Abstract
The microRNAs (miRNAs) are small non-coding RNA comprising approximately 19–25 nucleotides. miRNAs can act as tumour suppressors or oncogenes, and aberrant expression of miRNAs has been reported in several human cancers and has been associated with cancer initiation and progression. Recent evidence suggests that miRNAs play a major role in thyroid carcinogenesis. In this review, we summarize the role of miRNAs in thyroid cancer and describe the oncogenic or tumour suppressor function of miRNAs as well as their clinical utility as prognostic or diagnostic markers in thyroid cancer.
Keywords: microRNA, thyroid cancer, diagnosis, prognosis, treatment, circulating microRNA
Introduction
MicroRNAs (miRNA) are small non-coding RNAs that are approximately 19–25 nucleotides in length. The miRNAs regulate gene expression at the posttranscriptional level in both eukaryotic and prokaryotic cells. The discovery of miRNA has opened new insights into the biological processes leading to cancer initiation and progression. Although most miRNAs target the 3′ untranslated region of the mRNA targets, some miRNA can also interact with the coding sequence of the target gene. A single miRNA can bind to a large number of different gene mRNAs. Thus, miRNA regulation of mRNA plays a major role in several biological functions in normal and pathological conditions [1]. Although the expression of miRNAs is tightly regulated, cancer initiation and progression have been associated with dysregulated miRNA expression [2–6]. In several cancers, miRNAs have been shown to be good diagnostic and prognostic markers. Studies have also shown that the aberrant expression of miRNAs plays an important role in cancer cell proliferation, differentiation, invasion, migration and cell death. Upregulated miRNAs are considered as OncomiR, and downregulated miRNA act as tumor suppressors. However, the function of each miRNA is tissue- and context-dependent. Genetic alterations in the genes coding for miRNAs or epigenetic changes such as methylation contribute to the aberrant expression of miRNAs in some cancers and diseases.
Thyroid cancer is the most common endocrine malignancy [7]. There are four histological subtypes of thyroid cancer: papillary thyroid cancer (PTC), follicular thyroid cancer (FTC), poorly differentiated thyroid cancer (PDTC) and anaplastic or undifferentiated thyroid cancer (ATC) [8]. PTC is the most prevalent type of tumor among thyroid malignancies, accounting for ~80% of all thyroid cancer cases [9]. Medullary thyroid cancer, unlike the remainder of the thyroid lesions, is a neuroendocrine tumor that derives from C cells (parafollicular cells) and accounts for 3–5% of all thyroid cancers [10]. There is growing evidence that miRNAs play an important role in thyroid carcinogenesis and may have a role as diagnostic and prognostic biomarkers of thyroid neoplasms. In this review, we summarize recent studies that have explored the role of miRNA in thyroid cancer and describe the recent findings about the potential clinical application of miRNA as diagnostic and prognostic markers.
miRNA signature in thyroid cancer
Papillary thyroid cancer
Although the majority of PTC cases have excellent prognoses and responses to therapy, 10–15% of patients show signs of disease recurrence during follow up, which is associated with shorter overall survival [11]. PTC progression is often associated with the dedifferentiation of cancer cells and, as such, with increased resistance to radioactive iodine (RAI) therapy. Non-RAI avid disease is associated with a poor prognosis [12–14]. Therefore, a better understanding of the mechanisms involved in PTC progression and metastasis are very important and may have clinical ramifications. Several studies have analyzed miRNA profiles in PTC. However, only a few miRNAs were commonly dysregulated among the studies. The most consistently upregulated miRNAs are miR-146b, miR-222, miR-221 and miR-181b [15–19]. miR-221 and miR-222share the same seed sequence, and many studies have reported abnormal expression of these two miRNA in several cancers. The tumor suppressor and cell-cycle regulator p27 has been identified as a direct target of miR-221 and miR-222 in many cancers, including thyroid cancer [16]. Moreover, several studies have reported a significant downregulation of p27 in PTC compared to normal thyroid tissue [20–22]. Reduced expression of p27 in PTC due to overexpression of miR-221 and miR-222 might explain the proliferation rate observed in PTC cells. Microarray expression analysis of miRNA in BRAF V600E mutant versus wild-type tumors showed a higher level of miR-221 in BRAF mutant samples, which were characterized by aggressive behavior, advanced disease stage, extrathyroidal invasion and the presence of lymph node metastasis [23]. It has been proposed that BRAF V600E, by activating the NFkB pathway, promotes the expression of the oncogenic miR-221 [24,25].
The miR-146b is one of the most overexpressed miRNA in PTC compared to normal tissue, and its expression positively correlates with tumor aggressiveness as well as the presence of extrathyroidal tumor extension [26,27]. Analysis of the predicted targets of miR-146b showed that retinoic acid receptor beta (RARβ) has a putative miR-146b binding site in the 3′UTR region and that RARβ mRNA expression is significantly downregulated in PTC compared to normal tissue. RARβ expression is reduced in several cancers and showed a tumor suppressive function in several studies [28]. A pilot study in a cohort of patients with advanced PTC who were treated with retinoic acid, RARβ ligand, showed that 38% of the patients experienced a reduction in tumor size. Furthermore, 26% of the patients showed an increase in radioiodine uptake [29]. These findings suggest that miR-146b, by targeting RARβ, is involved in thyroid cancer initiation and progression. miR-222, miR-221 and miR-146b were found to be overexpressed in aggressive PTC, as their expression was associated with lymph nodes and distant metastases, risk of recurrence and the presence of BRAF V600E mutation.
Some studies have also observed overexpression of miR-181b in PTC compared to normal thyroid tissue. Analysis of the mechanism by which miR-181b regulates cell transformation and cancer initiation showed that miR-181b directly binds to the 3′UTR of CYLD and inhibits its expression. CYLD is downregulated in several cancers and inhibits the NFkB pathway [17,30]. CYLD also induces apoptosis and has been found to be underexpressed in several human cancers [31]. Studies focusing on down-regulated miRNAs in thyroid cancer showed that miR-145, miR-451, miR-613 and miR-137 were underexpressed in PTC compared to normal thyroid tissue [32–35]. In vitro and in vivo studies of thyroid cancer cells suggest that miR-145 is a master regulator of thyroid cancer growth and that it mediates its effect through the PI3K/Akt pathway [35]. Reporter assay experiments also revealed that miR-145 targets DUSP6 [34]. DUSP6 is a mitogen-activated protein kinase (MAPK) phosphatase enzyme that inactivates ERK1/2, p38 and FOXO1 [36–40]. DUSP6 is upregulated in PTC and PDTC [41], and it has been suggested that DUSP6 plays a tumor-promoting role in some human cancers, such as glioblastoma [42]. Taken together, these findings suggest that miR-145 has several targets and acts a tumor suppressor in thyroid cancer. miR-613 is downregulated in PTC compared to normal thyroid tissue [32]. In vitro and in vivo studies identified SPHK2 as a direct target of miR-613 in ATC [32]. Overexpression of SPHK2 rescued the anti-tumor effects of miR-613. Moreover, immunohistochemistry staining showed that SPHK2 is highly expressed in PTC compared to adjacent normal tissue, suggesting the inverse correlation between SPHK2 and miR-613 expression. These finding suggest that miR-613-SPHK2 is involved in PTC cell proliferation and invasion [32].
In a miRNA expression profiling study in PTC, miR-137 expression has been shown to be down-regulated [33]. In thyroid cancer cells, miR-137 inhibits cellular proliferation, invasion and migration and targets CXCL12 [33]. CXCL12 is a chemokine that was found to be upregulated in PTC and inversely correlated with miR-137 expression [33,43]. Binding of CXCL12 on its receptor CXCR4 leads to the activation of the oncogenic pathways ERK1/2, MAPK, JNK and Akt [44,45]. These data suggest that miR-137 has anti-tumor effects in PTC, partially by targeting CXCL12.
miR-451a has been studied in many human cancers and is downregulated in PTC [46–49]. Studies have observed inverse associations between miR-451a expression level and tumor aggressiveness, TNM stage and the presence of extrathyroidal invasion [49]. The miR-451 targets AKT1, a major component of the PI3k/AKT signaling pathway, which is known to be altered in thyroid cancer [49–51]. Thus, by targeting the PI3/AKT pathway, miR-451 acts as a tumor suppressor in PTC. A comparison of miRNA expression profiles between patients with PTC both with and without lymph node metastasis showed that the patients with loco-regional disease had higher miR-451 expression [52]. This finding suggests an oncogenic role for miR-451 in PTC. Further studies in larger cohorts are needed to explain the discrepant results on the role of miR-451 in thyroid cancer.
Follicular thyroid cancer
FTC is characterized by activating RAS mutations and PAX8/PPARγ rearrangement in 40–53% and 25–63% of FTCs, respectively [53,54]. Data regarding miRNA expression profiles in FTC are very limited, and to our knowledge there is no study that correlates the genetic alteration of FTC with abnormal miRNA expression. Downregulation of miR-199a-5p and miR-144 has been observed in FTC, whereas miR-197 and miR-346 are overexpressed in FTC compared to follicular adenoma. A comparison of the miRNA expression in two histological types of FTC (conventional and oncocytic variants) revealed an upregulation of miR-182/-183/-221/-222/-125a-3p and downregulation of miR-542-5p/-574-3p/-455/-199a in both histological types compared to normal thyroid tissue [55]. However, miR-885-5p was found to be upregulated in the oncocytic variant of FTC only. Of all the microRNA that showed a significant up or downregulation in FTC, only miR-199a-5p was studied in vitro. This miRNA targets CTGF, which leads to the inhibition of cell cycle progression [56]. miR-146b and miR-221 were found to be significantly upregulated in FTC compared to normal thyroid tissue, suggesting that these two miRNA are not specific for PTC but are common to well-differentiated thyroid cancers [57].
