SUMMARY
Introduction:
Several genetic mutations affect the first-line triple therapy for Helicobacter pylori. We aimed to study the most common genetic mutations affecting the metronidazole and clarithromycin therapy for H. pylori-infected Egyptian patients.
Patients and Methods:
In our study, we included 100 successive dyspeptic patients scheduled for diagnosis through upper gastroscopy at Cairo's University Hospital, Egypt. Gastric biopsies were tested for the presence of H. pylori by detection of the 16S rRNA gene. Positive biopsies were further studied for the presence of the rdxA gene deletion by Polymerase Chain Reaction (PCR), while clarithromycin resistance was investigated by the presence of nucleotide substitutions within H. pylori 23S rRNA V domain using MboII and BsaI to carry out a Restricted Fragment Length Polymorphism (RFLP) assay.
Results:
Among 70 H. pylori positive biopsies, the rdxA gene deletion was detected in 44/70 (62.9%) samples, while predominance of the A2142G mutations within the H. pylori 23S rRNA V domain was evidenced in 39/70 (55.7%) of the positive H. pylori cases. No statistically significant difference was found between the presence of gene mutations and different factors such as patients 'age, gender, geographic distribution, symptoms and endoscopic findings.
Conclusion:
Infection with mutated H. pylori strains is considerably high, a finding that imposes care in the use of the triple therapy to treat H. pylori in Egypt, since the guidelines recommend to abandon the standard triple therapy when the primary clarithromycin resistance rate is over 20%1.
KEYWORDS: 16s rRNA, Metronidazole resistance, rdxA gene, Clarithromycin resistance.
INTRODUCTION
Eradication of Helicobacter pylori infection is challenging. The recommended first-line eradication therapy, especially in patients with peptic ulcer disease is based on the combination of a proton pump inhibitor, clarithromycin, together with amoxicillin or metronidazole 1 . However, recent data showed that the combination therapy has lost its efficacy due to the emergence of resistant strains. The rate of resistance is increasing worldwide with variations according to the geographic area 2 .
The molecular mechanism of metronidazole resistance is mainly due to inactivation of an oxygen-insensitive NADPH nitroreductase (rdxA) responsible for metronidazole resistance as a result of a deletion in the rdxA encoding gene, and may also be boosted by mutations in the frxA gene that encodes a NAD(P)H-flavin oxidoreductase 3 , 4 .
Furthermore, the major cause of clarithromycin resistance in H. pylori is the lack of binding of the macrolide to the 23S rRNA components of the bacterial ribosome due to modification of the target site by point mutations in the peptidyl transferase region of domain V of the 23S rRNA 5 , 6 . The A2142G and A2143G nucleotide substitutions are the most common mutations causing clarithromycin resistance worldwide 6 , 7 .
In Egypt, the majority of clinicians prescribe the classical triple regimen composed of clarithromycin; metronidazole and a proton pump inhibitor for seven days as the first line therapy to treat H. pylori infections. The high failure rate of the first line triple therapy encourages the investigation of the prevalence and genetic background of clarithromycin and metronidazole resistance in H. pylori-infected patients.
PATIENTS AND METHODS
Patients and specimens
This study included 100 dyspeptic patients attended at the endoscopy unit of the Kaser Al Aini, Cairo's University Hospital, to undergo upper gastrointestinal (GI) endoscopy, from January 2013 to November 2013. The study included 54% males and 46% females, aged 21-60 years old (mean 44 +/- 11.1) and 53% of the studied patients lived in urban areas. The patients had not received the first-line triple therapy for H. pylori (clarithromycin, metronidazole and proton pump inhibitors), i.e., they were naive regarding all of the eradication therapy drugs so as to determine the primary resistance rate to clarithromycin and metronidazole. An informed consent was obtained from each patient.
Patients who were less than 30 years or more than 60 years old were excluded from the study, as well as patients with any contraindications to undergo upper GI endoscopy, or had been submitted to upper GI endoscopy for other reasons (e.g. cirrhotic patients with varices).
Upper GI endoscopy was conducted by means of an Olympus endoscope video, and 100 gastric biopsies from 100 patients were performed from the stomach corpus or antrum, placed in glycerol solution and kept at -80 0C and then sent to the clinical pathology department laboratory for further laboratory work up.
DNA extraction
DNA was extracted from gastric biopsies using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to manufacturer recommendations.
16s rRNA-PCR
PCR was performed on extracted DNA targeting the H. pylori 16s rRNA gene (Hp16s). The following cycling conditions were used: 35 cycles of 95 °C for 30 sec, 60 °C for 30 sec and 72 °C for 30 sec and an extension time of 72 °C for 5 min according to Secka et al. 2011 8 . All of the primers used in this study are listed in Table 1.
