutions (Table 1). These substitutions were subsequently detected as early as week 68 after the start of ETV therapy. Substitution rtI169T causes an F161L substitution in HBV surface antigen (HBsAg), while substitution rtM250V is outside the HBsAg open reading frame.
Patient B (AI463015-B) profile.
Patient B is a 55-year-old male who developed HBV disease recurrence following liver transplantation and who was HBV e antigen negative due to infection with precore-negative HBV (G1896A mutation resulting in a stop codon substitution for the codon corresponding to tryptophan at residue 28). Posttransplantation antiviral therapy for HBV recurrence included 63 weeks of ganciclovir, 13 weeks of foscarnet, 194 weeks of famciclovir, and 190 weeks of 3TC, and immunosuppressive therapy included >9 years of cyclosporine. The early case history and the selection of famciclovir resistance had been reported previously (3). Immediately prior to ETV therapy, the patient had received >3.5 years of 3TC therapy, during which a viral load of 9.69 log10 copies/ml at the start of therapy was only modestly reduced to 8.27 log10 copies/ml upon completion, with isolated virus having 3TCr substitutions. Treatment with 1.0 mg of ETV resulted in a further decrease in the viral load from 8.34 to 5.99 log10 copies/ml after 24 weeks (Fig. 1B). However, an increase in the viral load of >3 log10 copies/ml was noted after 76 weeks of ETV therapy, and the patient was withdrawn after 92 weeks following the development of sustained elevations in both ALT levels and the viral load.
The genotype D serotype ayw2 baseline virus isolated from patient B contained the 3TCr and famciclovir resistance substitutions rtS78T, rtV173L, rtL180M, rtT184S, and rtM204V (3; reviewed in reference 13). By week 76 of ETV treatment, increases in the viral load and ALT levels were detected in association with the emergence of substitutions rtS184G and rtS202I within the preexisting 3TCr background. These changes were detected in isolates from as early as week 68 of therapy, with substitution rtI169T appearing at week 84. Substitutions rtT184G and rtS202I resulted in sL176V and sV194F changes in HBsAg, respectively.
Phenotypic analysis.
Patient RT substitutions that emerged during therapy were evaluated for their effects on ETV susceptibility in cell cultures. Patient-specific wt viruses, constructed by site-directed mutagenesis of baseline viruses to remove resistance-associated substitutions, were also tested. Recombinant HBV genomes containing patient RT domains or specific substitutions were used to transfect HepG2 cells, which were tested for the production of extracellular HBV in the presence of ETV. Reconstructed recombinant wt viruses for both patients were >100-fold to >500-fold more susceptible to ETV than to 3TC, ADV, or l-dT (Tables 3 and 4). As expected, baseline viruses with the 3TCr signature substitutions L180M and M204V were completely resistant to both 3TC and l-dT, while ETV still retained nanomolar potency against these viruses. As previously reported (20), however, HBV containing the signature resistance substitutions encoding 3TCr displayed somewhat reduced ETV susceptibility. Accordingly, for patient A, the baseline 3TCr recombinant virus was 10-fold less susceptible to ETV than the reconstructed wt virus, while an HBV recombinant containing the RT domain with additional substitutions rtI169T and rtM250V from the week 106 patient A sample showed a >1,000-fold reduction in ETV susceptibility compared to the wt construct (Tables 3 and 4). These mutations had no effect on the susceptibility to ADV. In accordance with these observations, subsequent treatment of patient A with ADV after discontinuation of ETV markedly reduced the viral load (data not shown).
TABLE 3.
Cell culture antiviral susceptibility of patient A recombinant viruses
| Virusa | RT substitution atb:
|
EC50 (nM)c of:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| I169 | V173 | L180 | M204 | M250 | ETV | ADV | 3TC | l-dT | |
| wt | — | — | — | — | — | 3.0 ± 1.5 | 1,600d | 1,600d | 1,700 ± 700 |
| Baseline | — | L | M | V | — | 30 ± 22 | 1,400 ± 600 | >500,000 | >200,000 |
| wk 106 | T | L | M | V | V | >4,000 | 1,300 ± 400 | >500,000 | >200,000 |
wt HBV clones were derived from site-directed mutagenesis of patient baseline virus to remove antiviral resistance substitutions.
—, wt residue.
Values represent the mean and standard deviation from three or more independent experiments unless otherwise indicated.
Values represent the mean from two independent experiments.
TABLE 4.