Anaplastic thyroid cancer
ATC is the most aggressive type of thyroid cancer. Although ATC represents less than 2% of all thyroid tumors, this neoplasm causes 14%–39% of thyroid cancer–related deaths [58–60]. ATC is resistant to most of the conventional therapies and is refractory to radioiodine treatment [61]. Thus, ATC patients display a rapidly progressive disease that may cause death within six months [59,61,62]. ATC can originate from a preexisting well-differentiated cancer. This hypothesis is supported by studies documenting the presence of mutations typical for differentiated thyroid cancer, such as BRAF and RAS, in substantial proportion of ATC [63,64]. However, the TP53 mutation is found exclusively in ATC [65,66]. Many miRNAs have been found to be dysregulated in thyroid cancer, but only a few miRNAs (e.g., miR-200a, b and c) are exclusively dysregulated in ATC, which suggests a role in cancer progression. Downregulation of miR-200 family members has been described in many metastatic tumors. The miR-200c is transcriptionally regulated by TP53, and TP53 inactivating mutations in ATC lead to miR-200c downregulation [67]. The two main targets of miR-200s are the epithelial-mesenchymal-transition (EMT) transcription factors ZEB1 and ZEB2 [68]. EMT is a mechanism that enables cells to migrate and invade [69,70]. This process is mediated by the EMT transcription factors (such as ZEB1, ZEB2, SNAIL and SNAI2) that repress the epithelial features of cancer cells and disrupt the cell–cell adhesion mediated by loss of E-cadherin [71]. On the other hand, the EGF/EGFR pathway leads to silencing of miR-200s and an increase in Rho/Rho kinase activity as well as EMT and cancer cell invasion [72]. EGFR expression is greater in ATC compared to PTC and normal thyroid tissue, and the inhibition of EGFR pathways leads to cell death [73]. Taken together, these data suggest that the EGFR pathway and a TP53 mutation in ATC lead to a loss of miR-200 expression, which in turn increases ZEB1 and ZEB2 transcripts and protein levels and blocks E-cadherin expression.
It has been reported that BRAF V600E oncogene activation stimulates the expression of the miR-17-92 cluster [74]. The primary transcript of miR-17-92 is processed into seven different mature miRNAs: miR-17 (miR-17-5p and miR-17-3p), miR-18a, miR-19a, miR-19b, miR-20a and miR-92a [75,76]. An important target of the miR-17-92 cluster is the tumor suppressor PTEN [77]. Reporter assays revealed the existence of several binding sites for miR-19a/b, miR-17 and miR-20a in the 3′-UTR of PTEN. In fact, downregulation of PTEN levels is a key step during tumorigenesis that potentiates the activation of AKT/mTOR growth and survival signaling. The data suggest that these miRNAs play a role in cancer initiation and progression in ATC. Another important tumor-suppressive pathway targeted by several members of miR-17-92 is the TGFβ signaling pathway. By activating the NOTCH pathway, BRAF V600E increases the transcriptional activity of the main oncogenic component of the 17-92 cluster, which is miR-19 [74]. In turn, miR-19 inhibits SMAD4 expression. SMAD4 mediates the tumor-inhibitory effect of the TGFβ signaling [74]. The TGFβ signaling pathway is an important regulator of epithelial cell proliferation, with an anti-proliferative effect mediated by p21. Thus, the BRAF V600E–mediated increase of miR-19 leads to a loss of the tumor inhibitory effect of TGFβ in thyroid cancer cells, which enhances cell proliferation.
miR-30 family members clearly distinguish between ATC and differentiated thyroid cancer, as downregulation of miR-30 was found only in ATC [78,79], suggesting that miR-30 plays a role in thyroid cancer differentiation and progression. Overexpression of miR-30 family members led to a change in cell morphology and decreased vimentin expression in ATC cells [79,80]. Overexpression of all miR-30 family members decreases SMAD2 expression [79]. A reporter assay experiment showed that miR-30e binds directly to the SMAD2 3′UTR region and thus inhibits its expression. SMAD2 is significantly upregulated in ATC compared to normal thyroid tissue and well-differentiated thyroid cancer, and it regulates the invasive potential of ATC cells. miR-30a also plays a tumor-suppressive role in ATC. Low expression of miR-30a was associated with poor prognosis, extrathyroidal invasion and shorter survival [80]. Analysis of the predicted targets of miR-30a showed that overexpression of miR-30a significantly decreases lysyl oxidase (LOX) expression. LOX is strongly expressed in ATC and aggressive thyroid cancers and is also associated with higher mortality rate [80]. The miRNA signature in ATC suggests that miR-200 and miR-30 family members are specific to undifferentiated thyroid tumors and are involved in cancer progression by regulating the EMT process.
miR-146a-5p and miR-146b-5p, two of the most upregulated miRNAs in PTC, are also overexpressed in ATC. A comparison of the expression of miR-146a in PTC foci within ATC showed a much higher expression in ATC compared to the differentiated areas in tumor samples [81]. In vitro studies have shown that NFkB controls the transcription of miR-146a in lymphoma and ATC cells [82,83]. NFkB is overactivated in ATC and controls the expression of anti-apoptotic genes. These results suggest that the pro-tumorigenic role of NFkB in ATC may be mediated, at least in part, by miR-146b.
Poorly differentiated thyroid cancer
The clinical behavior of PDTC falls between differentiated thyroid cancer and ATC. Patients diagnosed with PDTC have a higher risk of recurrence. Approximately 15% and 35% of PDTCs harbor BRAF and RAS mutations, respectively, and 17–30% have a TP53 mutation [84–87]. This suggests that PDTC may originate from differentiated tumors. However, the biology and molecular mechanisms leading to PDTC are not fully understood. Analysis of the miRNA profile in PDTC tumors showed that miR-183-3p is upregulated in PDTC compared to PTC and normal tissue [88]. miR-150 and miR-23b were both downregulated in PDTC. These miRNAs have been associated with poor prognosis in many human cancers and are significantly associated with tumor relapse and mortality in PDTC [89,90]. In vitro studies showed that the EMT transcription factor ZEB1 is a direct target of miR-150. Analysis of the epigenetics alteration of miR-23b showed that the gene promoter is silenced by hypermethylation. Moreover, the SRC oncogene was identified as a direct target of miR-23b [90]. These data provide new insights into the mechanism leading to cancer progression and metastasis in aggressive thyroid cancers.
Medullary thyroid cancer
Medullary thyroid cancer (MTC) is a neuroendocrine tumor that originates from the parafollicular cells (C cells). Approximately 25% of MTCs are caused by germline mutation of the RET proto-oncogene [91,92]. It has been shown that miR-129-5p was significantly downregulated in MTC compared to normal tissue, and overexpression of miR-129-5p in vitro decreased cellular invasion and migration and inhibited the phosphorylation of AKT [93]. Furthermore, a reporter vector assay showed that miR-129-5p directly targets RET in MTC cells [93]. Another study analyzing the miRNA signature in six pairs of primary tumors as well as lymph node metastasis from patients with sporadic MTC revealed that miR-7, miR-10a, miR-29c and miR-200b/-200c were significantly downregulated in metastatic sites compared to primary tumors. Additionally, five miRNAs were upregulated in metastatic tumors: miR-130a, miR-138, miR-193a-3p, miR-373 and miR-498. The miR-200 family has been shown to be a key regulator of the EMT process, as it targets EMT transcription factors such as ZEB1, ZEB2, SNAI1, SNAI2 and TWIST-1. In fact, inhibition of the miR-200 family in MTC cell lines led to downregulation of E-cadherin and upregulation of vimentin, TGFβ1 and TGFβ2 [94]. Further studies on a larger cohort of MTC patients are needed to fully understand the role of miRNA in the biology of MTC and their interaction with the RET proto-oncogene.
Biogenesis of miRNA in thyroid cancer
The biogenesis of miRNA starts with miRNA genes that are transcribed with RNA polymerase II into long, non-coding primary RNA (pri-miRNA) [95]. The pri-miRNA is further processed by a complex, formed from the RNase III enzyme DROSHA and DGCR8 protein, that generates the pre-miRNA [96]. The pre-miRNA is then exported into the cytoplasm by Exportin-5 (XPO5). Pre-miRNA is cleaved by DICER, generating a short duplex molecule. The mature miRNA strand is then loaded into an RNA-induced silencing complex (RISC) that will guide the mature miRNA to the target mRNA sequence [97]. Dysregulation of DICER and DROSHA expression has been observed in several tumors and has been associated with oncogenic transformation of epithelial cells [98–100]. In fact, decreased DICER mRNA has been found in thyroid cancer and is associated with tumor aggressiveness, extrathyroidal invasion, lymph nodes and distant metastasis, recurrence and BRAF V600E mutation [101]. Furthermore, miRNA abundance was lower in malignant tissue compared to normal thyroid tissue. These data suggest that abnormal DICER expression leads to dysregulated miRNA expression, which contributes to follicular cell transformation and cancer initiation. MTC is characterized by a general increase in miRNA levels [102]. Analysis of the expression profile of four genes involved in miRNA biogenesis (DROSHA, DICER, DGCR8 and XPO8) in a large cohort of MTC cases showed that DICER, DCGR8 and XPO8 mRNA were significantly upregulated in MTC with a RET mutation. Among the MTC group with a RET mutation, tumors with a codon 634 mutation had more pronounced increased in DICER1 and DGCR8 compared to wild-type tumors. However, tumors with RAS mutations did not show any significant difference compared to wild-type tumors [103]. It is still not known whether abnormal expression of the enzymes involved in miRNA biogenesis is responsible for miRNA dysregulation and cancer initiation, and further studies are needed.