Table 1. Sequence of primers used in this study.
| Gene | Primers '5-3' | Expected fragment (bp) | References |
| Hp1 Hp2 | CTGGAGAGACTAAGCCCTCC ATTACTGACGCTGATTGTGC | 109bp | [8] |
| rdxA1 rdxA2 | AATTTGAGCATGGGGCAGA GAAACGCTTGAAAACACCCCT | 850bp | [3] |
| Hp23Sr6 Hp23Sr7 | CACACAGGTAGATGAGATGAGTA CACACAGAACCACCGGATCACTA | 768bp | [9] |
Hp: H. pylori
rdxA gene deletion-PCR
All of the H. pylori-positive samples for the 16s rRNA-PCR were submitted to the detection of metronidazole-resistance gene rdxA gene by PCR. By this method, the expected molecular weight of the PCR fragment for the wild type rdxA gene is 850 bp, and for the mutated allele 650 bp 3 . PCR reactions were carried out in 25 µL mixtures containing 12.5 µL of the master mix (QIAGEN, Hilden, Germany) 9.5 µL of sterile deionized water, 1 µL of the template DNA and 1 µL of each of the oligonucleotide primers. The thermos cycler conditions were as follows: an initial denaturation at 94 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 1 min, annealing for 1 min at 55 °C, extension at 72 °C for 1 min. The final extension step was extended to 10 min at 72 °C 3 .
Detection of 23S rRNA mutations by PCR-RFLP analysis
DNA amplification was carried out using Taq PCR Master Mix Kit supplied by (QIAGEN, Hilden, Germany) on the extracted DNA using the primers Hp23Sr6 (sense) and Hp23Sr7 (antisense). The amplified DNA product corresponded to the domain V of the H. pylori 23S rDNA. The cycling conditions were as follows; denaturation at 94 °C 5 min followed by 40 cycles at 94 °C for 30 sec; 60 °C for 30 sec; 72 °C for 30 sec and one final extension cycle at 72 °C for 7 min, in a total volume of 25 µL containing 1× PCR buffer, 200 µM dNTPs, 2.0 mM MgCl2, 1 µM of each oligonucleotide primer, 1.25 U Taq DNA according to Suzuki et al. 2013 9 .
The amplified DNA fragments were digested with MboII and BsaI (New England Biolabs) according to the manufacturer instructions. These restriction enzymes are able to discriminate mutations within the H. pylori domain V of the 23S rDNA at positions 2,142 and 2,143, respectively 9 .
In all the PCR reactions a negative and a positive control were used corresponding to, sterile water and H. pylori positive gastric biopsies, respectively.
The amplified fragments were detected by electrophoresis in a 2% agarose gel stained with ethidium bromide and visualized using Gel Doc XR documentation system (Bio-Rad, Hercules, CA, USA).
Statistical analyses
Data were statistically described in terms of mean +/- standard deviation (+/- SD), median and range, or frequencies (number of cases) and percentages when appropriate. Comparison of quantitative variables between the study groups used the Mann-Whitney U test for independent samples. For comparison of qualitative variables, the Chi-square (X2) test was performed. The Fisher's exact test was used when the expected frequency was less than 5. The agreement was calculated using the Kappa test. Sensitivity, specificity, positive predictive value, negative predictive value, and overall accuracy were calculated and p-values less than 0.05 were considered statistically significant. All the statistical calculations were performed using the SPSS software, Statistical Package for Social Sciences (IBM, NY, USA) version 15 for Windows.
RESULTS
From the total of 100 extracted DNA samples, 70% were positive by the H. pylori 16S rRNA gene-PCR, yielding the 109 bp product. Demographic features of the Hp16S-positive patients are shown in Table 2, and the clinical features of Hp16S-positive patients as well as the endoscopic findings of Hp16S-positive patients are shown in Tables 3 and 4 , respectively.
