Abstract
Neocarzilins (NCZs) are antitumor chlorinated polyenones produced by “Streptomyces carzinostaticus” var. F-41. The gene cluster responsible for the biosynthesis of NCZs was cloned and characterized. DNA sequence analysis of a 33-kb region revealed a cluster of 14 open reading frames (ORFs), three of which (ORF4, ORF5, and ORF6) encode type I polyketide synthase (PKS), which consists of four modules. Unusual features of the modular organization is the lack of an obvious acyltransferase domain on modules 2 and 4 and the presence of longer interdomain regions more than 200 amino acids in length on each module. Involvement of the PKS genes in NCZ biosynthesis was demonstrated by heterologous expression of the cluster in Streptomyces coelicolor CH999, which produced the apparent NCZ biosynthetic intermediates dechloroneocarzillin A and dechloroneocarzilin B. Disruption of ORF5 resulted in a failure of NCZ production, providing further evidence that the cluster is essential for NCZ biosynthesis. Mechanistic consideration of NCZ formation indicates the iterative use of at least one module of the PKS, which subsequently releases its product by decarboxylation to generate an NCZ skeleton, possibly catalyzed by a type II thioesterase encoded by ORF7. This is a novel type I PKS system of bacterial origin for the biosynthesis of a reduced polyketide chain. Additionally, the protein encoded by ORF3, located upstream of the PKS genes, closely resembles the FADH2-dependent halogenases involved in the formation of halometabolites. The ORF3 protein could be responsible for the halogenation of NCZs, presenting a unique example of a halogenase involved in the biosynthesis of an aliphatic halometabolite.
Polyketides are well known as the components of a variety of microbial and plant secondary metabolites, including clinically valuable antibiotics and anticancer agents. An essential polyketide carbon skeleton is constructed by the repeated condensation of acyl units, mostly malonyl- or methylmalonyl coenzyme A (CoA), on an acyl carrier protein (ACP) by the catalytic activity of ketosynthase (KS). The responsible enzyme, polyketide synthase (PKS), provides subsequent optional steps after each condensation, catalyzed by ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER) functions. The remarkable structural diversity of polyketides is derived from variations in the choice of starter and extending units, the number of condensations, and the extent of reductive cycles. Because of the mechanistic analogy to fatty acid synthases, two types of PKSs were designated on the basis of their protein structures. These PKSs are multifunctional type I, which is involved in macrolide biosynthesis, and type II, which consists of discrete monofunctional proteins involved in aromatic polyketide biosynthesis (see reference 21 and references cited therein).
Later, rather unusual bacterial PKS genes were identified from actinomycetes. An example is aviM from Streptomyces viridochromogenes, which encodes a type I PKS that functions iteratively to produce an aromatic compound, orsellinic acid (15). More recently, iterative type I PKS genes have been shown to be involved in the production of enediyne antibiotics, C-1027 from Streptomyces globisporus (30) and calicheamicin from Micromonospora echinospora (1). Furthermore, characterization of rppA from Streptomyces griseus, which encodes a homologue of a plant chalcone synthase, as a tetrahydroxynaphthalene synthase led to the realization that type III PKSs also occur in bacteria (14). The actinomycetes thus present promising opportunities for the discovery of novel types of PKS genes potentially useful for drug development.
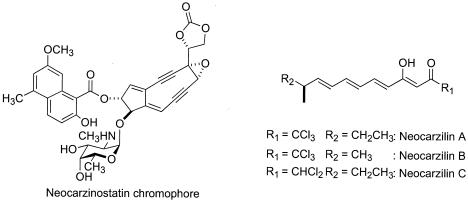
Here, we focus on “Streptomyces carzinostaticus” var. F-41, a producer of the antitumor antibiotics neocarzinostatin (12, 23) and neocarzilins (NCZs) (34, 35) (Fig. 1). Neocarzinostatin is a typical nine-membered enediyne derivative whose biosynthesis is predictably catalyzed by an iterative type I PKS. Antitumor polyenones, NCZs, are characterized for their chloromethyl groups, which are required for biological activity. Although a large number of halogenated natural products have been reported (17), information on their biosynthesis is limited to aromatic halometabolites. Molecular genetic studies (9) identified the chl gene as being responsible for the chlorination of tetracycline in Streptomyces aureofaciens. Biochemical studies showed that a FADH2-dependent halogenase (PrnA) is involved in the biosynthesis of the tryptophan-derived halometabolite pyrrolnitrin in Pseudomonas fluorescens (20, 27). Biosynthetic studies of NCZs could allow us to characterize a novel type of PKS for the polyenone skeleton and an as yet unknown halogenase involved in the biosynthesis of an aliphatic halometabolite. In this study, we describe the cloning, sequencing, and functional analysis of the gene cluster for the biosynthesis of the NCZs in “S. carzinostaticus.”
FIG. 1.