Circulating miRNAs in thyroid cancer
Recent findings showed that miRNA can be detected in blood, saliva, urine and milk, suggesting secretion of miRNA by the cells and potential implications in cell–cell communication [104,105]. Furthermore, circulating miRNA can also be altered in several pathological conditions, including cancer. Circulating miRNAs can either be secreted by passive leakage from dead cells, by active secretion in exosomes or with RNA-binding proteins (RBP) such as AGO2 or NPM1. miRNAs are protected from RNase degradation either through active secretion in vesicles or by binding to RBP. In the past few years, studies have attempted to elucidate the role of circulating miRNA in cancer [106–110]. It has been proposed that miRNA secreted by cancer cells may modify the tumor micro-environment [111]. The expression profile of miRNA in tumor cells and in the exosomes is the same, suggesting that circulating miRNA can be used as diagnostics markers in cancer [112,113]. In thyroid cancer, circulating miR-146b and miR-155 discriminated between benign lesions and PTC [114]. miR-221 and miR-222 were found to be elevated in serum samples from patients with PTC. Furthermore, total thyroidectomy and lymph node dissection decreased the levels of circulating miR-221 and miR-222 levels [115]. Comparing the secretion of miRNA in the serum of patients with PTC to those with benign lesions showed that the level of circulating miR-25-3p and onco-miR-451 were significantly higher in plasma from PTC patients [116]. Both miRNAs were significantly reduced after surgical removal. Taken together, these data suggest that miRNAs are secreted by cancer cells and can be used as non-invasive diagnostic markers.
There is no direct evidence that secreted miRNA plays a role in cell communication that promotes the transformation of adjacent normal cells or that it promotes metastasis. Interestingly, analysis of the circulating tumor suppressor miR-145 in PTC showed higher levels in the exosomes from serum in patients with PTC compared to patients with benign lesions or healthy controls [35]. This suggests that cancer cells may use active secretion in exosomes to eliminate miRNAs with anti-cancer effects. Given the emerging role of circulating miRNAs in thyroid cancer, further studies are needed to elucidate the role of circulating miRNAs and their potential use as diagnostic markers.
All of the above findings and observations suggest an important role of miRNA in thyroid cancer initiation, proliferation and differentiation (Table 1, Fig. 1). The next step is to assess miRNA’s potential utility as a diagnostic and prognostic tool.
Table 1.
Dysregulated miRNA and target genes in thyroid cancer.
| miRNA | Gene target(s) | Studies |
|---|---|---|
| miR-145 |
DUSP6 Akt3 |
Gu Y et al., 2015 Boufraqech M et al., 2014 |
| miR-30a | LOX | Boufraqech M et al., 2015 |
| miR-30e | SMAD2 | |
| miR-144 | ZEB1 and ZEB2 | Guan H et al., 2015 |
| miR-222 |
p27 PTEN |
Zhong C et al., 2015 Li B et al., 2016 |
| miR-221 | PTEN | Li B et al., 2016 |
| miR-146a | RARB | Czajika A et al., 2016 |
| miR-26a | CKS2 | Lv M et al., 2013 |
| miR-34b |
MET C-MYC |
Migliore C et al., 2008 Leucci E et al., 2008 |
| miR-21 |
THRB PDCD4 PTEN BCL2 CDC25A |
Jazdzewski K et al., 2011 Lu Z et al., 2008 Meng F et al., 2007 Wang P et al., 2009 |
| Let-7 |
RAS HMGA1, HMGA2 MYC CCNB1 CDK1 CDC25C |
Johnson M et al., 2005 Lee Y.S et al., 2007 Sampson V.B. et al., 2007 Johnson C.D et al., 2007 Johnson C.D et al., 2007 Johnson C.D et al., 2007 |
| miR-200 | ZEB1 | Sun J et al., 2015 |
| miR-199a | CTGF | Sun D et al., 2016 |
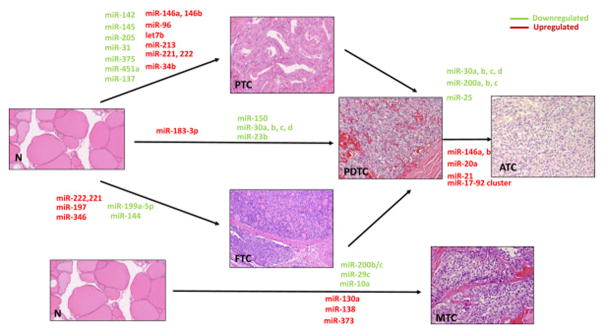
Fig. 1.
miRNAs signature in different histological types of thyroid cancer.
miRNA analysis in thyroid cancer diagnosis
The reporting system and cytological characteristics of benign versus malignant thyroid lesions were variable until the National Cancer Institute Thyroid Fine-Needle Aspiration State of the Science Conference in 2007 provided consensus recommendations known as the Bethesda System for Reporting Thyroid Cytopathology. The Bethesda system recognizes six diagnostic categories and provides an estimation of cancer risk within each category:
Category I, Nondiagnostic or Unsatisfactory: Estimated risk of malignancy 1–4%
Category II, Benign: Risk of malignancy 0–3%
Category III, Atypia of Undetermined Significance or Follicular Lesion of Undetermined Significance (AUS/FLUS): Cancer risk 5–15%
Category IV, Follicular Neoplasm or Suspicious for a Follicular Neoplasm (FN/SFN), Hürthle Cell Neoplasm or Suspicious for Hürthle Cell Neoplasm: Risk of malignancy 15–30%
Category V, Suspicious for Malignancy (SUSP): Risk of thyroid cancer 60–75%
Category VI, Malignant – Risk of thyroid cancer 97–99% [117].
The cytopathology evaluation is characterized by high accuracy when the nodules belong to Bethesda II (benign) with a negative predictive value (NPV) > 95% or Bethesda VI (malignant) with a positive predictive value (PPV) > 98%. Unfortunately, approximately 30–35% of nodules are characterized by indeterminate cytology. In a meta-analysis of 25,445 thyroid fine-needle aspiration (FNA) samples, 9.6% of all samples were diagnosed as AUS/FLUS with cancer risk of 15.9%, while 10.1% were diagnosed as FN/SFN with cancer risk of 26.1% and 2.7% of the nodules were diagnosed as SUSP with an average cancer risk of 75.2% [118]. According to the 2015 American Thyroid Association guidelines, diagnosis of AUS/FLUS occurs in 2–18% of nodules, FN/SFN in 2–25% of nodules and SUSP in 1–6% of nodules [1]. In addition to traditional cytological evaluation, molecular adjunct testing has been implemented to help refine FNA diagnosis for inconclusive results.
Initial studies describing miRNAs profiles in FNA biopsy focused on the distinction between malignant and benign thyroid lesions in general and documented a panel of miRNAs significantly over-expressed in thyroid cancer compared with benign cells, including miR-221, miR-222 and miR-146b, all of which are involved in regulation of the KIT oncogene responsible for cell growth and differentiation [15] (Table 2). Pallante et al. documented significant overexpression of miR-221, miR-222 and miR-181b in FNA samples derived from PTC versus normal thyroid cells [17] (Table 2). Chen and colleagues found that miRNAs 221, 222 and 146b were significantly differentially expressed between 20 benign and 20 malignant thyroid tumors with ex vivo FNA samples (Table 2) [119]. Interestingly, in this study, miR-221 and miR-222 were only significantly different between PTC and follicular adenoma but not between PTC versus hyperplastic nodule [119]. Mazeh et al. found that the most accurate determinants of malignant lesions were miR-146b, miR-221 and miR-222, with an accuracy rate of 96%, 98% and 92%, respectively (Table 2) [120]. Nikiforova and associates performed miRNA profiling using a probe designed to detect 158 human miRNAs and analyzed miR expression levels in both thyroid tumors and normal thyroid tissue. This study resulted in the selection of a set of seven miRs (miR-187, -221, -222, -146b, -155, -224 and -197) that were the most accurate in differentiating between thyroid cancer and hyperplastic nodules. The team then applied this set to a test sample set for validation in 62 consecutive patients undergoing FNA biopsy of a thyroid nodule(s). Unfortunately, the data could not be fully validated, as only 13 out of 62 patients underwent surgery and thus had a histologic diagnosis (Table 2) [19]. Moreover, the clear distinction between the malignant (Bethesda category VI) and benign nodules (Bethesda II) is achieved with extremely high accuracy by cytology evaluation alone, without the need for implementation of molecular diagnostics. Thus, subsequent studies should focus on the utility of miRNA profiling as a tool for improving thyroid cancer risk estimation in the most clinically challenging indeterminate nodules (AUS/FLUS, FN/SFN). Based on a screening study of 850 miRNAs performed on 104 benign and malignant tissue samples, Vriens et al. identified four miRs significantly differently expressed between malignant and benign lesions (miR-100, miR-125b, miR-138 and miR-768-3p). The results were validated on 125 indeterminate FNA biopsy samples, of which 37 turned out to be malignant at histologic evaluation. In this study, miR-138 had an accuracy of 75% for distinguishing between benign and malignant indeterminate nodules. However, the NPV was 81%, meaning this test can’t be used to rule out malignancy (Table 2) [121].