Table 2. Demographic features of H. pylori positive patients.
| PCR for H. Pylori | p value | ||||
| Positive (n=70) | Negative (n=30) | ||||
| Sex Male Female | N 40 30 | % 57.1 42.9 | N 14 16 | % 46.7 53.3 | 0.4 NS |
| Residence Urban Rural | N 36 34 | % 51.4 48.6 | N 17 13 | % 56.7 43.3 | 0.7 NS |
| Age years Mean ± SD | 43.6 ±11.3 | 44.9 ±10.6 | 0.6 NS | ||
NS: non-significant
Table 3. Clinical features of H. pylori positive patients.
| PCR for H. Pylori | p value | ||||
| Positive (n=70) | Negative (n=30) | ||||
| N | % | N | % | ||
| Epigastric pain | 61 | 87% | 25 | 83% | 0.8 NS |
| Vomiting | 50 | 71.4% | 21 | 70% | 1.0 NS |
| Heartburn | 36 | 51.4% | 21 | 70% | 0.1 NS |
| Regurgitation | 23 | 32.9% | 13 | 43.3% | 0.4 NS |
| Bleeding | 22 | 31.4% | 8 | 26% | 0.8 NS |
NS: non-significant
Table 4. Endoscopic findings of H. pylori positive patients.
| PCR for H. Pylori | p value | ||||
| Positive (n=70) | Negative (n=30) | ||||
| N | % | N | % | ||
| Diffuse gastritis | 33 | 47.1% | 9 | 30% | 0.1 NS |
| Reflux esophagitis | 23 | 32.9% | 9 | 30% | 0.8 NS |
| Bulb duodenitis | 19 | 27.1% | 4 | 13.3% | 0.2 NS |
| Antral gastritis | 10 | 14.3% | 6 | 20% | 0.6 NS |
| Duodenal ulcer | 7 | 10% | 0 | 0 % | 0.1 NS |
| Gastric mass | 5 | 7.1% | 5 | 16.7% | 0.2 NS |
| Esophageal varices or PHG | 4 | 5.7% | 2 | 6.7% | 1.0 NS |
| Others | 4 | 5.7% | 2 | 6.7% | 1.0 NS |
| Gastric ulcer | 3 | 4.3% | 2 | 6.7% | 0.6 NS |
| Normal | 0 | 0% | 1 | 3.3% | 0.3 NS |
NS: non-significant
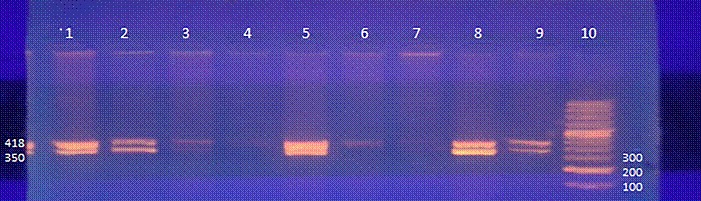
Among the seventy positive H. pylori samples, 44 (62.9 %) were also positive by the rdxA gene-PCR yielding a 650 bp product, whereas in 19 (27.1%) the rdxA gene was positive, but the amplification product had a molecular weight of 850 pb. The RFLP pattern showed that 39/70 (55.7%) samples had the A2142G mutation as shown in Figures 1 and 2 . Moreover, none of the tested samples had the A2143G mutation.
Fig. 1. Results of the rdxA gene-PCR. Lanes 1, 2, 6, 7, 9, 10 and 11 contain rdxA gene-positive amplification products of 650 bp; lanes 3, 4, 5 and 8 are rdxA-negative; lane 12 is the 100 bp molecular weight marker.
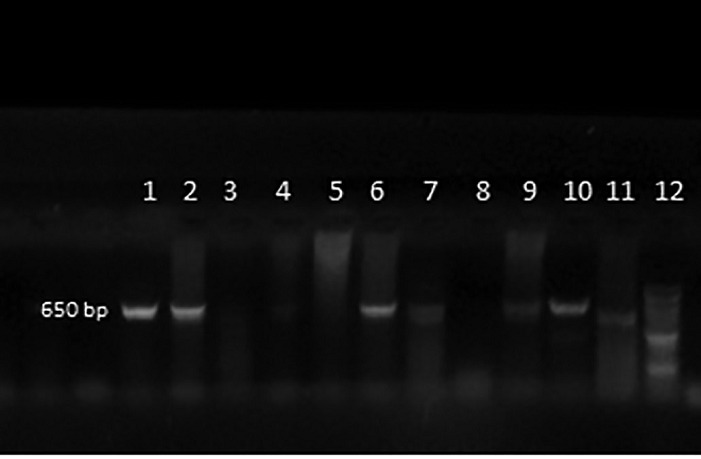
Fig. 2. Detection of mutations associated with clarithromycin resistance in Helicobacter pylori strains by PCR-RFLP. Electrophoresis in a 3% agarose gel of 23S rDNA-PCR-RFLP. Lane 10 is the 100-bp molecular weight marker. Lane 9 is the positive control. Lanes 1, 2, 3, 5, 7 and 8 show the amplifications products of samples containing the A2142G mutation after digestion with the MboII restriction enzyme. Lanes 4 and 6 are the 23S-PCR negative controls.