Structures of “S. carzinostaticus” products, neocarzinostatin chromophore and NCZs.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
“S. carzinostaticus” var. F-41 was obtained from Kayaku Co. Ltd. “S. carzinostaticus” and Streptomyces coelicolor CH999 (proA1 argA1 redE60 Δact::ermE SCP− SCP2−) (32) were maintained on GYM agar medium (44). For protoplast preparation, “S. carzinostaticus” and S. coelicolor were grown in liquid YEME for 40 h by the standard procedure (28). Protoplasts were regenerated on R2YE medium. For the production of NCZs, spores of “S. carzinostaticus” were transferred to liquid R2YE medium (28) and grown at 28°C in 500 ml of baffled flasks filled with 100 ml of medium at 200 rpm. For the expression of gene clusters, Streptomyces transformants were grown in liquid medium (100 ml in a 500-ml Erlenmeyer flask) as described previously (50). Escherichia coli strain DH5α (supE44 hdfR17 recA1 endA1 gyrA96 thi-1 relA1) was used for standard cloning experiments. The E. coli strains (Stratagene) used for cosmid manipulations were XL-1 Blue MRF′ {Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lac1q ZDM 15Tn10 (Tet)r]}. Plasmids were passed through E. coli ET12567 (with dam, dcm, and hadS mutations) to generate unmethylated DNA before they were used to transform “S. carzinostaticus” and S. coelicolor CH999. pBluescript II SK(+) and pT7Blue(R) T-Vector were from Stratagene and Novagen, respectively. Cosmid pOJ446 was described previously (3). Plasmid pTST59.1 was a generous gift from Josef Altenbuchner, University of Stuttgart.
DNA manipulations.
Plasmid isolation, DNA endonuclease restriction analysis, ligation, transformation, and colony and Southern hybridizations were performed by standard methods. Genomic DNA of “S. carzinostaticus” var. F-41 was isolated by a modified procedure described previously (28). PCR was performed in a final volume of 50 μl with AmpliTaqGold (Perkin Elmer) and “S. carzinostaticus” genomic DNA as the template and with the following primers and thermal cycler conditions: primer KSMA-F (designed from the conserved sequence LAMDPQQ; 5′-TSGCSATGGACCCSCAGCAG-3′), primer KSMB-R (designed from the conserved sequence VEAHGTG; 5′-CCSGTSCCGTGSGCCTCSAC-3′), and thermal cycler conditions of 0.5 min at 95°C for 30 s, 0.5 min at 60°C, and 1 min at 72°C for 45 cycles. The primers used for the colony PCR in this study were as follows: primer Neo-F (5′-AGACAATCGGCTGCTCTGATG-3′), primer Neo-R (5′-TAAAGCACGAGGAAGCGGTCAGCCC-3′), primer KAN2-Fa (5′-GGTTGATGAGAGCTTTGTTGTAGGT-3′), primer KAN2-Ra, 5′-CTCAAAATCTCTGATGTTACATTGC-3′, primer PKS-F (5′-ACATCATCCTCGACCCGATGGCCTC-3′), primer PKS-R (5′-GTTGGGTGCCGCGAAGTGGAGGTTG-3′), primer Halo-F (5′-CTGTTCACCCACATGATCGGGGTCG-3′), and primer Halo-R (5′-ACGAGTTCCATCGTGTTGGTCAGGC-3′). The synthetic oligonucleotides used for the PCR primers were obtained from Nihon Bioservice (Saitama, Japan).
Spectroscopic analysis.
Nuclear magnetic resonance (NMR) and mass spectra were measured on JEOL Alpha-500 and Shimadzu GCMS-QP2010 instruments, respectively.
Construction and screening of cosmid library.
For the construction of a cosmid library from “S. carzinostaticus” var. F-41, chromosomal DNA was partially digested with Sau3AI, and fragments of 25 to 40 kb were ligated with pOJ446 digested with HpaI, followed by shrimp alkaline phosphatase treatment and BamHI digestion. In vitro packaging was performed with Gigapack III Gold (Stratagene), according to the protocol of the manufacturer. The phages were used to transduce E. coli XL-1 Blue MRF′. For screening of the cosmid library, the PCR product obtained with primers KSMA and KSMB was used as the probe, which was labeled with digoxigenin (DIG) by using a DIG labeling and detection kit (Roche Biomedical). Two clones (clones pMO3aD6 and pMO4aH3) were identified as positive by screening of approximately 10,000 independent clones.
DNA sequencing and computer-assisted sequence analysis.
Templates for sequencing were prepared as follows: cosmids pMO3aD6 and pMO4aH3 were further characterized by restriction mapping, and 5- to 10-kb overlapping fragments were subcloned into pBluescript II SK(+). Primer binding sites were randomly introduced into each clone by using the EZ::TN<KAN-2>Insertion kit (Epicentre). Clones with random transposon insertions were prepared by using the GFX microplasmid preparation kit (Amersham Pharmacia Biotech) and sequenced. The DNA sequence was determined by the dideoxy-chain termination method with double-stranded plasmid DNA. Sequencing was performed on automated DNA sequencers (models 4000L and 4200L; LI-COR Inc., Lincoln, Nebr.) with a Thermo Sequenase cycle sequencing kit (Amersham Pharmacia Biotech). DNA sequence data were analyzed with the DNASIS programs (version 3.7; Hitachi Software Engineering Co., Ltd., Tokyo, Japan). Frame plot software (2, 24) was used to identify potential protein-coding regions by using a World Wide Web-based version (http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl). Database searches for homologous genes and proteins were performed by using the National Center for Biotechnology Information (NCBI) BLAST server. A conserved-domain database search was performed with the reverse position BLAST software provided by NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Phylogenetic tree analysis was carried out with a version of the CLUSTAL W program (http://www.ddbj.nig.ac.jp/search/clustalw-e.html), provided by DDBJ, based on a neighbor-joining method. The phylogenetic tree was created with the TreeView program (version 1.6.2; freely available from the Taxonomy and Systematics server at the University of Glasgow).