Table 2.
The diagnostic accuracy of miRNA profiling in differentiating between malignant and benign thyroid nodules.
| Study | #FNA samples | miR | Accuracy | Sensitivity | Specificity | NPV | PPV |
|---|---|---|---|---|---|---|---|
| Malignant vs. benign FNAs | |||||||
| Pallante et al., 2006, malignant vs. benign | FNA SUSP n = 8 | 221, 222,181b over-expressed in cancer | n/a | n/a | n/a | n/a | n/a |
| Normal thyroid n = 8 | |||||||
| Chen et al., 2008, PTC vs. benign | Ex vivo FNAs n = 40 | 146b, 221, 222 over-expressed in PTC | n/a | n/a | n/a | n/a | n/a |
| Hyperplastic nodule n = 10 | |||||||
| Follicular adenoma n = 10 | |||||||
| Classic PTC n = 10 | |||||||
| FVPTC n = 10 | |||||||
| Mazeh et al., 2011, PTC vs. benign | FNAs of PTC n = 20 | 21 | 79% | 50% | 100% | 100% | 73% |
| benign n = 7 | 31 | 77% | 45% | 100% | 100% | 71% | |
| 146b | 91% | 80% | 100% | 100% | 87% | ||
| 187 | 79% | 50% | 100% | 100% | 73% | ||
| 221 | 98% | 90% | 100% | 100% | 93% | ||
| 222 | 96% | 90% | 100% | 100% | 93% | ||
| Nikiforova et al., 2008, malignant vs. benign | Consecutive FNA n = 62, 13/62 with hist-pat diagnosis | 187,221, 222, 146b, 155, 224, 197 | |||||
| >1 miR upregulated | 95% | 100% | 94% | n/a | n/a | ||
| >3 miRs upregulated | 98% | 88% | 100% | n/a | n/a | ||
| Indeterminate FNAs | |||||||
| Vriens et al., 2012, malignant vs. benign | Indeterminate FNA n = 115 (benign n = 78, malignant n = 37) | miR 138 | 75% | 81% | |||
| Kitano et al., 2012, malignant vs. benign | Indeterminate FNAs study group n = 47/95, validation group n = 24/59 | 7, 126 down-regulated | 100% | 20% | 100% | 25% | |
| Keutgen et al., 2012, malignant vs. benign | Indeterminate FNAs study group n = 29, validation group n = 72 | 222, 328, 197, 21 | 90% | 100% | 86% | ||
| Shen R et al., 2012, malignant vs. benign | FNAs study group n = 60, validation group n = 68 | 146b, 221, 187, 30d | 85.3% | 88.9% | 78.3% | 89% | 78% |
| Mazeh et al., 2013, malignant vs. benign | Prospective cohort study 11 indeterminate FNAs | 21, 31, 146b, 187, 221, 222 | 90% | 89% | 100% | 66% | 100% |
| Labourier et al., 2015, malignant vs. benign | Indeterminate FNAs n = 109 | Multiplatform miR + mutation test | 89% | 85% | 94% | 74% | |
| AUS/FLUS n = 58 | AUS/FLUS | 94% | 80% | 97% | 68% | ||
| FN/SFN n = 51 | FN/SFN | 82% | 91% | 91% | 82% | ||
n/a – not applicable.
FNA – fine-needle aspiration.
SUSP – suspicious for thyroid cancer.
PTC – papillary thyroid cancer.
FVPTCFV – follicular variant of papillary thyroid cancer.
AUS/FLUS – atypia of undetermined significance or follicular lesion of undetermined significance.
FN/SFN – follicular neoplasm or suspicious for a follicular neoplasm.
Keutgen and colleagues utilized an ex vivo FNA biopsy approach in 29 thyroid nodules with indeterminate cytology, 14 of which were malignant on final histopathology. The authors performed a targeted analysis of miRNA expression known to be deregulated in malignant versus benign thyroid lesions (miR-222, miR-181, miR-146b, miR-328, miR-197 and miR-21) and used statistical modeling in order to find out which set of miRNAs had the highest diagnostic accuracy. They found a set of four miRNAs (miR-222, miR-328, miR-197 and miR-21) that have an 86% accuracy in the training set and 90% accuracy in a validation set of 72 indeterminate thyroid FNA biopsy samples (Table 2) [122]. Mazeh et al. implemented an even more practical approach. The authors utilized miRNA amplification from the residual cells left in 77 FNA biopsy samples and analyzed a set of six miRNAs (miR-21, miR-31, miR-146b, miR-187, miR-221 and miR-222) [123]. The analysis included only 11 out of 77 collected samples, which were characterized by the indeterminate cytology and with final pathology data available. Although the authors describe very high accuracy for the test (90% with 100% specificity and 100% PPV), the small study sample size does not allow for a strong conclusion. Moreover, the high PPV in this study is most likely a reflection of the prevalence of thyroid cancer in this cohort being 81% (9/11 patients).
Kitano et al. analyzed expression levels of four miRNAs (miR-7, miR-126, miR-374a and miR-let-7g) in 95 Bethesda I–VI category FNA biopsy samples as the training set and 59 FNA biopsy samples in the validation set. The application of the model to non-satisfactory and indeterminate FNA biopsy samples (47/95 in the study set and 24/59 in the validation FNABs) yielded a sensitivity of 100%, specificity of 20%, PPV of 25%, and NPV of 100%, suggesting that miR-7 and miR-126, significantly different between benign and malignant tumors, can be used as a “rule-out” test sparing diagnostic thyroidectomy [15].
Shen et al. performed a targeted analysis of eight miRNAs with well-established associations with thyroid cancer (miR-146b, -221, -187, -197, -346, -30d, -138 and -302c) in 60 indeterminate FNA biopsy samples. Based on this analysis, a set of four miRNAs (miR-146b, miR-221, miR-187 (upregulated) and miR-30d (downregulated)) correctly predicted the histologic diagnosis in 56 out of 60 cases. The diagnostic accuracy was 93.3%, while sensitivity and specificity were 93.2% and 93.8%, respectively. Nevertheless, in the validation sample including 30 AUS/FLUS cases, the diagnostic accuracy was low—only 73.3%—with a sensitivity of 63.6%, specificity of 78.9%, PPV of 64% and NPV of 79%. The miR prediction method in this study was accurate for papillary thyroid cancer cases, with malignant prediction rates of 35/36 (97.2%) and 34/36 (94.4%) in the training and validation sample sets, respectively. Unfortunately, the malignant prediction rates for FTC were relatively low—4 out of 5 (80%) in the training sample set and 5 out of 8 (62.5%) in the validation sample set—suggesting that this miR profiling is not useful in discriminating between follicular adenoma and FTC.
Several studies have tried to identify the molecular miRNA profile that distinguishes FTC from benign thyroid lesions. Weber and colleagues analyzed tissue samples derived from 23 FTCs, 20 follicular adenomas (FA) and 4 normal thyroid tissues and found miR-197 and miR-346 to be significantly overexpressed in FTC. Moreover, functional studies documented that in vitro overexpression of either miR induced proliferation and repressed the expression of their predicted target genes in vitro and in vivo, whereas inhibition led to growth arrest [124]. Nikiforova et al. confirmed differential upregulation of miR-187 and miR-197 in FTC [19]. However, there was an overlap between the large panel of upregulated miRNAs in FA and FTC. Kitano and associates profiled the expression of 1263 human miRNAs in 47 tumor samples (21 benign and 26 malignant) and focused on candidates discriminating between FA versus FTC, Hürthle cell adenoma (HA) versus Hürthle cell cancer (HCC) and FA/HA versus FTC/HTC/follicular variant of papillary thyroid cancer (FVPTC). The analysis revealed significant downregulation of miR-126 and miR-7 in malignant tumors [125]. The above mentioned studies analyzed tissue samples, not cytology samples obtained after FNA biopsy. To our knowledge, there are no data reported from prospective cohort studies yet.
An interesting strategy to increase the diagnostic accuracy of adjunct molecular testing is combining testing for common somatic mutations with miRNA profiling. Labourier et al. combined the seven-gene mutational testing with expression of a set of 10 miRNAs in preoperative FNA samples from 109 patients with AUS/FLUS or FN/SFN cytology and observed an 89% sensitivity and 85% specificity with a 73% PPV, 94% NPV and 32% prevalence of malignancy [126]. It is important to realize that the predictive value of any diagnostic procedure, whether based on pathology review or molecular testing, varies according to the pretest probability of the condition of interest in the evaluated population. Therefore, the authors used the Bayes’ theorem and modeled the accuracy of the test based on different cancer prevalence. The PPV was predicted to be greater than 50% for any prevalence greater than 15% and the NPV would range from 93 to 98% for a cancer probability greater than 15% and less than 35%. Thus, the utilization of this approach may result in an 85% decrease in the number of diagnostic thyroidectomies that may be performed in the absence of molecular testing [126].