DISCUSSION
The World Gastroenterology Organization (WGO) reported that the H. pylori prevalence in Egypt was 90% in adults. Regarding the Middle East, H. pylori prevalence ranges from 60-90% as documented by WGO10. The first line triple therapy regimen has proved to become inefficient worldwide, mainly as a result of the emergence of several genetic mutations reducing H. pylori susceptibility to clarithromycin and metronidazole.
H pylori is a relatively fastidious slowly growing microaerophilic bacterium a characteristic that makes the conventional culture method as well as the vitro antibiotic susceptibility tests (disk diffusion, agar dilution, and Epsilometer tests) time consuming, requiring 10-14 days to release a final report on the isolated H. pylori sensitivity. Molecular diagnostic techniques constitute attractive alternative methods to determine antibiotic susceptibilities with better accuracies and shorter turnaround time when performed directly on gastric biopsies, without culture 11 .
Molecular assays for the detection of resistance to metronidazole are still controversial. A large number of mutations have been described in rdxA, frxA and frxB genes, but their true role in metronidazole resistance is not clear. In our study, the prevalence of rdxA gene mutation was 62.9%. Spreading of the rdxA gene mutation can be explained by the empiric use of metronidazole for the treatment of parasitic and anaerobic infections in Egypt. The drug dosage in H. pylori infections has contributed to the emergence of resistant mutants, but is not able to eradicate the organism, leading a selective pressure effect.
In another Egyptian study conducted by Sherif et al. (2004) the phenotypic metronidazole resistance was found in 100% of the 48 H. pylori isolates. The difference of results may be explained by several factors. Firstly, different age groups were included in the two studies. In our study, we included patients > 18 years old 12 . In the study of Sherif et al. patients were 2-17 years old and were referred to the Abu-Resh Children's Hospital 12 . This age group is more frequently exposed to metronidazole for the treatment of parasitic infections and sometimes are highly exposed to this drug during frequent treatment schedules 13 . The second factor is the study design. In the study of Sherif et al. the gastric biopsies were incubated and the in vitro susceptibility was performed. In our study, metronidazole resistance was detected through a rdxA gene mutation. The finding of resistant strains to metronidazole by the in vitro methods is not clinically reliable as in vitro susceptibility tests have a tendency to overestimate resistance. For this reason and also due to the lack of a clinical-bacteriological correlation, the Maastricht IV Consensus Report has discouraged the routine metronidazole susceptibility testing 1 . The existence of other mechanisms associated with the rdxA gene inactivation such as the insertion of a transposable element called Mini-IS6053 is also noteworthy.
Regarding clarithromycin resistance 55.7% of H. pylori positive cases harbored a 2142G mutation known to confer clarithromycin resistance with a rise in the resistance percentage from 4% in Sherif et al. study. 12 Macrolides are widely used to treat upper respiratory tract infections in Egypt.
In our study the A2142G mutation was the commonest among the tested samples while the A2143G mutation was not found among the isolates, contrasting with other authors who reported that the commonest mutations were A2143G and A2142G found in equal percentages among the tested isolates 14 . Acosta et al. 2014 reported that the A2143G mutation was more frequent than the other mutation A2142G, and both were the most frequent mutations, while Hansomburana et al. (2012) documented that the A2142G mutation was more frequent than the A2143G one 15 , 16 .
Although clarithromycin resistance was found in 55.7% of H. pylori-positive cases, this percentage could be still underestimated due to the presence of other mutations which were not detected in our study. The eradication of H. pylori strains presenting high resistance rates to macrolides, or metronidazole could be reached by the use of approved tetracycline- or amoxicillin-containing regimens replacing the standard triple therapy 17 .
In conclusion, the discovery of this high prevalence of genetic mutations that may confer resistance to metronidazole and clarithromycin, is most likely due to the misuse of macrolides and metronidazole. The clinicians' awareness of the isolates current antimicrobial susceptibility profiles will improve the treatment strategies and the patients' outcomes.