Construction of expression plasmid.
pMO1 is a derivative of pOJ446 constructed by insertion of the tipA promoter and the thiostrepton resistance gene fragment amplified from pPM927 (48) into the BamHI site of pOJ446. pTST59.1 is a derivative of SUPERCOS1 (Stratagene) carrying the attP site and integrase derived from bacteriophage ΦC31 (31). pMO1 was digested with HpaI, followed by calf intestine alkaline phosphatase (CIAP) treatment and BamHI digestion. pMO4aH3 was digested with EcoRV, followed by ligation with BamHI-SmaI and pSmaI linkers (Takara) to both ends of the EcoRV fragments and ligation with the vector mentioned above. This mixture was packaged, followed by transfection into E. coli XL-1 Blue MRF′, and an optimal clone (pMO8) was selected. pTST59.1 was digested with XbaI, followed by CIAP treatment and BamHI digestion. A SpeI-XbaI fragment that included the EcoRV fragment of pMO4aH3, the tipA promoter, and the thiostrepton resistance gene was excised from pMO8 and blunt ended with the Klenow fragment, followed by ligation with BamHI-SmaI and pSmaI linkers. The resulting fragment was ligated with pretreated pTST59.1, and the mixture was packaged, followed by transfection into E. coli XL-1 Blue MRF′. The final expression plasmid was designated pMO11, which carries the 27-kb fragment covering open reading frames (ORFs) ORF4 to ORF12 of the cluster.
Heterologous expression of the gene cluster in S. coelicolor CH999.
Transformation of S. coelicolor CH999 was carried out by standard procedures (28). Genome integration was confirmed by colony PCR with the Neo-F and Neo-R primer set for the detection of kanamycin-resistant clones, as described previously (25). Transformants were cultured in liquid medium under inducing conditions with 5 μg of thiostrepton per ml. The medium was separated from the culture broth by centrifugation and was directly subjected to reversed-phase high-pressure liquid chromatography (HPLC) analysis under the following conditions: column, TSK gel ODS-80Ts (4.6 mm [inner diameter] by 150 mm; TOSOH Co., Ltd.); column temperature, 40°C; gradient elution, solvent A (0.5% acetic acid in acetonitrile) and solvent B (0.5% acetic acid in distilled H2O); gradient profile, 10% solvent A from 0 to 5 min, 10 to 95% solvent A from 5 to 20 min, and 95% solvent A from 20 to 25 min; flow rate, 0.75 ml/min; and photodiode array detector (PD-8020; TOSOH Co., Ltd.), 220 to 460 nm. The mycelium harvested by centrifugation was extracted with acetone at room temperature. After removal of the mycelia by filtration, the aqueous extract was evaporated to remove the acetone, followed by extraction with n-hexane. After evaporation of the solvent, the residue was subjected to reversed-phase HPLC analysis under the following conditions: column, TSK gel ODS-80Ts (4.6 mm [inner diameter] by 150 mm; TOSOH, Co., Ltd.); column temperature, 40°C; gradient elution, acetonitrile and distilled H2O, gradient profile, 50% acetonitrile from 0 to 5 min, 50 to 95% acetonitrile from 5 to 20 min, and 95% acetonitrile from 20 to 25 min; flow rate, 0.75 ml/min; and photodiode array detector (PD-8020; TOSOH Co., Ltd.), 220 to 460 nm. Mycelial extracts were also analyzed by gas chromatography (GC)-mass spectrometry (MS) (GCMS-QP2010; Shimadzu) under the following conditions: column, DB-5MS 0.25 mm [inner diameter] by 30 m; film (Agilent Technologies) thickness, 0.25 μm; helium flow rate, 50 ml/min; and temperature, maintained at 50°C for 3 min, elevated to 150°C at 10°C/min, and then held at 150°C for 10 min, followed by elevation to 300°C at 10°C/min.
Isolation, purification, and determination of structures of transformant metabolites.
S. coelicolor CH999/pMO11 mycelia were collected by centrifugation from 1.5 liters of a 5-day-old culture broth and extracted with acetone at room temperature. After removal of the mycelia by filtration, the extracts were evaporated to remove the acetone, followed by extraction with hexane. The combined extracts were dried over Na2SO4. After removal of the solvent, the residue was subjected to chromatography on silica gel (oxalic acid treated; Wako-gel C200; Wako) in hexane-benzene (20:1). Target fractions were combined, concentrated, and subjected to preparative reversed-phase HPLC to give compounds 1 (27 mg) and 2 (14 mg). The HPLC conditions were as follows: column, TSK gel ODS-80TM (7.6 mm [inner diameter] by 300 mm; TOSOH); column temperature, 40°C; isocratic elution, 80% CH3CN; and flow rate, 1.5 ml/min.
Compound 1 (dechloroneocarzilin A): yellow oil; electron ionization mass spectrum (EIMS) m/z 220 (M+); 1H NMR (CDCl3) δ 0.84 (3H, t, J = 7.5 Hz), 0.99 (3H, d, J = 6.5), 1.33 (2H, dq), 2.10 (3H, s), 2.17 (1H, dt), 5.50 (1H, s), 5.79 (1H, dd, J = 8 and 15.5 Hz), 5.85 (1H, d, J = 15 Hz), 6.09 (1H, dd, J = 10.5 and 15 Hz), 6.21 (1H, dd, J = 11.5 and 15 Hz), 6.50 (1H, dd, J = 11 and 15 Hz), 7.22 (1H, dd, J = 11 and 15 Hz), 15.29 (1H, s); 13C NMR (CDCl3) δ 11.7, 19.7, 27.0, 29.5, 38.8, 100.7, 124.9, 128.4, 128.7, 140.4, 140.7, 146.0, 147.3, 177.1, 197.5.