Cell culture antiviral susceptibility of patient B recombinant viruses
| Virusa | RT substitution atb:
|
EC50 (nM)c of:
|
||||||
|---|---|---|---|---|---|---|---|---|
| L180 | T184 | S202 | M204 | ETV | ADV | 3TC | l-dT | |
| wt | — | — | — | — | 3.7 ± 1.7 | 2,500 ± 900 | 400d | 2,900 ± 1,800 |
| Baseline | M | S | — | V | 120 ± 73 | 2,600 ± 1,000 | >500,000 | >200,000 |
| wk 92 | M | G | I | V | >4,000 | 4,200 ± 2,100 | >500,000 | >200,000 |
wt HBV clones were derived from site-directed mutagenesis of patient baseline virus to remove antiviral resistance substitutions.
—, wt residue.
Values represent the mean and standard deviation from three or more independent experiments unless otherwise indicated.
Values represent the mean from two independent experiments.
The baseline virus from patient B, containing substitutions rtS78T and rtT184S in addition to 3TCr substitutions rtV173L, rtL180M, and rtM204V, showed a twofold reduction in ETV susceptibility compared to that observed for 3TCr HBV (see Table 6) (20). Substitution rtS78T is associated with famciclovir therapy (25) but had no impact on ETV susceptibility, as recombinant viruses with and without the substitution had similar ETV EC50s (data not shown). Interestingly, this change results in a stop codon in the overlapping HBsAg gene, but no diminution of extracellular virions was observed (data not shown). An HBV recombinant containing the RT domain with substitutions rtS184G and rtS202I from the week 92 patient B sample was >1,000-fold less susceptible to ETV than the reconstructed wt HBV recombinant (Tables 3 and 4). As with patient A, substitutions that arose in viruses from patient B during ETV therapy had little effect on the susceptibility to ADV. These results suggest that the acquisition of additional substitutions in 3TCr HBV during ETV therapy can result in phenotypic resistance to ETV.
TABLE 6.
Recombinants containing specific substitutions from patient B
| RT sourcea | Genotypeb | Patient B RT substitution atc:
|
ETV EC50 (nM)d | Fold increase in EC50e | |||
|---|---|---|---|---|---|---|---|
| L180 | T184 | S202 | M204 | ||||
| Patient | wt | — | — | — | — | 3.7 ± 1.7 | |
| Baseline | M | S | — | V | 120 ± 73 | 32 | |
| wk 92 | M | G | I | V | >4,000 | >1,100 | |
| del 184S/G, 202I | M | — | — | V | 60 ± 29 | 16 | |
| del 184S/G | M | — | I | V | 330 ± 71 | 89 | |
| del 202I | M | G | — | V | 320 ± 190 | 86 | |
| del 3TCr | — | G | I | — | 2.5 ± 1.5 | 1 | |
| Laboratory strain | wt | — | — | — | — | 5.1 ± 2.4 | |
| add 3TCr | M | — | — | V | 27 ± 17 | 5 | |
| add 3TCr, 184G | M | G | — | V | 210 ± 120 | 41 | |
| add all | M | G | I | V | >4,000 | >784 | |
RT domain was derived from patient or laboratory genotype D strains.
Patient wt HBV clone was derived by removal of antiviral resistance substitutions from baseline virus. del, deletion; add, addition. All clones were wt with respect to positions 78, 169, and 173.
—, wt residue.
Values are the mean and standard deviation from three or more independent experiments.
Fold increase in EC50 over that of the wt rounded to the nearest whole number.
Substitutions responsible for ETV resistance.
To determine the precise role of individual substitutions in ETV susceptibility, recombinant HBV clones were constructed by site-directed mutagenesis and tested in cultures. For patient A, recombinant HBV clones containing 3TCr mutations plus rtM250V were all shown to be phenotypically resistant to ETV, with EC50s of greater than 1,000 nM (Table 5). Substitution rtI169T selected during ETV therapy did not result in decreased susceptibility to ETV by itself or in combination with 3TCr substitutions. However, the possibility that rtI169T plays a secondary role in ETV resistance and/or replication adaptation cannot be ruled out, since slightly lower EC50s were observed for several clones containing all of the patient A emergent substitutions except for rtI169T. Furthermore, as discussed below, in vitro endogenous HBV polymerase assays suggest a possible role for substitution rtI169T. Substitution rtM250V in the absence of all other substitutions conferred a low level of ETV resistance in cell cultures similar to that seen with HBV containing 3TCr substitutions. However, it is unclear whether this change is clinically relevant, given the predicted intracellular ETV-TP levels achieved during therapy (20) and the fact that substitution rtM250V was seen only in patient A isolates when 3TCr substitutions were present.
TABLE 5.