In summary, there is currently no validated miRNA test available in clinical practice. Since the reproducibility of molecular tests is highly variable in clinical practice, as both NPV and PPV are affected by the prevalence of thyroid cancer, there is a need to validate the implementation of miRNA profiling in FNA biopsy in a large multicenter prospective study that includes patients from tertiary referral centers as well as community centers.
miRNAs as prognostic markers and therapeutic targets in thyroid cancer
Given the important role of miRNAs in cancer initiation and/or progression, several investigators have proposed that miRNAs could serve as prognostic markers and therapeutic targets. Several researchers have documented upregulation of miRNA-146b, miRNA-221, miRNA-222, miRNA-155 and miRNA-31 and downregulation of miRNA-1, miRNA-34b, miRNA-130b and miRNA-138 as associated with aggressive forms of PTC [127–129]. Based on a study of miRNA expression in tissue samples from patients with thyroid cancer recurrence and long-term remission (average follow-up of 120 months), Sonderman and associates observed that miRNA-9 and miRNA-21 could serve as prognostic markers for PTC recurrence [130]. Dettmer and colleagues found an independent and significant association between miR-23b levels and thyroid cancer recurrence and miRNA-150 levels with thyroid cancer–related death [88]. Abraham et al. found that upregulation of miRNA-183 and miRNA-375 in MTC predicted lateral lymph node metastasis and was associated with residual disease, distant metastasis and mortality [131].
There is a growing body of evidence that the expression of certain miRNA is associated with sensitivity to certain chemotherapeutic agents. Zhang et al. documented that deregulation of miR-30d, which is involved in beclin1 expression in ATC cells, is responsible for the insensitivity to cisplatin by promoting autophagy survival [132]. Lankenau et al. examined the mechanisms of resistance to the BRAF inhibitor vemurafenib in BRAF V600E mutant PTC cells. The authors found that concomitant targeting of BRAF with vemurafenib and miRNA-3151 either with proteasome inhibitors, to limit its binding capacity to SP1/NF-κBs, or with antagomiRNA-3151 has synergistic pro-apoptotic effects [133]. Varmeh et al. identified a set of miRNAs that could be used as biomarkers of resistance to BRAF inhibitors in patients with thyroid cancer [134]. Lasalle and colleagues demonstrated that overexpression of miRNA-375 leads to downregulation of SEC23A—a protein involved in transport between the endoplasmic reticulum and Golgi apparatus—as well as increased sensitivity of MTC cells to vandetanib [135].
Several miRNAs (e.g., miR-339-5p and miR-195) are involved in the regulation of thyroid-specific genes, in particular the sodium-iodine (NIS) symporter, a key protein involved in the response to radioactive iodine (RAI) treatment [136]. Overexpression of miRNA-146b is commonly observed in thyroid cancer and may play a role in dedifferentiation and resistance RAI therapy. miRNA-146b impairs NIS mRNA translation and subsequently decreases protein levels and, consequently, uptake of iodine via NIS. Interestingly, TSH treatment results in a strong downregulation of miR-146b and induction of NIS expression, which is reversed by IGFI and TGFβ, two well-established repressors of TSH-induced NIS expression [137]. Li et al. also showed that inhibition of miR-146b by histone deacetylase 3 (HDAC3) in vitro in FTC cells leads to upregulation of NIS [138]. Knockdown of miRNA-21 is another potential mechanism of induction of NIS expression, which leads to induction of differentiation in ATC cell lines [139].
The studies described above used in vitro models of targeting regulatory miRNAs, which begs the question: Is there a chance to translate these finding from bench to bedside? Several strategies are employed to target miRNA expression in vivo. The majority of the attempts to develop therapeutic agents have involved synthesis of expression vectors (miRNA sponges), small-molecule inhibitors and antisense oligonucleotides (ASOs). The molecular biology techniques enabled the synthesis of various chemically modified and conjugated oligonucleotides targeted to efficiently block miR function in vitro and in vivo. In fact, the first miRNAs-targeting therapeutic (anti-sense inhibitor of miRNA-122) is now in clinical trials for hepatitis C virus (HCV) infection (clinicaltrials.gov NCT01727934). The initial phase 1 clinical trial in solid tumors using siRNAs as the therapeutic agent targeted tissues of interest with a special nanoparticles delivery system. That led to the reduction of mRNA and protein expression of the product of the gene of interest in cancer tissue [140].
There are currently ongoing pre-clinical studies on miRNA-221 antisense inhibitors in hepatocellular carcinoma. Since miRNA-221 is commonly overexpressed in thyroid cancer and plays a role in thyroid cancer proliferation, testing this compound in pre-clinical thyroid cancer models may prove useful.
Another way to use miRNAs therapeutically is compound library screening. This approach identifies small molecules specifically inhibiting miRNA expressions, such as azobenzene, which affects miRNA-21 expression [141], as well as several other small molecules that inhibit miRNA-122 [142]. However, the therapeutic potential of these molecules is limited by their high EC50 values, which are in the micromolar range, making it difficult to achieve in human plasma and tissues of interest [143]. Another approach focuses on reinstating certain miRNAs that serve as tumor suppressors. Currently, there is one ongoing phase 1 clinical trial focused on miRNA-34 replacement in liver cancer or liver metastases of other cancers (clinicaltrials.gov NCT01829971).
Let-7 was shown to induce growth arrest in multiple cancer cell lines and to suppress tumor growth in a xenograft model of human lung cancer [144,145]. Interestingly, Let-7 miRNA was found to reduce MAPK/ERK signaling, inhibit proliferation and induce differentiation in human thyroid cancer cell lines with RET/PTC rearrangement [146]. Using Let-7 as a potential miRNA replacement for cancer treatment is currently being investigated [143].
Therapeutic utilization of miRNA faces a challenge when it comes to optimal drug delivery, as all forms of miRNA-targeting oligonucleotides have been found to localize primarily to the liver, spleen and kidney [143]. Moreover, there is a significant risk of off-target effects related to nonspecific RNA binding as well as immunostimulatory response, leading to unpredictable toxicities. In addition, thyroid and other cancers are driven by multiple cellular pathways. Therefore, combining miRNA-based therapy with well-established therapies (e.g., tyrosine kinase inhibitors) might be of interest for novel therapeutic options in thyroid cancer.
To summarize, we have documented a pivotal role of miRNA in thyroid carcinogenesis, progression and differentiation and its potential utility in thyroid cancer diagnosis and prognosis. Since deregulation of miRNAs is not the only molecular event associated with thyroid cancer, it is reasonable to utilize a more comprehensive approach in thyroid cancer diagnosis, prognosis and treatment. Future studies should focus on combining somatic mutations with miRNA profiling to improve the diagnostic accuracy of FNAB of indeterminate thyroid nodules. In order to improve the accuracy of the predictive models, it might be worthwhile to incorporate well-established clinic–pathological features such as age, tumor size, presence of lymph node and distant metastases, extrathyroid extension and completeness of the surgery with molecular characteristics such as somatic mutation status and miRNAs profiling. Importantly, miRNA profiling may serve as a useful tool for predicting responses to certain therapeutics (e.g., vendetanib or vemurafenib), thus enabling a more personalized therapeutic approach.
Practice points.
Differentially expressed MiRNAs may be helpful adjunct diagnostic markers to refine thyroid nodule fine needle aspiration biopsy.
MiRNAs in tissue and blood samples that are markers of aggressive thyroid cancer may provide helpful prognostic information when planning adjuvant therapy and follow up, and as biomarkers of treatment response.
MiRNAs with tumor suppressive function in thyroid cancer could provide potential therapeutic alternatives in patients with advanced/metastatic thyroid cancer.
Research agenda.
Large, prospective and well-designed studies are needed to validate diagnostic and prognostic miRNA markers in thyroid neoplasm.
Further analysis of the mechanism of dysregulated miRNAs in thyroid cancer and their target genes is needed to better understand their function in thyroid cancer imitation/progression.
Development of strategies for efficient delivery of miRNAs into the tumour site is required.
Acknowledgments
Grant support
Supported by the intramural research program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Footnotes
Disclosure statement
The authors have nothing to disclose.