Compound 2 (dechloroneocarzilin B): yellow oil; EIMS m/z 206 (M+); 1H NMR (CDCl3) δ 1.01 (6H, d, J = 7 Hz), 2.11 (3H, s), 2.38 (1H, dq), 5.50 (1H, s), 5.85 (1H, d, J = 15.5 Hz), 5.87 (1H, dd, J = 7 and 15 Hz), 6.09 (1H, dd, J = 11 and 15.5 Hz), 6.22 (1H, dd, J = 11 and 14.5 Hz), 6.49 (1H, dd, J = 11 and 15 Hz), 7.22 (1H, dd, J = 11.5 and 15 Hz), 15.28 (1H, s); 13C NMR (CDCl3) δ 22.0, 27.0, 31.5, 100.7, 124.9, 128.4, 128.8, 140.4, 140.7, 147.1, 177.1, 197.5.
Construction of ORF5 and ORF3 disruptants by insertional inactivation.
The 9.3-kb SacI fragment from pMO3aD6 containing ORF5 was subcloned into pBSIISK(+). The kanamycin resistance gene (km) cassette was introduced into this plasmid by using the EZ::TN<KAN-2>Insertion kit, and a clone with the km cassette inserted in the center of the KS region of ORF5 module 3 was selected. This plasmid, pMO5, provides 5.4-kb (upstream) and 3.9-kb (downstream) regions flanking km. In the same manner, pMO7, which carries 3.7-kb (upstream) and 5.8-kb (downstream) regions flanking km inserted in the center of ORF3, was used for ORF3 disruption. Protoplast formation and transformation of “S. carzinostaticus” were carried out by standard procedures, with minor modifications. Insertion of the km cassette by double crossing over was confirmed by colony PCR (25) with the primer sets used for amplification of the kanamycin resistance genes and the KS region of ORF5 module 3 or ORF3. The expected size of a PCR product derived from double crossing over is 1.7 kb, whereas the wild type gives a 0.5-kb fragment. To assess NCZ productivity, “S. carzinostaticus” strains were cultured in R2YE liquid medium for 48 h, and the mycelial extracts were subjected to reversed-phase HPLC analysis under the conditions described above.
Feeding experiment with [2-13C]acetate.
At day 1 and day 3 of the production culture, 100 mg of sodium [2-13C]acetate (99 atom% 13C; ISOTEC, Inc.) was added to a 1-liter culture of S. coelicolor CH999/pMO11 (50). Isolation of the labeled compounds as described above yielded 19 mg of dechloroneocarzilin A and 13 mg of dechloroneocarzilin B. Isotope enrichments were evaluated by analysis of the 13C NMR spectra.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to the DDBJ, EMBL, and GenBank databases under accession number AB097904.
RESULTS
Cloning of type I PKS genes.
The partial polyene structures of NCZs, indicating their reduced polyketide origin, led us to assume that a type I PKS is involved in NCZ biosynthesis. A strategy was initiated to isolate PCR fragments possibly involved in NCZ biosynthesis on the basis of sequence conservation in the essential KS regions encoded by the known type I PKS genes for erythromycin (GenBank accession nos. M63676 and M63677), rapamycin (GenBank accession no. X86780), tylosin (GenBank accession no. U78289), and epothilone (GenBank accession no. AF210843). Degenerate primers KSMA-F and KSMB-R were used for PCR with genomic DNA of “S. carzinostaticus” as a template. The PCR product of the expected size (700 bp) was subcloned and sequenced to reveal a unique 681-bp gene. The genome library was screened by using the PCR product as a hybridization probe to identify two positive clones, pMO3aD6 and pMO4aH3, for further characterization.
DNA sequence analysis.
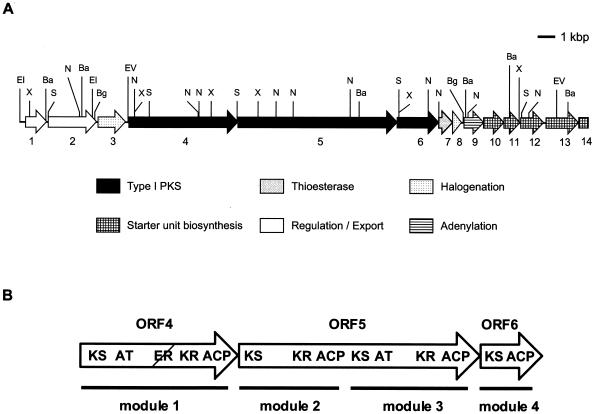
Restriction mapping was performed for the pMO3aD6 and pMO4aH3 inserts, and their overlapping subclones were sequenced to cover a 33-kb region of the pMO4aH3 insert (33,079 bp; overall G+C content, 74.4 mol%). Probable ORFs were detected with Frame plot software (2, 24) and by the presence of potential ribosome-binding sites (49). The deduced ORFs were functionally designated on the basis of database searches, as shown in Fig. 2. We identified 13 complete ORFs with an incomplete ORF at the right-hand end (Table 1; Fig. 2).
FIG. 2.
(A) Organization of the ncz cluster of “S. carzinostaticus” and (B) modular organization of type I PKS ORF4, ORF5, and ORF6. Abbreviations for restriction sites used for mapping: Ba, BamHI; Bg, BglII; EI, EcoRI; EV, EcoRV; N, NotI; S, SacI; X, XhoI. The slash indicates a putative nonfunctional domain.