Recombinants containing specific substitutions from patient A
| RT sourcea | Genotypeb | Patient A RT substitution atc:
|
ETV EC50 (nM)d | Fold increase in EC50e | ||||
|---|---|---|---|---|---|---|---|---|
| I169 | V173 | L180 | M204 | M250 | ||||
| Patient | wt | — | — | — | — | — | 3.0 ± 1.5 | |
| Baseline | — | L | M | V | — | 30 ± 22 | 10 | |
| wk 106 | T | L | M | V | V | >4,000 | >1,333 | |
| del 169T | — | L | M | V | V | 1,000 ± 800 | 333 | |
| del 250V | T | L | M | V | — | 24 ± 22 | 8 | |
| del 3TCr | T | L | — | — | V | 22 ± 9 | 7 | |
| del 173L | T | — | M | V | V | >4,000 | >1,333 | |
| del 173L, 169T | — | — | M | V | V | >4,000 | >1,333 | |
| del 173L, 250V | T | — | M | V | — | 21 ± 6 | 7 | |
| del 173L, 3TCr | T | — | — | — | V | 18 ± 8 | 6 | |
| Laboratory strain | wt | — | — | — | — | — | 5.1 ± 2.4 | |
| add 169T | T | — | — | — | — | 3.3 ± 0.7 | 1 | |
| add 173L, 3TCr | — | L | M | V | — | 40 ± 21 | 8 | |
| add 250V | — | — | — | — | V | 47 ± 24 | 9 | |
| add all but 169T | — | L | M | V | V | 1,300 ± 630 | 255 | |
| add all | T | L | M | V | V | >4,000 | >784 | |
RT domain was derived from patient or laboratory genotype D strains.
Patient wt HBV clone was derived by removal of antiviral resistance substitutions from baseline virus. del, deletion; add, addition.
—, wt residue.
Values are the mean and standard deviation from three or more independent experiments.
Fold increase in EC50 over that of the wt rounded to the nearest whole number.
For patient B, prolonged ETV therapy resulted in the selection of two novel amino acid substitutions, rtT184G and rtS202I (Table 2). The ETV susceptibility of recombinant clones containing either substitution in combination with 3TCr substitutions was reduced relative to that of the baseline isolate. However, HBV containing all four of these substitutions (rtT184G, rtS202I, rtL180M, and rtM204V) showed the highest level of resistance (Table 6). Unlike what was seen with substitution rtM250V identified in patient A, there was no change in ETV susceptibility for HBV containing substitutions rtT184G and rtS202I in the absence of 3TCr substitutions.
Effects of substitutions on viral replicative capacity.
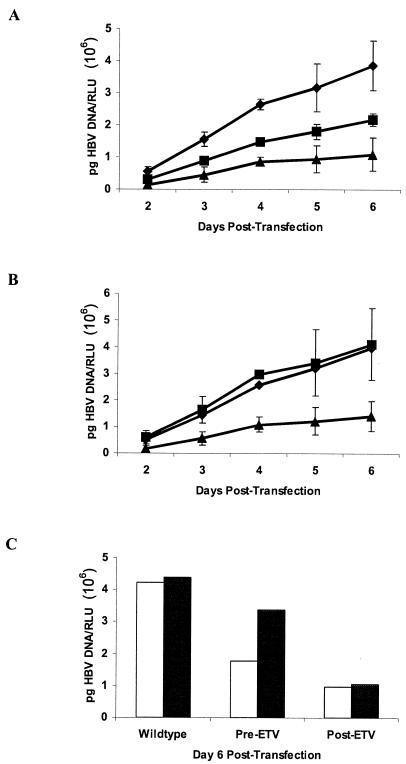
The replication levels in recombinant viruses from patients A and B were measured after transfection to assess the effect of emergent substitutions on replicative capacity. To control for variations in transfection efficiencies, all values were normalized to luciferase levels expressed from a cotransfected control plasmid. As shown in Fig. 2A, the patient A baseline recombinant virus displayed a reduced replication rate relative to the wt virus that is characteristic of HBV with 3TCr substitutions. The addition of substitution rtM250V resulted in significant replication impairment, a result which may explain the observation that substitution rtM250V was always found mixed with the wt residue at this position (Table 1). Patient B recombinant virus with mutations also displayed impaired replication characteristics compared to the baseline virus. Intracellular replicative intermediates were also quantitated to ensure that the reduced replication rates observed for the released extracellular virus were a result of RT substitutions and were not due to concomitant changes in the overlapping HBsAg open reading frame that could affect virion secretion. As shown in Fig. 2C, the accumulation of intracellular replicative intermediates reflected the results for extracellular virus. Taken together, these results suggest that viruses with substitutions which decrease ETV susceptibility display reduced replication rates as a direct result of changes in the polymerase protein.
FIG. 2.
Viral replication with ETV resistance substitutions. Extracellular (A and B) and intracellular (C) HBV DNA from recombinant HBV clones transfected in HepG2 cells. (A) wt (♦), baseline (▪), and week 106 (▴) viruses for patient A. (B) wt (♦), baseline (▪), and week 92 (▴) viruses for patient B. Virus genotypes in transfected clones are represented in Tables 3 and 4. Values are reported as the mean and standard deviation from three independent experiments. (C) Intracellular replicative intermediates for patient A (white bars) and patient B (black bars). RLU, relative light units.