REFERENCES
- 1.Malfertheiner P, Bazzoli F, Delchier JC, Celiñski K, Giguère M, Rivière M. Helicobacter pylori eradication with a capsule containing bismuth subcitrate potassium, metronidazoleand tetracycline given with omeprazole versus clarithromycin-based triple therapy a randomised, open-label, non-inferiority, phase 3 trial. Lancet. 2011;377:905–913. doi: 10.1016/S0140-6736(11)60020-2. [DOI] [PubMed] [Google Scholar]
- 2.Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut. 2010;59:1143–1153. doi: 10.1136/gut.2009.192757. [DOI] [PubMed] [Google Scholar]
- 3.Debets-Ossenkopp YJ, Pot RG, van Westerloo DJ, Goodwin A, Vandenbroucke-Grauls CM, Berg DE. Insertion of mini-IS605 and deletion of adjacent sequences in the nitroreductase (rdxA) gene cause metronidazole resistance in Helicobacter pylori NCTC11637. Antimicrob Agents Chemother. 1999;43:2657–2662. doi: 10.1128/aac.43.11.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten SJ, Berg DE, Hoffman PS. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 5.Goldman RC, Zakula D, Flamm R, Beyer J, Capobianco J. Tight binding of clarithromycin, its 14-(R)-hydroxy metabolite and erythromycin to Helicobacter pylori ribosome. Antimicrob Agents Chemother. 1994;38:1496–1500. doi: 10.1128/aac.38.7.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verlasovic J, Osato M, Spakovsky , Dore M, Reddy R, Stone G. Point of mutations in the 23S rRNA gen of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997;40:283–286. doi: 10.1093/jac/40.2.283. [DOI] [PubMed] [Google Scholar]
- 7.Taylor D, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23 S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S RNAr mutations. Antimicrob Agents Chemother. 2008;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Secka O, Antonio M, Tapgun M, Berg DE, Bottomley C, Thomas V. PCR-based genotyping of Helicobacter pylori of Gambian children and adults directly from biopsy specimens and bacterial cultures. Gut Pathog. 2011;3:5–5. doi: 10.1186/1757-4749-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki RB, Lopes RA, da Camara Lopes GA, Hung Ho T, Speranca MA. Low Helicobacter pylori primary resistance to clarithromycin in gastric biopsy specimens from dyspeptic patients of a city in the interior of São Paulo, Brazil. BMC Gastroenterol. 2013;13:164–164. doi: 10.1186/1471-230X-13-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt RH, Xiao SD, Megraud F, Leon-Barua R, Bazzoli F, van der Merwe S. Helicobacter pylori in developing countries World Gastroenterology Organization Global Guideline. J Gastrointestin Liver Dis. 2011;20:299–304. [PubMed] [Google Scholar]
- 11.Owen RJ. Molecular testing for antibiotic resistance in Helicobacter pylori. Gut. 2002;50:285–289. doi: 10.1136/gut.50.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherif M, Mohran Z, Fathy H, Rockabrand DM, Rozmajzl PJ, Frenck RW. Universal high-level primary metronidazole resistance in Helicobacter pylori isolated from children in Egypt. J Clin Microbiol. 2004;42:4832–4834. doi: 10.1128/JCM.42.10.4832-4834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siavoshi F, Saniee P, Latifi-Navid S, Massarrat S, Sheykholeslami A. Increase in resistance rates of H pylori isolates to metronidazole and tetracycline-comparison of three 3-year studies. Arch Iran Med. 2010;13:177–187. [PubMed] [Google Scholar]
- 14.Klesiewicz K, Nowak P, Karczewska E, Skiba I, Wojtas-Bonior I, Sito E. PCR-RFLP detection of point mutations A2143G and A2142G in 23S rRNA gene conferring resistance to clarithromycin in Helicobacter pylori strains. Acta Biochim Pol. 2014;61:311–315. [PubMed] [Google Scholar]
- 15.Acosta CP, Hurtado FA, Trespalacios AA. Determination of single nucleotide mutations in the 23S rRNAgene of Helicobacter pylori resistance to clarithromycin related in a population of Cauca, Colombia. Biomédica. 2014;34(1):156–162. doi: 10.1590/S0120-41572014000500018. [DOI] [PubMed] [Google Scholar]
- 16.Hansomburana P1, Anantapanpong S, Sirinthornpunya S, Chuengyong K, Rojborwonwittaya J. Prevalence of single nucleotide mutation in clarithromycin resistant gene of Helicobacter pylori a 32-months prospective study by using hybridization real time polymerase chain reaction. J Med Assoc Thai. 2012;95(3):S28–S35. [PubMed] [Google Scholar]
- 17.Ghaith D, Elzahry M, Mostafa G, Mostafa S, Elsherif R, Ramzy I. Mutations affecting domain V of the 23SrRNA gene in Helicobacter pylori from Cairo, Egypt. J Chemother. 2016;28:367–370. doi: 10.1179/1973947815Y.0000000067. [DOI] [PubMed] [Google Scholar]