TABLE 1.
Deduced functions of ORFs.
| ORF | Putative function | Size (no. of amino acids) | Homologues
|
||||
|---|---|---|---|---|---|---|---|
| Gene | Deduced role | SM/ID of producta | Origin | GenBank nucleotide accession no. | |||
| 1 | Transport | 417 | Integral membrane ion antiporter | 54/42 | Amycolatopsis orientalis | AL078635 | |
| 2 | Regulation | 934 | pikD | Transcriptional activator | 44/33 | Streptomyces venezuelae | AF079139 |
| 3 | Halogenase | 547 | pmC | Halogenase | 59/43 | Myxococcus fulvus | AF161185 |
| 4 | PKS | 2,057 | epoD | PKS | 42/32 | Polyangium cellulosum | AF217189 |
| 5 | PKS | 3,112 | mxaC | PKS | 50/37 | Stigmatella aurantiaca | AF319998 |
| 6 | PKS | 828 | stiG | PKS | 58/45 | Stigmatella aurantiaca | AJ421825 |
| 7 | Thioesterase | 268 | rifR | Thioesterase | 44/32 | Amycolatopsis mediterranei | AF040570 |
| 8 | Flavin reductase | 173 | rebF | Flavin reductase | 55/47 | Lechevalieria aerocolonigenes | AJ414559 |
| 9 | Adenylation enzyme | 377 | tycA | NRPSb adenylation domain | 59/43 | Brevibacillus parabrevis | P09095 |
| 10 | Starter unit for biosynthesis | 437 | SCGD3.18c | BCDH E1 α subunit | 73/61 | S. coelicolor A3(2) | AL939117 |
| 11 | Starter unit for biosynthesis | 381 | SCGD3.17c | BCDH E1 β subunit | 84/77 | S. coelicolor A3(2) | AL939117 |
| 12 | Starter unit for biosynthesis | 442 | SCGD3.16c | BCDH E2 subunit | 57/50 | S. coelicolor A3(2) | AL939117 |
| 13 | Starter unit for biosynthesis | 628 | SCAH10.35c | Oxidoreductase α subunit | 89/86 | S. coelicolor A3(2) | AL939127 |
| 14 | Starter unit for biosynthesis | >210 | SCD20.12c | Oxidoreductase β subunit | 97/96 | S. coelicolor A3(2) | AL939120 |
SM/ID, percent similarity/percent identity of amino acid sequences.
NRPS, nonribosomal peptide synthase.
Identification of four ORFs as type I PKSs and related genes.
Three large ORFs, ORF4, ORF5, and ORF6, were identified as genes encoding type I PKSs on the basis of localized similarity to known KS, acyltransferase (AT), and ACP domains. ORF4 carries module 1, ORF5 carries modules 2 to 3, and ORF6 carries module 4. A typical type I PKS module for macrolide biosynthesis consists of KR, DH, and ER, in addition to the essential components KS, AT, and ACP. In contrast to such architectures, the present PKS genes displayed rather unusual domain organizations. Each module carried the following obvious functional domains in the indicated order: module 1, KS, AT, KR, and ACP; module 2, KS, KR, and ACP; module 3, KS, AT, KR, and ACP; and module 4, KS and ACP. The initial PCR product turned out to be the KS domain of module 1.
Some interdomain regions for which functions were unassigned by the database search also exist, and they may possibly be functional (see Discussion). Another unusual feature is the lack of the loading module (LM) and thioesterase (TE) domains; both are normally found in the first and terminal modules of type I PKSs, respectively. Frame analysis indicated potential translational coupling of the four genes, together with the downstream gene, ORF8, presumed to encode a flavin reductase. We therefore attempted functional expression of the cloned genes to characterize the PKS products.
Heterologous expression and chemical characterization.
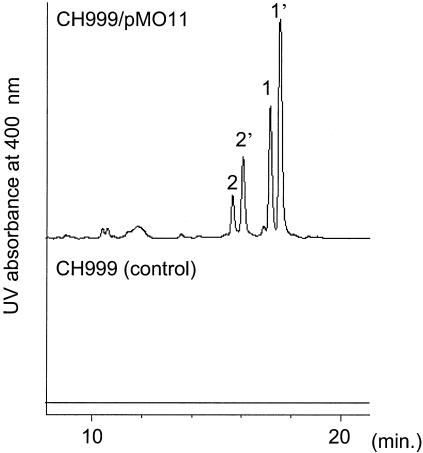
Functional analysis of the gene cluster was attempted by heterologous expression using an integrative vector (pTST59.1) for stable replication of the large DNA fragment derived from cosmid clones. The 27-kb EcoRV fragment from pMO4aH3, including ORF4 to ORF12, was placed under the direct control of the tipA promoter for efficient cotranscription of the target genes. The resultant gene segment, flanked by the thiostrepton resistance gene, was inserted into pTST59.1 to construct pMO11. This plasmid was then introduced into S. coelicolor CH999. Liquid cultures of the recombinants were analyzed for metabolites from the broth as well as the mycelium. A significant difference in the metabolic profiles of mycelial extracts of CH999/pMO11 and CH999 (control) was detected by HPLC (Fig. 3). Four peaks (peaks 1, 1′, 2, and 2′) turned out to be two related pairs of compounds: the compounds with peaks 1 and 2 are enol forms and the compounds with peaks 1′ and 2′ are the corresponding keto forms, as determined by chemical characterization, including NMR and GC-MS analyses. The overall structures of compounds 1 and 2 were determined to be nonchlorinated derivatives of neocarzilin A (NCZ-A) and neocarzilin B (NCZ-B), respectively. The present results suggest that the PKS gene cluster is involved in NCZ biosynthesis.