Phenotypes of recombinant enzymes.
HBV polymerase activity was assessed directly to better understand the impact of emergent RT substitutions. The most authentic in vitro assay of HBV polymerase activity uses isolated HBV nucleocapsids in which the polymerase protein is specifically primed for replication through covalent linkage to its HBV DNA template. Therefore, recombinant intracellular HBV nucleocapsids were isolated after cell transfection and assayed for susceptibility to ETV-TP. Tables 7 and 8 show the susceptibility of enzymes derived from recombinant viruses from patients A and B. As expected, all nucleocapsids containing the 3TCr substitutions rtL180M and rtM204V displayed 350- to 1,000-fold resistance to 3TC triphosphate, with IC50s above 5 μM (range, 6.4 to 18.3 μM), while the laboratory wt virus IC50 was 18.4 ± 4.4 nM (mean and standard deviation). 3TCr changes generally conferred an ∼10- to 30-fold decrease in susceptibility to ETV in cell culture assays (Tables 5 and 6) (20), although previous in vitro enzyme assays with recombinant nucleocapsids produced in insect cells showed >50-fold cross-resistance to ETV-TP (20). The recombinant 3TCr HBV capsids produced in mammalian HepG2 cells in this report showed similar levels of cross-resistance to ETV-TP (Tables 7 and 8). The results obtained for polymerases from patient A viruses correlated with the cell culture assay results in that substitutions rtI169T and rtM250V decreased ETV-TP susceptibility more than did 3TCr substitutions alone. In addition, deletion of either the 3TCr substitutions or substitution rtM250V eliminated the observed resistance phenotype, confirming the need for all three substitutions. Another congruence with the cell culture assay results was the modest reduction in the ETV-TP IC50 when substitution rtI169T was omitted from the recombinant clone (IC50 for the clone with the deletion, 140 nM; IC50 for the week 106 clone, 240 nM). Although the levels of nucleocapsids produced by the patient A recombinant viruses varied (Fig. 2A), the specific polymerase activities deviated by less than 50% (data not shown).
TABLE 7.
In vitro endogenous HBV polymerase sensitivity to ETV-TP for patient A
| Genotypea | Patient A RT substitution atb:
|
ETV IC50 (nM)c | Fold increase in IC50d | ||||
|---|---|---|---|---|---|---|---|
| I169 | V173 | L180 | M204 | M250 | |||
| wt | — | — | — | — | — | 0.5 ± 0.1 | 1 |
| Baseline (3TCr) | — | L | M | V | — | 27 ± 6 | 54 |
| wk 106 | T | L | M | V | V | 240 ± 71 | 480 |
| del 173L | T | — | M | V | V | 240 ± 11 | 480 |
| del 169T | — | L | M | V | V | 140 ± 16.4 | 280 |
| del 250 | T | L | M | V | — | 26 ± 0.4 | 52 |
| del 173L, 250V | T | — | M | V | — | 32 ± 9.3 | 64 |
| del 173L, 3TCr | T | — | — | — | V | 37 ± 3.7 | 74 |
wt values are for the laboratory genotype D clone pCMV-HBV 180B3. del, deletion.
—, wt residue.
Values are the mean and standard deviation from three or more independent experiments.
Fold increase in IC50 over that of the wt control.
TABLE 8.
In vitro endogenous HBV polymerase sensitivity to ETV-TP for patient B
| Genotypea | Patient B RT substitution atb:
|
ETV IC50 (nM)c | Fold increase in IC50d | |||
|---|---|---|---|---|---|---|
| L180 | T184 | S202 | M204 | |||
| wt | — | — | — | — | 0.5 ± 0.1 | 1 |
| Baseline (3TCr) | M | S | — | V | 31 ± 2.7 | 62 |
| wk 92 | M | G | I | V | 640 ± 19 | 1,280 |
| del 184G | M | — | I | V | 780 ± 85 | 1,560 |
| del 202I | M | G | — | V | 340 ± 26 | 680 |
| del 3TCr | — | G | I | — | 30 ± 3.3 | 60 |
wt values are for the laboratory genotype D clone pCMV-HBV 180B3. del, deletion.
—, wt residue.
Values are the mean and standard deviation from three or more independent experiments.
Fold increase in IC50 over that of the wt control.