References
- 1.Ardekani AM, Naeini MM. The role of MicroRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2(4):161–79. [PMC free article] [PubMed] [Google Scholar]
- 2.Eiring AM, Harb JG, Neviani P, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140(5):652–65. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 4.Mutharasan RK, Nagpal V, Ichikawa Y, et al. microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am J Physiol Heart Circ Physiol. 2011;301(4):H1519–30. doi: 10.1152/ajpheart.01080.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Y, Nazari-Jahantigh M, Chan L, Zhu M, et al. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation. 2013;127(15):1609–19. doi: 10.1161/CIRCULATIONAHA.112.000736. [DOI] [PubMed] [Google Scholar]
- 6.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 7.Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer. 2005;12(2):245–62. doi: 10.1677/erc.1.0978. [DOI] [PubMed] [Google Scholar]
- 8.Kondo T, Ezzat S, Asa SL. Pathogenetic mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer. 2006;6(4):292–306. doi: 10.1038/nrc1836. [DOI] [PubMed] [Google Scholar]
- 9.LiVolsi VA. Papillary neoplasms of the thyroid. Pathologic and prognostic features. Am J Clin Pathol. 1992;97(3):426–34. doi: 10.1093/ajcp/97.3.426. [DOI] [PubMed] [Google Scholar]
- 10.Cote GJ, Grubbs EG, Hofmann MC. Thyroid C-Cell biology and oncogenic transformation. Recent Results Cancer Res. 2015;204:1–39. doi: 10.1007/978-3-319-22542-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baudin E, Schlumberger M. New therapeutic approaches for metastatic thyroid carcinoma. Lancet Oncol. 2007;8(2):148–56. doi: 10.1016/S1470-2045(07)70034-7. [DOI] [PubMed] [Google Scholar]
- 12.Sciuto R, Romano L, Rea S, et al. Natural history and clinical outcome of differentiated thyroid carcinoma: a retrospective analysis of 1503 patients treated at a single institution. Ann Oncol. 2009;20(10):1728–35. doi: 10.1093/annonc/mdp050. [DOI] [PubMed] [Google Scholar]
- 13.Pacini F, Castagna MG. Approach to and treatment of differentiated thyroid carcinoma. Med Clin North Am. 2012;96(2):369–83. doi: 10.1016/j.mcna.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Xing M, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet. 2013;381(9871):1058–69. doi: 10.1016/S0140-6736(13)60109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102(52):19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.Visone R, Russo L, Pallante P, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14(3):791–8. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 17.Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13(2):497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 18.Tetzlaff MT, Liu A, Xu X, et al. Differential expression of miRNAs in papillary thyroid carcinoma compared to multi-nodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol. 2007;18(3):163–73. doi: 10.1007/s12022-007-0023-7. [DOI] [PubMed] [Google Scholar]
- 19.Nikiforova MN, Tseng GC, Steward D, et al. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93(5):1600–8. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldassarre G, Belletti B, Bruni P, et al. Overexpressed cyclin D3 contributes to retaining the growth inhibitor p27 in the cytoplasm of thyroid tumor cells. J Clin Investig. 1999;104(7):865–74. doi: 10.1172/JCI6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tallini G, Garcia-Rostan G, Herrero A, et al. Downregulation of p27KIP1 and Ki67/Mib1 labeling index support the classification of thyroid carcinoma into prognostically relevant categories. Am J Surg Pathol. 1999;23(6):678–85. doi: 10.1097/00000478-199906000-00007. [DOI] [PubMed] [Google Scholar]
- 22.Saltman B, Singh B, Hedvat CV, et al. Patterns of expression of cell cycle/apoptosis genes along the spectrum of thyroid carcinoma progression. Surgery. 2006;140(6):899–905. doi: 10.1016/j.surg.2006.07.027. discussion 905-6. [DOI] [PubMed] [Google Scholar]
- 23.Zhou YL, Liu C, Dai XX, et al. Overexpression of miR-221 is associated with aggressive clinicopathologic characteristics and the BRAF mutation in papillary thyroid carcinomas. Med Oncol. 2012;29(5):3360–6. doi: 10.1007/s12032-012-0315-8. [DOI] [PubMed] [Google Scholar]
- 24.Pacifico F, Leonardi A. Role of NF-kappaB in thyroid cancer. Mol Cell Endocrinol. 2010;321(1):29–35. doi: 10.1016/j.mce.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Bommarito A, Richiusa P, Carissimi E, et al. BRAFV600E mutation, TIMP-1 upregulation, and NF-kappaB activation: closing the loop on the papillary thyroid cancer trilogy. Endocr Relat Cancer. 2011;18(6):669–85. doi: 10.1530/ERC-11-0076. [DOI] [PubMed] [Google Scholar]
- 26.Chou CK, Chen RF, Chou FF, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20(5):489–94. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Zhang H, He L, et al. Association between the expression of four upregulated miRNAs and extrathyroidal invasion in papillary thyroid carcinoma. Onco Targets Ther. 2013;6:281–7. doi: 10.2147/OTT.S43014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oridate N, Lotan D, Xu XC, et al. Differential induction of apoptosis by all-trans-retinoic acid and N-(4-hydroxyphenyl) retinamide in human head and neck squamous cell carcinoma cell lines. Clin Cancer Res. 1996;2(5):855–63. [PubMed] [Google Scholar]
- 29.Simon D, Korber C, Krausch M, et al. Clinical impact of retinoids in redifferentiation therapy of advanced thyroid cancer: final results of a pilot study. Eur J Nucl Med Mol Imaging. 2002;29(6):775–82. doi: 10.1007/s00259-001-0737-6. [DOI] [PubMed] [Google Scholar]
- 30.Schwertheim S, Sheu SY, Worm K, et al. Analysis of deregulated miRNAs is helpful to distinguish poorly differentiated thyroid carcinoma from papillary thyroid carcinoma. Horm Metab Res. 2009;41(6):475–81. doi: 10.1055/s-0029-1215593. [DOI] [PubMed] [Google Scholar]
- 31.Li D, Jian W, Wei C, et al. Down-regulation of miR-181b promotes apoptosis by targeting CYLD in thyroid papillary cancer. Int J Clin Exp Pathol. 2014;7(11):7672–80. [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu W, Yang Z, Fan Y, et al. MicroRNA-613 inhibits cell growth, migration and invasion of papillary thyroid carcinoma by regulating SphK2. Oncotarget. 2016 doi: 10.18632/oncotarget.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong S, Jin M, Li Y, et al. miR-137 acts as a tumor suppressor in papillary thyroid carcinoma by targeting CXCL12. Oncol Rep. 2016;35(4):2151–8. doi: 10.3892/or.2016.4604. [DOI] [PubMed] [Google Scholar]
- 34.Gu Y, Li D, Luo Q, et al. MicroRNA-145 inhibits human papillary cancer TPC1 cell proliferation by targeting DUSP6. Int J Clin Exp Med. 2015;8(6):8590–8. [PMC free article] [PubMed] [Google Scholar]
- *35.Boufraqech M, Zhang L, Jain M, et al. miR-145 suppresses thyroid cancer growth and metastasis and targets AKT3. Endocr Relat Cancer. 2014;21(4):517–31. doi: 10.1530/ERC-14-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Z, Jiao P, Huang X, et al. MAPK phosphatase-3 promotes hepatic gluconeogenesis through dephosphorylation of forkhead box O1 in mice. J Clin Investig. 2010;120(11):3901–11. doi: 10.1172/JCI43250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Chi Y, Gao K, et al. p53 protein-mediated up-regulation of MAP kinase phosphatase 3 (MKP-3) contributes to the establishment of the cellular senescent phenotype through dephosphorylation of extracellular signal-regulated kinase 1/2 (ERK1/2) J Biol Chem. 2015;290(2):1129–40. doi: 10.1074/jbc.M114.590943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groom LA, Sneddon AA, Alessi DR, et al. Differential regulation of the MAP, SAP and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J. 1996;15(14):3621–32. [PMC free article] [PubMed] [Google Scholar]
- 39.Camps M, Nichols A, Gillieron C, et al. Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science. 1998;280(5367):1262–5. doi: 10.1126/science.280.5367.1262. [DOI] [PubMed] [Google Scholar]
- 40.Arkell RS, Dickinson RJ, Squires M, et al. DUSP6/MKP-3 inactivates ERK1/2 but fails to bind and inactivate ERK5. Cell Signal. 2008;20(5):836–43. doi: 10.1016/j.cellsig.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 41.Degl’Innocenti D, Romeo P, Tarantino E, et al. DUSP6/MKP3 is overexpressed in papillary and poorly differentiated thyroid carcinoma and contributes to neoplastic properties of thyroid cancer cells. Endocr Relat Cancer. 2013;20(1):23–37. doi: 10.1530/ERC-12-0078. [DOI] [PubMed] [Google Scholar]
- 42.Messina S, Frati L, Leonetti C, et al. Dual-specificity phosphatase DUSP6 has tumor-promoting properties in human glioblastomas. Oncogene. 2011;30(35):3813–20. doi: 10.1038/onc.2011.99. [DOI] [PubMed] [Google Scholar]
- 43.Zhu X, Bai Q, Lu Y, et al. Expression and function of CXCL12/CXCR4/CXCR7 in thyroid cancer. Int J Oncol. 2016;48(6):2321–9. doi: 10.3892/ijo.2016.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu DY, Tang CH, Yeh WL, et al. SDF-1alpha up-regulates interleukin-6 through CXCR4, PI3K/Akt, ERK, and NF-kappaB-dependent pathway in microglia. Eur J Pharmacol. 2009;613(1–3):146–54. doi: 10.1016/j.ejphar.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Roland J, Murphy BJ, Ahr B, et al. Role of the intracellular domains of CXCR4 in SDF-1-mediated signaling. Blood. 2003;101(2):399–406. doi: 10.1182/blood-2002-03-0978. [DOI] [PubMed] [Google Scholar]
- 46.Bandres E, Bitarte N, Arias F, et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15(7):2281–90. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- 47.Zhang F, Huang W, Sheng M, et al. MiR-451 inhibits cell growth and invasion by targeting CXCL16 and is associated with prognosis of osteosarcoma patients. Tumour Biol. 2015;36(3):2041–8. doi: 10.1007/s13277-014-2811-2. [DOI] [PubMed] [Google Scholar]