FIG. 3.
HPLC profiles (UV absorption at 360 nm) of S. coelicolor CH999 (control) and CH999/pMO11.
Gene disruption.
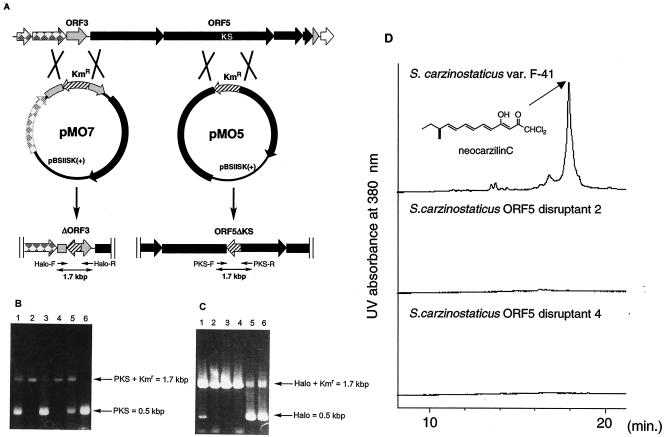
To confirm the expected involvement of the cluster in NCZ biosynthesis, we inactivated ORF5 and ORF3 by insertional disruption by double crossing over (Fig. 4). HPLC analysis of the metabolites indicated that the disruptants were deficient in NCZ production (Fig. 4). The combined results provide further evidence that the gene cluster is responsible for NCZ biosynthesis, thus allowing it to be designated the ncz cluster.
FIG. 4.
Disruption of ORF3 and ORF5 by double crossing over. (A) ORF maps of “S. carzinostaticus” var. F-41 and disruptants showing the predicted insertion position of the kanamycin resistance marker and fragment sizes amplified with the illustrated primer set. (B) Colony PCR results with the PKS-specific primer set and pMO5 transformants as templates; transformants 1, 3, and 5 (lanes 1, 3, and 5, respectively) are disruptants obtained by single crossing over, whereas transformants 2 and 4 (lanes 2 and 4, respectively) are disruptants obtained by double crossing over. Lane 6, control for the wild type. (C) Colony PCR results with the Halo primer set and pMO7 transformants as templates; transformants 1, 5, and 6 (lanes 1, 5, and 6, respectively) are disruptants obtained by single crossing over, whereas transformants 2, 3, and 4 (lanes 2, 3, and 4, respectively) are disruptants obtained by double crossing over. (D) HPLC analysis with UV detection at 380 nm of mycelial extracts from “S. carzinostaticus” var. F-41, ORF5 disruptants 2 and 4.
Feeding experiments with [2-13C]acetate.
13C-labeling experiments were carried out to clarify the biosynthetic origin of the basic carbon skeleton in the products derived from the ncz PKS. Highly specific incorporation of the label at the C-1, C-3, C-5, C-7, and C-9 positions of compounds 1 and 2 (dechloroneocarzilin A and B) at comparable levels indicated that their origin is a single polyketide chain (Table 2).
TABLE 2.
13C NMR results from incorporation of [2-13C]acetate
| Carbon atom | Dechloroneocarzilin A
|
Dechloroneocarzilin B
|
||
|---|---|---|---|---|
| Chemical shifta | Relative intensityb | Chemical shifta | Relative intensityb | |
| 1 | 27.0 | 4.2 | 27.0 | 3.7 |
| 2 | 197.5 | 1.0 | 197.5 | 1.0 |
| 3 | 100.7 | 5.0 | 100.7 | 3.5 |
| 4 | 177.1 | 1.0 | 177.2 | 0.52 |
| 5 | 124.9 | 4.9 | 124.9 | 3.8 |
| 6 | 140.4 | 1.1 | 140.4 | 0.73 |
| 7 | 128.7 | 4.8 | 128.8 | 3.4 |
| 8 | 140.7 | 1.3 | 140.7 | 1.1 |
| 9 | 128.4 | 4.1 | 127.2 | 4.6 |
| 10 | 146.0 | 1.9 | 147.1 | 1.0 |
| 11 | 38.8 | 1.3 | 31.5 | 0.84 |
| 12 | 29.5 | 0.59 | 22.0 | 0.61 |
| 13 | 11.7 | 1.1 | 22.0 | 0.61 |
| 14 | 19.7 | 0.92 | ||
In parts per million in CDCl3, 125 MHz.
Peak intensities were normalized to the signal of C-2.
DISCUSSION
Using degenerate primers designed from highly conserved KS regions of type I PKSs, we obtained a novel PKS gene fragment from “S. carzinostasiticus” var. F-41 by PCR amplification. Subsequent screening of the library and sequencing analysis identified a 33-kb cluster carrying 13 complete ORFs and an incomplete ORF14. Expression of the 27-kb fragment, including genes encoding an unusual type I PKS (ORF4, ORF5, and ORF6) and a type II TE (ORF7), in S. coelocolor CH999 produced dechloroneocarzilins A and B, which are apparent biosynthetic intermediates of NCZs. The present results, together with the fact that ORF5 disruption abolished NCZ production, strongly suggest the genes (the ncz cluster) responsible for NCZ biosynthesis.