The ETV-TP susceptibility of recombinant enzymes from patient B viruses also largely reflected the findings from cell culture studies in that substitutions rtT184G and rtS202I each contributed some resistance when combined with the 3TCr substitutions. While the deletion of substitution rtT184G from the fully resistant virus did not have a marked effect on the ETV EC50, the resulting virus was severely replication impaired, producing 5% of the intracellular nucleocapsids of the baseline virus (data not shown). Despite this impairment, the specific polymerase activity of recombinant virus lacking either substitution rt184G or substitution rt202I was similar to that of the baseline virus, while that of the week 92 virus was two- to threefold lower (data not shown). Like those from patient A, patient B recombinant viruses also showed an absolute requirement for 3TCr substitutions with either rtT184G or rtS202I or both for reduced ETV susceptibility.
DISCUSSION
This report describes virologic breakthrough after prolonged ETV therapy in two patients from phase II clinical trials. The results of genotypic and phenotypic analyses of samples from these patients suggest that clinically relevant resistance to ETV requires at least three different concomitant amino acid substitutions in HBV RT, in contrast to the single amino acid substitutions that lead to viral resistance to 3TC (1) or ADV (2, 28).
Both patients had failed 3TC therapy prior to receiving ETV, and baseline viral isolates showed genotypic evidence of the signature 3TCr substitutions rtL180M and rtM204V. Levine et al. previously reported that the intracellular accumulation of ETV-TP was such that potency against viruses with 3TCr substitutions was retained (20), a finding that was confirmed in clinical studies (4, 26). However, patient A had an unusually poor response to ETV, with a reduction in viral load of only about 2 log10 copies/ml after 1 year in comparison with similar phase II trial patients (4). Further treatment of patient A with combined ETV (0.5 mg) and 3TC (100 mg) resulted in only modest fluctuations in viral load. From week 108 to week 133 (>2.5 years of therapy), virologic breakthrough was noted, with an increase in HBV DNA of 1.2 log10 copies/ml, along with a corresponding increase in ALT levels. The atypical clinical picture that emerged for patient A may have favored the development of viral resistance; the viral load was not reduced below 6 log10 copies/ml despite ETV treatment for more than 2.5 years. It is important to note that the currently indicated ETV dose of 1.0 mg for patients with 3TCr HBV was established subsequent to the enrollment of patient A into clinical studies to further enhance the suppression of 3TCr HBV. The ETV resistance genotype for patient A, however, first emerged during treatment with 0.5 mg of ETV (Table 1).
Evaluation of samples from patient A resulted in the identification of a unique HBV RT substitution, rtM250V, that resides in conserved domain E of viral RT and appears to play a critical role in the expression of an ETV resistance phenotype. In a three-dimensional homology model of HBV and human immunodeficiency virus RTs, rtM250 lies in a position corresponding to the “primer grip” component that interacts with the 3′ terminus of the DNA template (8). While the 3TCr substitution rtM204V within the YMDD motif of RT likely affects the initial polymerase binding of deoxynucleoside triphosphate analog inhibitors, substitutions at residue 250 may affect other polymerization events, such as elongation. In vitro HBV cell culture and enzyme experiments with recombinant viruses from patient A revealed that the combination of 3TCr substitutions rtL180M and rtM204V with substitution rtM250V is required for high levels of ETV resistance. It is intriguing to postulate that changes in more than one functional stage of the polymerase, involving multiple substitutions, operated to bring about ETV resistance in patient A.
Patient B was a liver transplant recipient who had failed previous HBV therapies that had been administered over the course of 8.5 years (3). Multiple substitutions were noted in the RT domain of the baseline viral isolate, including those associated with failed 3TC or famciclovir therapy (1, 25; reviewed in reference 13). Patient B initially responded to ETV treatment with a reduction in viral DNA levels of 2.35 log10 copies/ml by week 24, a decrease that was maintained through week 60. However, as with patient A, viral DNA levels were never sustained below 6 log10 copies/ml, and elevations in both viral DNA and ALT levels were noted at week 76 (1.5 years of therapy). Importantly, the basis for a suboptimal initial response to ETV in this patient, despite the use of the 1.0-mg dose, may lie in the combination of baseline substitution rtT184S and other 3TCr substitutions. In keeping with this proposed model, cell culture results for patient B revealed an ∼2-fold increase in the EC50 of the baseline virus relative to an identical virus carrying the wt threonine at position 184 (Table 6). Substitution rtT184S has been found in some 3TCr isolates in combination with changes at residues 180 and 204 (1).