- 48.Zeng T, Peng L, Chao C, et al. miR-451 inhibits invasion and proliferation of bladder cancer by regulating EMT. Int J Clin Exp Pathol. 2014;7(11):7653–62. [PMC free article] [PubMed] [Google Scholar]
- *49.Minna E, Romeo P, Dugo M, et al. miR-451a is underexpressed and targets AKT/mTOR pathway in papillary thyroid carcinoma. Oncotarget. 2016;7(11):12731–47. doi: 10.18632/oncotarget.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saji M, Ringel MD. The PI3K-Akt-mTOR pathway in initiation and progression of thyroid tumors. Mol Cell Endocrinol. 2010;321(1):20–8. doi: 10.1016/j.mce.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ringel MD, Hayre N, Saito J, et al. Overexpression and overactivation of Akt in thyroid carcinoma. Cancer Res. 2001;61(16):6105–11. [PubMed] [Google Scholar]
- 52.Wang Z, Zhang H, Zhang P, et al. Upregulation of miR-2861 and miR-451 expression in papillary thyroid carcinoma with lymph node metastasis. Med Oncol. 2013;30(2):577. doi: 10.1007/s12032-013-0577-9. [DOI] [PubMed] [Google Scholar]
- 53.Nikiforova MN, Lynch RA, Biddinger PW, et al. RAS point mutations and PAX8-PPAR gamma rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab. 2003;88(5):2318–26. doi: 10.1210/jc.2002-021907. [DOI] [PubMed] [Google Scholar]
- 54.Nikiforov YE. Thyroid carcinoma: molecular pathways and therapeutic targets. Mod Pathol. 2008;21(Suppl 2):S37–43. doi: 10.1038/modpathol.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dettmer M, Vogetseder A, Durso MB, et al. MicroRNA expression array identifies novel diagnostic markers for conventional and oncocytic follicular thyroid carcinomas. J Clin Endocrinol Metab. 2013;98(1):E1–7. doi: 10.1210/jc.2012-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun D, Han S, Liu C, et al. Microrna-199a-5p functions as a tumor suppressor via suppressing connective tissue growth factor (CTGF) in follicular thyroid carcinoma. Med Sci Monit. 2016;22:1210–2117. doi: 10.12659/MSM.895788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wojtas B, Ferraz C, Stokowy T, et al. Differential miRNA expression defines migration and reduced apoptosis in follicular thyroid carcinomas. Mol Cell Endocrinol. 2014;388(1–2):1–9. doi: 10.1016/j.mce.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 58.Ain KB. Anaplastic thyroid carcinoma: behavior, biology, and therapeutic approaches. Thyroid. 1998;8(8):715–26. doi: 10.1089/thy.1998.8.715. [DOI] [PubMed] [Google Scholar]
- 59.Kebebew E, Greenspan FS, Clark OH, et al. Anaplastic thyroid carcinoma. Treatment outcome and prognostic factors. Cancer. 2005;103(7):1330–5. doi: 10.1002/cncr.20936. [DOI] [PubMed] [Google Scholar]
- 60.Gilliland FD, Hunt WC, Morris DM, et al. Prognostic factors for thyroid carcinoma. A population-based study of 15,698 cases from the Surveillance, Epidemiology and End Results (SEER) program 1973–1991. Cancer. 1997;79(3):564–73. doi: 10.1002/(sici)1097-0142(19970201)79:3<564::aid-cncr20>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 61.Keutgen XM, Sadowski SM, Kebebew E. Management of anaplastic thyroid cancer. Gland Surg. 2015;4(1):44–51. doi: 10.3978/j.issn.2227-684X.2014.12.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smallridge RC, Copland JA. Anaplastic thyroid carcinoma: pathogenesis and emerging therapies. Clin Oncol R Coll Radiol. 2010;22(6):486–97. doi: 10.1016/j.clon.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charles RP, Silva J, Iezza G, et al. Activating BRAF and PIK3CA mutations cooperate to promote anaplastic thyroid carcinogenesis. Mol Cancer Res. 2014;12(7):979–86. doi: 10.1158/1541-7786.MCR-14-0158-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santarpia L, El-Naggar AK, Cote GJ, et al. Phosphatidylinositol 3-kinase/akt and ras/raf-mitogen-activated protein kinase pathway mutations in anaplastic thyroid cancer. J Clin Endocrinol Metab. 2008;93(1):278–84. doi: 10.1210/jc.2007-1076. [DOI] [PubMed] [Google Scholar]
- 65.Fagin JA, Matsuo K, Karmakar A, et al. High prevalence of mutations of the p53 gene in poorly differentiated human thyroid carcinomas. J Clin Investig. 1993;91(1):179–84. doi: 10.1172/JCI116168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moretti F, Nanni S, Farsetti A, et al. Effects of exogenous p53 transduction in thyroid tumor cells with different p53 status. J Clin Endocrinol Metab. 2000;85(1):302–8. doi: 10.1210/jcem.85.1.6295. [DOI] [PubMed] [Google Scholar]
- 67.Chang CJ, Chao CH, Xia W, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13(3):317–23. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun J, Ding W, Zhi J, et al. MiR-200 suppresses metastases of colorectal cancer through ZEB1. Tumour Biol. 2015 doi: 10.1007/s13277-015-3822-3. [DOI] [PubMed] [Google Scholar]
- 69.Savagner P. Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 2001;23(10):912–23. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- 70.Fuchs IB, Lichtenegger W, Buehler H, et al. The prognostic significance of epithelial-mesenchymal transition in breast cancer. Anticancer Res. 2002;22(6A):3415–9. [PubMed] [Google Scholar]
- 71.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–94. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 72.Xue L, Su D, Li D, et al. miR-200 regulates epithelial-mesenchymal transition in anaplastic thyroid cancer via EGF/EGFR signaling. Cell Biochem Biophys. 2015;72(1):185–90. doi: 10.1007/s12013-014-0435-1. [DOI] [PubMed] [Google Scholar]
- 73.Schiff BA, McMurphy AB, Jasser SA, et al. Epidermal growth factor receptor (EGFR) is overexpressed in anaplastic thyroid cancer, and the EGFR inhibitor gefitinib inhibits the growth of anaplastic thyroid cancer. Clin Cancer Res. 2004;10(24):8594–602. doi: 10.1158/1078-0432.CCR-04-0690. [DOI] [PubMed] [Google Scholar]
- 74.Fuziwara CS, Kimura ET. High iodine blocks a Notch/miR-19 loop activated by the BRAF(V600E) oncoprotein and restores the response to TGFbeta in thyroid follicular cells. Thyroid. 2014;24(3):453–62. doi: 10.1089/thy.2013.0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 77.Jin HY, Oda H, Lai M, et al. MicroRNA-17~92 plays a causative role in lymphomagenesis by coordinating multiple oncogenic pathways. EMBO J. 2013;32(17):2377–91. doi: 10.1038/emboj.2013.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fuziwara CS, Kimura ET. MicroRNA deregulation in anaplastic thyroid cancer biology. Int J Endocrinol. 2014;2014:743450. doi: 10.1155/2014/743450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *79.Braun J, Hoang-Vu C, Dralle H, et al. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29(29):4237–44. doi: 10.1038/onc.2010.169. [DOI] [PubMed] [Google Scholar]
- *80.Boufraqech M, Nilubol N, Zhang L, et al. miR-30a inhibits LOX expression and progression of anaplastic thyroid cancer. Cancer Res. 2014;10 doi: 10.1158/0008-5472.CAN-14-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aherne ST, Smyth PC, Flavin RJ, et al. Geographical mapping of a multifocal thyroid tumour using genetic alteration analysis & miRNA profiling. Mol Cancer. 2008;7:89. doi: 10.1186/1476-4598-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kluiver J, van den Berg A, de Jong D, et al. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26(26):3769–76. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- 83.Pacifico F, Crescenzi E, Mellone S, et al. Nuclear factor-{kappa}B contributes to anaplastic thyroid carcinomas through up-regulation of miR-146a. J Clin Endocrinol Metab. 2010;95(3):1421–30. doi: 10.1210/jc.2009-1128. [DOI] [PubMed] [Google Scholar]
- 84.Xing M. BRAF V600E mutation and papillary thyroid cancer. JAMA. 2013;310(5):535. doi: 10.1001/jama.2013.8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xing M, Alzahrani AS, Carson KA, et al. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA. 2013;309(14):1493–501. doi: 10.1001/jama.2013.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nikiforov YE. Molecular analysis of thyroid tumors. Mod Pathol. 2011;24(Suppl 2):S34–43. doi: 10.1038/modpathol.2010.167. [DOI] [PubMed] [Google Scholar]
- 87.Nikiforov YE. Genetic alterations involved in the transition from well-differentiated to poorly differentiated and anaplastic thyroid carcinomas. Endocr Pathol. 2004;15(4):319–27. doi: 10.1385/ep:15:4:319. [DOI] [PubMed] [Google Scholar]
- 88.Dettmer MS, Perren A, Moch H, et al. MicroRNA profile of poorly differentiated thyroid carcinomas: new diagnostic and prognostic insights. J Mol Endocrinol. 2014;52(2):181–9. doi: 10.1530/JME-13-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yokobori T, Suzuki S, Tanaka N, et al. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104(1):48–54. doi: 10.1111/cas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Majid S, Dar AA, Saini S, et al. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Res. 2012;72(24):6435–46. doi: 10.1158/0008-5472.CAN-12-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hofstra RM, Landsvater RM, Ceccherini I, et al. A mutation in the RET proto-oncogene associated with multiple endocrine neoplasia type 2B and sporadic medullary thyroid carcinoma. Nature. 1994;367(6461):375–6. doi: 10.1038/367375a0. [DOI] [PubMed] [Google Scholar]
- 92.Eng C, Clayton D, Schuffenecker I, et al. The relationship between specific RET proto-oncogene mutations and disease phenotype in multiple endocrine neoplasia type 2. International RET mutation consortium analysis. JAMA. 1996;276(19):1575–9. [PubMed] [Google Scholar]
- 93.Duan L, Hao X, Liu Z, et al. MiR-129-5p is down-regulated and involved in the growth, apoptosis and migration of medullary thyroid carcinoma cells through targeting RET. FEBS Lett. 2014;588(9):1644–51. doi: 10.1016/j.febslet.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 94.Santarpia L, Calin GA, Adam L, et al. A miRNA signature associated with human metastatic medullary thyroid carcinoma. Endocr Relat Cancer. 2013;20(6):809–23. doi: 10.1530/ERC-13-0357. [DOI] [PubMed] [Google Scholar]