The deduced products of ncz ORF4, ORF5, and ORF6 are type I PKSs with unusual features. They lack an obvious AT domain, which is mechanistically essential for polyketide chain extension, in modules 2 and 4. A few type I PKSs that lack the obvious AT domain have been reported (7, 11, 13, 37, 40); however, the discrete ATs or AT domains are found in those clusters. The leinamycin biosynthetic gene cluster (lnm) contains no cognate AT domain but contains a discrete AT in the same cluster (7). Cheng and coworkers (8) demonstrated by in vitro biochemical characterization that the AT protein loads malonyl-CoA to six of seven ACP domains, reasonably suggesting its iterative role in loading extender units into each module. The NCZ PKS, which does not contain a discrete AT domain in the sequenced region, was demonstrated to be sufficient to synthesize the NCZ skeleton, indicating that a distinct AT mechanism is involved in NCZ biosynthesis.
The NCZ PKS showed significant similarity to other type I PKSs, especially to proteins from myxobacteria, rather than the typical actinomycete type I PKSs. The ncz ORF4 product resembles EpoD for epothilone biosynthesis in Sorangium cellulosum. EpoD consists of four modules, each of which carries combinations of KS, AT, KR, DH, ER, and ACP, which provide a domain organization consistent with that of the epothilone structure (51). In contrast, ncz ORF4 harbors module 1 with KS, AT, KR, and ACP and a substantial of interdomain (ID) region length (ca. 550 amino acids), which has no meaningful similarity to known catalytic domains of type I PKSs. Similarly, ncz ORF5 and ORF6 encode modules 2, 3, and 4; and each module carries an ID region of ca. 400, 500, and 200 amino acids, respectively. The NCZ PKS homologues from myxobacteria are involved in the biosynthesis of metabolites of mixed origin, such as epothilone (the epo cluster in S. cellulosum), myxalamid (the mxa cluster in Stigmatella aurantiaca) (45), and stigmatellin (the sti cluster in S. aurantiaca) (16). The mxa cluster includes a combined PKS-nonribosomal peptide synthase consisting of nine modules, each of which carries large spacer (S) regions more than 300 amino acids in length between the AT and KR domains. Although no definite roles have been determined for the S regions, they appear to be common to all myxobacterial PKSs (45). The cluster resemblance of the NCZ PKSs with the mxa protein PKS counterparts might mean that their S-region functionalities are important for maintenance of the catalytic folds of the enzyme secondary structure. Another possibility is that the ID region acts as an AT domain or some other functional domain, such as DH, despite the lack of sequence similarity, since the triene structural element of NCZ is derived from reductive steps after polyketide chain extensions catalyzed by KR and DH. DH domains are relatively less well conserved among the known examples whose apparent homologues were not identified in the NCZ PKS modules. The ER domain recognized in module 1 appears to be nonfunctional for NCZ biosynthesis, and it has no nucleotide binding motif (43) necessary for cofactor association.
In the conventional organization of type I PKSs, core extending modules are preceded by an LM to supply an activated starter unit to the polyketide assembly line. LM is characterized by an AT and ACP set (ATL and ACPL, respectively); an example of the use of this LM is in erythromycin biosynthesis, in which propionyl-CoA is transferred (29, 39). Another type of LM (KSQ/ATL/ACPL) contains an extra KSQ domain, which is a mutated KS that acts as chain initiation factor (4). The terminal structures of NCZs suggest that their biosynthetic chain extensions initiate with isobutyryl-CoA and (S)-2-methylbutyryl-CoA as starter units. This is evidently the case for avermectin biosynthesis in Streptomyces avermitilis (22), in which an ATL/ACPL type of LM is involved. No LM was discovered upstream of ncz ORF4, which represents another unusual feature of the type I PKS.
At the end of the type I PKS assembly line there is usually a TE domain (TE I), which releases the completed acyl chain from its covalent linkage to the synthase. The terminal module 4 of the ncz cluster lacks this domain and is flanked by a separate ORF7, which likely encodes a monofunctional TE classified as type II (TE II). Recent genetic and biochemical studies suggest that TE II typically has a kind of editing role by removing aberrant intermediates that would otherwise interfere with PKS functions (19). TE II genes are occasionally discovered as an extra TE in type I PKS gene clusters, as in the case of the tyl cluster for tylosin biosynthesis in Streptomyces fradiae (5) and the pik cluster in Streptomyces venezuelae (53). Interestingly, a TE II, PikAV in S. venezuelae, was demonstrated to play a role in producing different macrocylic lactones with either 12- or 14-membered rings (53). This can be interpreted as a positive function at a branch point for the generation of structural diversity in antibiotic metabolites. Although no definite role for ORF7 can be deduced from the available information, ORF7 could be cotranscribed with the upstream PKS genes to encode a TE for the efficient release of the PKS product.
As mentioned earlier, isobutyryl-CoA and (S)-2-methylbutyryl-CoA are postulated to be used as starter units in NCZ biosynthesis. They also function as primers for branched-chain fatty acids and are derived from valine, leucine, and isoleucine by the action of the branched-chain amino acid transaminase and branched-chain α-keto acid dehydrogenase (BCDH) complex (26). S. avermitilis, a producer of avermectin, has two operons, bkdABC and bkdFGH, for putative BCDH subunits E1α, E1β, and E2 (47). Gene disruption studies subsequently proved that bkdFGH is involved in the supply of starter units not only for avermectin production but also for the biosynthesis of branched-chain fatty acids (10). ncz ORF10, ORF11, and ORF12 are clear homologues of E1α, E1β, and E2 (Table 1), respectively, and are possibly responsible for starter unit formation. ORF13 and ORF14 (partial) appear downstream of ORF10 to ORF12 and are deduced to encode the 2-oxoacid ferredoxin oxidoreductase complex. Because this complex catalyzes the reaction that is the reverse from that catalyzed by BCDH, the finding of ORF13 and ORF14 would allow us to postulate that ORF10 to ORF14 might coordinately control the level of starter supply. Under the expression conditions used in the present study, starter units could be supplied by the host strain, S. coelicolor CH999, whose genome has homologues of the relevant genes (Table 1).