Genotypic analysis of patient B isolates showed simultaneous selection of substitutions rtT/S184G and rtS202I within the 3TCr background beginning at week 68 of ETV therapy. This observation is consistent with the results of the cell culture phenotypic analysis (Table 6), which showed that the quadruple mutant (rtL180M/rtT184G/rtS202I/rtM204V) exhibited the highest level of ETV resistance. We also observed that the recombinant virus containing 3TCr and rtS202I substitutions in the absence of the rtT184G substitution was severely replication impaired for the production of extracellular HBV and for the yield of intracellular nucleocapsids isolated for polymerase studies (both <5% wt values) (data not shown). Like the 3TCr substitutions rtL180M and rtM204V, rtT184G and rtS202I reside in domain B and domain C of polymerase, respectively. The rtT184 residue is conserved in alignments of 250 wt HBV genomes and among the human, woodchuck, and duck HBV RTs (9). While serine 202 is conserved among HBV wt sequences, the corresponding residues in woodchuck HBV and duck HBV are alanine and threonine, respectively (9). It is unclear what effects these differences would have in the context of 3TCr changes, although ETV is a very potent inhibitor of both woodchuck and duck HBV infections. A model of 3TCr HBV RT predicts that substitution rtL180M compensates structurally for substitution rtM204V, consistent with their coselection (1, 8). Perhaps the coselection of the changes at residues 184 and 202 in viruses from patient B represents similar compensatory changes in HBV polymerase.
Interestingly, substitution rtI169T, located in conserved domain B of polymerase, was selected in both patients, although it was found not to be responsible for the majority of resistance. Its role was tested by using recombinant viruses from patient A, and it was found to modestly increase the level of ETV resistance—more than two- to threefold—in two clones containing 3TCr and rtM250V substitutions while not affecting resistance in a third. Perhaps substitution rtI169T confers an ancillary or adaptive change and is analogous to substitution rtV173L found after the development of other 3TCr changes (10).
Discontinuation of 3TC therapy often results in the reappearance of the wt virus (5). This was not the case for patients A and B, despite treatment with ETV alone for at least 1 year (Tables 1 and 2). The virologic advantage of reduced ETV susceptibility may account for this finding, although shifts from 3TCr substitutions to wt residues during ETV therapy have been observed for other patients (unpublished observations). Nevertheless, retention of the 3TCr substitutions along with additional substitutions in viral RT was essential for the development of ETV resistance in these patients.
Substitutions in HBV polymerase are often accompanied by changes in HBsAg encoded in an overlapping reading frame, including those in response to antiviral therapy (27). The ETV-related RT substitutions rtI169T, rtT184G, and rtS202I seen in patients A and B result in HBsAg changes sF161L, sL176V, and sV194F, respectively. The results reported here support the concept that RT changes and not HBsAg substitutions conferred the decreased susceptibility to ETV seen in these patients. First, resistance was evident at the level of in vitro endogenous HBV polymerase activity, as determined by using cores that lack HBsAg. Second, resistance to ETV was seen in cultures when both extracellular (Tables 3 and 4) and intracellular (data not shown) nucleocapsids were assayed. Since the HepG2 transfection system is incapable of multiple infectious cycles involving virus release and reinfection, HBsAg arguably does not influence the levels of intracellular replicated HBV DNA within nucleocapsids. However, the experiments described here did not address the interaction of the resulting HBsAg substitutions and the patient's immune system in the control or selection of ETV-resistant viruses.
It is unclear which enzymatic functions of HBV polymerase are affected by the ETV resistance substitutions identified in the patients reported here. While phenotypic resistance to ETV in cell cultures was also evident in the in vitro enzyme assays, the magnitude of resistance appeared to differ. This result was also observed for viruses and enzymes with 3TCr substitutions and phenotypes (20). ETV-TP is a potent inhibitor of all three polymerase functions—priming and first (minus)- and second (plus)-strand DNA synthesis (23)—and all of these functional activities would be operational in the cell culture system. In contrast, the in vitro endogenous polymerase assay primarily evaluates functional steps postpriming. Nonetheless, it will be interesting to determine whether the various substitutions influence different polymerase activities.
In contrast to findings for 3TC, the overall results of evaluations to date suggest that the emergence of ETV-resistant variants likely will be infrequent in both patients who have not had treatment and patients who have been treated with 3TC (4; unpublished results). Of the two ETV-resistant variants identified, both were discovered in patients who had undergone treatment and contained key signature substitutions (rtL180M and rtM204V) associated with 3TCr prior to ETV treatment. Furthermore, the results described here indicate that resistance to ETV requires the presence of 3TCr substitutions, since the changes that emerged during ETV therapy had only modest effects on ETV susceptibility when tested alone.
Given its greater intrinsic antiviral potency and range of inhibitory activities, along with a predicted lower incidence of viral resistance over the course of long-term therapy, ETV offers the potential for an effective, safe, and durable treatment for HBV infections.
Acknowledgments
We thank Patricia Poundstone for excellent work in sequencing HBV DNA, Heidi Wang for help in preparing the manuscript, Sharon Zhang for help with initial experiments and methodology, Stephen Goff for the kind gift of plasmid pCMV-HBV, and Joe Baldick for comments on the manuscript.