- 95.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- *97.Gregory RI, Chendrimada TP, Cooch N, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 98.Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96(2):111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359(25):2641–50. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grelier G, Voirin N, Ay AS, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101(4):673–83. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Erler P, Keutgen XM, Crowley MJ, et al. Dicer expression and microRNA dysregulation associate with aggressive features in thyroid cancer. Surgery. 2014;156(6):1342–50. doi: 10.1016/j.surg.2014.08.007. discussion 1350. [DOI] [PubMed] [Google Scholar]
- *102.Mian C, Pennelli G, Fassan M, et al. MicroRNA profiles in familial and sporadic medullary thyroid carcinoma: preliminary relationships with RET status and outcome. Thyroid. 2012;22(9):890–6. doi: 10.1089/thy.2012.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Puppin C, Durante C, Sponziello M, et al. Overexpression of genes involved in miRNA biogenesis in medullary thyroid carcinomas with RET mutation. Endocrine. 2014;47(2):528–36. doi: 10.1007/s12020-014-0204-3. [DOI] [PubMed] [Google Scholar]
- 104.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- *106.Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 107.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zile MR, Mehurg SM, Arroyo JE, et al. Relationship between the temporal profile of plasma microRNA and left ventricular remodeling in patients after myocardial infarction. Circ Cardiovasc Genet. 2011;4(6):614–9. doi: 10.1161/CIRCGENETICS.111.959841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101(10):2087–92. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neviani P, Fabbri M. Exosomic microRNAs in the tumor microenvironment. Front Med (Lausanne) 2015;2:47. doi: 10.3389/fmed.2015.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 113.Rabinowits G, Gercel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–6. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 114.Lee YS, Lim YS, Lee JC, et al. Differential expression levels of plasma-derived miR-146b and miR-155 in papillary thyroid cancer. Oral Oncol. 2015;51(1):77–83. doi: 10.1016/j.oraloncology.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 115.Lee JC, Zhao JT, Clifton-Bligh RJ, et al. MicroRNA-222 and microRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer. 2013;119(24):4358–65. doi: 10.1002/cncr.28254. [DOI] [PubMed] [Google Scholar]
- 116.Li M, Song Q, Li H, et al. Circulating miR-25-3p and miR-451a may be potential biomarkers for the diagnosis of papillary thyroid carcinoma. PLoS One. 2015;10(7):e0132403. doi: 10.1371/journal.pone.0132403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haugen BR, Alexander EK, Bible KC, et al. 2015 American thyroid association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: the American thyroid association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid. 2016;26(1):1–133. doi: 10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bongiovanni M, Spitale A, Faquin WC, et al. The Bethesda system for reporting thyroid cytopathology: a meta-analysis. Acta Cytol. 2012;56(4):333–9. doi: 10.1159/000339959. [DOI] [PubMed] [Google Scholar]
- 119.Chen YT, Kitabayashi N, Zhou XK, et al. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod Pathol. 2008;21(9):1139–46. doi: 10.1038/modpathol.2008.105. [DOI] [PubMed] [Google Scholar]
- 120.Mazeh H, Mizrahi I, Halle D, et al. Development of a microRNA-based molecular assay for the detection of papillary thyroid carcinoma in aspiration biopsy samples. Thyroid. 2011;21(2):111–8. doi: 10.1089/thy.2010.0356. [DOI] [PubMed] [Google Scholar]
- 121.Vriens MR, Weng J, Suh I, et al. MicroRNA expression profiling is a potential diagnostic tool for thyroid cancer. Cancer. 2012;118(13):3426–32. doi: 10.1002/cncr.26587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Keutgen XM, Filicori F, Crowley MJ, et al. A panel of four miRNAs accurately differentiates malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin Cancer Res. 2012;18(7):2032–8. doi: 10.1158/1078-0432.CCR-11-2487. [DOI] [PubMed] [Google Scholar]
- 123.Mazeh H, Levy Y, Mizrahi I, et al. Differentiating benign from malignant thyroid nodules using micro ribonucleic acid amplification in residual cells obtained by fine needle aspiration biopsy. J Surg Res. 2013;180(2):216–21. doi: 10.1016/j.jss.2012.04.051. [DOI] [PubMed] [Google Scholar]
- 124.Weber F, Teresi RE, Broelsch CE, et al. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab. 2006;91(9):3584–91. doi: 10.1210/jc.2006-0693. [DOI] [PubMed] [Google Scholar]
- *125.Kitano M, Rahbari R, Patterson EE, et al. Expression profiling of difficult-to-diagnose thyroid histologic subtypes shows distinct expression profiles and identify candidate diagnostic microRNAs. Ann Surg Oncol. 2011;18(12):3443–52. doi: 10.1245/s10434-011-1766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Labourier E, Shifrin A, Busseniers AE, et al. Molecular testing for miRNA, mRNA, and DNA on fine-needle aspiration improves the preoperative diagnosis of thyroid nodules with indeterminate cytology. J Clin Endocrinol Metab. 2015;100(7):2743–50. doi: 10.1210/jc.2015-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *127.Yip L, Kelly L, Shuai Y, et al. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol. 2011;18(7):2035–41. doi: 10.1245/s10434-011-1733-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun M, Fang S, Li W, et al. Associations of miR-146a and miR-146b expression and clinical characteristics in papillary thyroid carcinoma. Cancer Biomark. 2015;15(1):33–40. doi: 10.3233/CBM-140431. [DOI] [PubMed] [Google Scholar]
- 129.Chou CK, Yang KD, Chou FF, et al. Prognostic implications of miR-146b expression and its functional role in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98(2):E196–205. doi: 10.1210/jc.2012-2666. [DOI] [PubMed] [Google Scholar]
- 130.Sondermann A, Andreghetto FM, Moulatlet AC, et al. MiR-9 and miR-21 as prognostic biomarkers for recurrence in papillary thyroid cancer. Clin Exp Metastasis. 2015;32(6):521–30. doi: 10.1007/s10585-015-9724-3. [DOI] [PubMed] [Google Scholar]
- 131.Abraham D, Jackson N, Gundara JS, et al. MicroRNA profiling of sporadic and hereditary medullary thyroid cancer identifies predictors of nodal metastasis, prognosis, and potential therapeutic targets. Clin Cancer Res. 2011;17(14):4772–81. doi: 10.1158/1078-0432.CCR-11-0242. [DOI] [PubMed] [Google Scholar]
- 132.Zhang Y, Yang WQ, Zhu H, et al. Regulation of autophagy by miR-30d impacts sensitivity of anaplastic thyroid carcinoma to cisplatin. Biochem Pharmacol. 2014;87(4):562–70. doi: 10.1016/j.bcp.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lankenau MA, Patel R, Liyanarachchi S, et al. MicroRNA-3151 inactivates TP53 in BRAF-mutated human malignancies. Proc Natl Acad Sci USA. 2015;112(49):E6744–51. doi: 10.1073/pnas.1520390112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Varmeh S, Vanden Borre P, Gunda V, et al. Genome-wide analysis of differentially expressed miRNA in PLX4720-resistant and parental human thyroid cancer cell lines. Surgery. 2016;159(1):152–62. doi: 10.1016/j.surg.2015.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lassalle S, Zangari J, Popa A, et al. MicroRNA-375/SEC23A as biomarkers of the in vitro efficacy of vandetanib. Oncotarget. 2016 doi: 10.18632/oncotarget.8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lakshmanan A, Wojcicka A, Kotlarek M, et al. microRNA-339-5p modulates Na+/I- symporter-mediated radioiodide uptake. Endocr Relat Cancer. 2015;22(1):11–21. doi: 10.1530/ERC-14-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Riesco-Eizaguirre G, Wert-Lamas L, Perales-Paton J, et al. The miR-146b-3p/PAX8/NIS regulatory circuit modulates the differentiation phenotype and function of thyroid cells during carcinogenesis. Cancer Res. 2015;75(19):4119–30. doi: 10.1158/0008-5472.CAN-14-3547. [DOI] [PubMed] [Google Scholar]
- 138.Li L, Lv B, Chen B, et al. Inhibition of miR-146b expression increases radioiodine-sensitivity in poorly differential thyroid carcinoma via positively regulating NIS expression. Biochem Biophys Res Commun. 2015;462(4):314–21. doi: 10.1016/j.bbrc.2015.04.134. [DOI] [PubMed] [Google Scholar]
- 139.Haghpanah V, Fallah P, Tavakoli R, et al. Antisense-miR-21 enhances differentiation/apoptosis and reduces cancer stemness state on anaplastic thyroid cancer. Tumour Biol. 2016;37(1):1299–308. doi: 10.1007/s13277-015-3923-z. [DOI] [PubMed] [Google Scholar]
- 140.Davis ME, Zuckerman JE, Choi CH, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–70. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gumireddy K, Young DD, Xiong X, et al. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47(39):7482–4. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Young DD, Connelly CM, Grohmann C, et al. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132(23):7976–81. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- 143.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–38. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 144.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105(10):3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 146.Ricarte-Filho JC, Fuziwara CS, Yamashita AS, et al. Effects of let-7 microRNA on cell growth and differentiation of papillary thyroid cancer. Transl Oncol. 2009;2(4):236–41. doi: 10.1593/tlo.09151. [DOI] [PMC free article] [PubMed] [Google Scholar]