REFERENCES
- 1.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. Tyrrell, N. Brown, L. D. Condreay, et al. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Angus, P., R. Vaughan, S. Xiong, H. Yang, W. Delaney, C. Gibbs, C. Brosgart, D. Colledge, R. Edwards, A. Ayres, A. Bartholomeusz, and S. Locarnini. 2003. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology 125:292-297. [DOI] [PubMed] [Google Scholar]
- 3.Aye, T. T., A. Bartholomeusz, T. Shaw, S. Bowden, A. Breschkin, J. McMillan, P. Angus, and S. Locarnini. 1997. Hepatitis B virus polymerase mutations during antiviral therapy in a patient following liver transplantation. J. Hepatol. 26:1148-1153. [DOI] [PubMed] [Google Scholar]
- 4.Chang, T.-T., S. Hadziyannis, J. Cianciara, M. Rizzeto, E. Schiff, G. Pastore, K. Klesczewski, G. Denisky, D. DeHertogh, and R. Hindes. 2002. Sustained viral load and ALT reduction following 48 weeks of entecavir treatment in subjects with chronic hepatitis B who have failed lamivudine. Hepatology 36:300A. [Google Scholar]
- 5.Chayama, K., Y. Suzuki, M. Kobayashi, A. Tsubota, M. Hashimoto, Y. Miyano, H. Koike, I. Koida, Y. Arase, S. Saitoh, N. Murashima, K. Ikeda, and H. Kumada. 1998. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology 27:1711-1716. [DOI] [PubMed] [Google Scholar]
- 6.Cihlar, T., D. C. Lin, J. B. Pritchard, M. D. Fuller, D. B. Mendel, and D. H. Sweet. 1999. The antiviral nucleotide analogs cidofovir and adefovir are novel substrates for human and rat renal organic anion transporter 1. Mol. Pharmacol. 56:570-580. [DOI] [PubMed] [Google Scholar]
- 7.Colonno, R. J., E. V. Genovesi, I. Medina, L. Lamb, S. K. Durham, M. L. Huang, L. Corey, M. Littlejohn, S. Locarnini, B. C. Tennant, B. Rose, and J. M. Clark. 2001. Long-term entecavir treatment results in sustained antiviral efficacy and prolonged life span in the woodchuck model of chronic hepatitis infection. J. Infect. Dis. 184:1236-1245. [DOI] [PubMed] [Google Scholar]
- 8.Das, K., X. Xiong, H. Yang, C. E. Westland, C. S. Gibbs, S. G. Sarafianos, and E. Arnold. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine and emtricitabine. J. Virol. 75:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaney, W., IV, A. Bartholomeusz, and S. A. Locarnini. 2002. Evolving therapies for the treatment of chronic hepatitis B virus infection. Expert Opin. Investig. Drugs 11:169-187. [DOI] [PubMed] [Google Scholar]
- 10.Delaney, W. E., IV, H. Yang, C. E. Westland, K. Das, E. Arnold, C. S. Gibbs, M. D. Miller, and S. Xiong. 2003. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J. Virol. 77:11833-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Man, R. A., L. M. Wolters, F. Nevens, D. Chua, M. Sherman, C. L. Lai, A. Gadano, Y. Lee, F. Mazzotta, N. Thomas, and D. DeHertogh. 2001. Safety and efficacy of oral entecavir given for 28 days in patients with chronic hepatitis B virus infection. Hepatology 34:578-582. [DOI] [PubMed] [Google Scholar]
- 12.Fallows, D., and S. Goff. 1995. Mutations in the epsilon sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J. Virol. 69:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain, M., and A. S. Lok. 1999. Mutations in the hepatitis B virus polymerase gene associated with antiviral treatment for hepatitis B. J. Viral Hepat. 6:183-194. [DOI] [PubMed] [Google Scholar]
- 14.Innaimo, S. F., M. Seifer, G. S. Bisacchi, D. N. Standring, R. Zahler, and R. J. Colonno. 1997. Identification of BMS-200475 as a potent and selective inhibitor of hepatitis B virus. Antimicrob. Agents Chemother. 41:1444-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai, C. L., J. Dienstag, E. Schiff, N. W. Leung, M. Atkins, C. Hunt, N. Brown, M. Woessner, R. Boehme, and L. Condreay. 2003. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin. Infect. Dis. 36:687-696. [DOI] [PubMed] [Google Scholar]
- 16.Lai, C. L., V. Ratziu, M. F. Yuen, and T. Poynard. 2003. Viral hepatitis B. Lancet 362:2089-2094. [DOI] [PubMed] [Google Scholar]
- 17.Lai, C.-L., M. Rosmawati, J. Lao, H. Van Vlierberghe, H. Anderson Frank, N. Thomas, and D. Dehertogh. 2002. Entecavir is superior to lamivudine in reducing hepatitis B virus DNA in patients with chronic hepatitis B infection. Gastroenterology 123:1831-1838. [DOI] [PubMed] [Google Scholar]
- 18.Lee, W. M. 1999. Hepatitis B therapy: the plot thickens. Hepatology 30:579-581. [DOI] [PubMed] [Google Scholar]
- 19.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 20.Levine, S., D. Hernandez, G. Yamanaka, S. Zhang, R. Rose, S. Weinheimer, and R. J. Colonno. 2002. Efficacies of entecavir against lamivudine-resistant hepatitis B virus replication and recombinant polymerases in vitro. Antimicrob. Agents Chemother. 46:2525-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marion, P. L., F. H. Salazar, M. A. Winters, and R. J. Colonno. 2002. Potent efficacy of entecavir (BMS-200475) in a duck model of hepatitis B virus replication. Antimicrob. Agents Chemother. 46:82-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mason, W. S., J. Cullen, G. Moraleda, J. Saputelli, C. E. Aldrich, D. S. Miller, B. Tennant, L. Frick, D. Averett, L. D. Condreay, and A. R. Jilbert. 1998. Lamivudine therapy of WHV-infected woodchucks. Virology 245:18-32. [DOI] [PubMed] [Google Scholar]
- 23.Seifer, M., R. K. Hamatake, R. J. Colonno, and D. N. Standring. 1998. In vitro inhibition of hepadnavirus polymerases by the triphosphates of BMS-200475 and lobucavir. Antimicrob. Agents Chemother. 42:3200-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seifer, M., R. Hamatake, M. Bifano, and D. N. Standring. 1998. Generation of replication-competent hepatitis B virus nucleocapsids in insect cells. J. Virol. 72:2765-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuyver, L. J., S. A. Locarnini, A. Lok, D. D. Richman, W. F. Carman, J. L. Dienstag, and R. F. Schinazi. 2001. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology 33:751-757. [DOI] [PubMed] [Google Scholar]
- 26.Tassopoulos, N., S. Hadziyannis, J. Cianciara, M. Rizzetto, E. Schiff, G. Pastore, V. Rutkiewicz, N. Thomas, G. Denisky, and S. Joshi. 2001. Entecavir is effective in treating patients with chronic hepatitis B who have failed lamivudine therapy. Hepatology 34:340A.11481619 [Google Scholar]
- 27.Torresi, J., L. Earnest-Silveira, G. Civitico, T. E. Walters, S. R. Lewin, J. Fyfe, S. A. Locarnini, M. Manns, C. Trautwein, and T. C. Bock. 2002. Restoration of replication phenotype of lamivudine-resistant hepatitis B virus mutants by compensatory changes in the “fingers” subdomain of the viral polymerase selected as a consequence of mutations in the overlapping S gene. Virology 299:88-99. [DOI] [PubMed] [Google Scholar]
- 28.Villeneuve, J. P., D. Durantel, S. Durantel, C. Westland, S. Xiong, C. L. Brosgart, C. S. Gibbs, P. Parvaz, B. Werle, C. Trepo, and F. Zoulim. 2003. Selection of a hepatitis B virus strain resistant to adefovir in a liver transplantation patient. J. Hepatol. 39:1085-1089. [DOI] [PubMed] [Google Scholar]
- 29.Wolters, L. M., B. E. Hansen, H. G. Niesters, D. DeHertogh, and R. A. de Man. 2002. Viral dynamics during and after entecavir therapy in patients with chronic hepatitis B. J. Hepatol. 37:137-144. [DOI] [PubMed] [Google Scholar]
- 30.Wong, D. K. H., A. M. Cheung, K. O'Rourke, C. D. Naylor, A. S. Detsky, and J. Heathcote. 1993. Effect of alpha-interferon treatment in patients with hepatitis B e antigen-positive chronic hepatitis B: a meta-analysis. Ann. Intern. Med. 119:312-323. [DOI] [PubMed] [Google Scholar]
- 31.Yan, J. H., M. Bifano, P. Nichola, E. O'Mara, E. Loizillon, D. Zhang, and S. Olsen. 2002. Entecavir pharmacokinetics after multiple doses in healthy subjects. J. Clin. Pharmacol. 42:1070. [DOI] [PubMed] [Google Scholar]
- 32.Zhou, T., J. Saputelli, C. E. Aldrich, M. Deslauriers, L. D. Condreay, and W. S. Mason. 1999. Emergence of drug-resistant populations of woodchuck hepatitis virus in woodchucks treated with the antiviral nucleoside lamivudine. Antimicrob. Agents Chemother. 43:1947-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]