Chlorination is one of the key structural features of NCZs. We found ORF3, deduced to encode a FADH2-dependent halogenase, together with a putative flavin reductase gene, ORF8. Disruption of ORF3 resulted in no accumulation of dechloroneocarzilins but completely abolished NCZ production (data not shown), indicating that the expression of the ncz PKS and halogenase is coordinated by an uncharacterized regulatory mechanism. The ORF3 product carries the highly conserved nucleotide binding motif GXGXXG toward the N terminus (18). Although chlorination in antibiotics is mechanistically unclear, all known examples of FADH2-dependent halogenases are involved in halogenation of indole, phenol, and pyrrole rings, such as pyrrolnitrin (18), chloroeremomycin (52), balhimycin (38), pyoluteorin (33), and rebeccamycin (36, 42). The present finding, together with the finding from a recent report (41) on the cloning of FADH2-dependent halogenase gene fragments from S. venezuelae ISP5230 that are possibly involved in chloramphenicol biosynthesis, could indicate a novel example of a halogenase involved in the chlorination of an aliphatic carbon.
Limited information is available on the chlorination of an aliphatic natural product. An interesting example is the biosynthetic origin of the trichloromethyl group of the lipopeptide barbamide from the marine cyanobacterium Lyngbya majuscula. Earlier feeding experiments (46) demonstrated the high level of incorporation of trichloroleucine. Subsequent genetic studies identified the two putative barbamide biosynthetic genes, barB1 and barB2, possibly responsible for leucine chlorination (6). Although the mutually homologous proteins BarB1 and BarB2 are sequentially unrelated to a family of halogenases, they are presumed to be involved in chlorination on the basis of their high degrees of similarity to the biosynthetic gene products of another chlorinated lipopeptide, syringomycin (6). The barbamide biosynthetic gene cluster (the bar cluster) provides a mixed PKS-nonribosomal peptide synthase system consisting of the four adenylation domains. One of the domains, AE, encoded by barE, was demonstrated (6) to activate trichloroleucine specifically, indicating the chlorination reaction of leucine prior to assembly of the peptide and polyketide chain. This is a marked difference from NCZ biosynthesis, for which we propose the involvement of chlorination in tailoring steps (see below). Further functional studies on ncz ORF3 as well as the other available putative halogenase genes are expected to shed light on the mechanistic aspects of halogenation of aliphatic carbons.
The regulatory and antibiotic export genes are ORF1 and ORF2, which are found farthest upstream of the ncz cluster. The ORF1 protein has an overall similarity to a putative integral membrane ion antiporter encoded by CZA382.28 (GenBank accession no. AL078635) and is possibly involved in biosynthesis of an antibiotic of the vancomycin group in Amycolatopsis orientalis. The ORF2 product resembles PikD, which is a putative transcriptional activator probably involved in mixed macrolide biosynthesis in S. venezuelae (53).
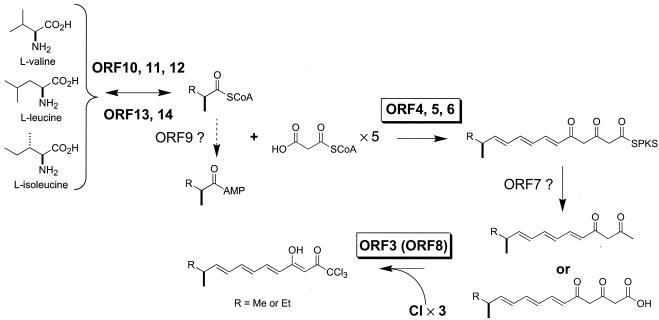
The combined experimental data and the deduced functions of the ncz genes allow a plausible biosynthetic pathway of NCZs in “S. carzinostaticus” to be proposed (Fig. 5). Starter units supplied by the products of ORF10 to ORF12 (or ORF10 to ORF14) initiate the assembly of the NCZ PK. Our feeding experiments with [2-13C]acetate gave highly specific incorporations at C-1, C-3, C-5, C-7, and C-9 of dechloroneocarzilins A and B, suggesting that all the carbons besides the starter units are derived from an extending unit, malonyl-CoA, by five condensations. The NCZ PKS genes encode four modules, two of which possess organizations that are unusual, in that they lack an obvious AT domain. The necessary number of condensations for NCZ biosynthesis apparently requires repeated use of the PKS functionality, which presents an example of an iterative type I PKS. Because the last two condensations do not require subsequent reductive steps, module 4, which consists of only one KS domain and one ACP domain, most likely functions iteratively to produce the NCZ skeleton. The only example known to be a mixture of modular and iterative type I PKS systems is the stigmatellin biosynthetic (sti) gene cluster in S. aurantiaca (16). The sti cluster has nine modules, despite the need for 10 condensations for stigmatellin biosynthesis, which would require one of the last two modules to function iteratively.
FIG. 5.
Proposed biosynthetic pathway of the NCZs and putative functional assignment of the ncz proteins. Me, methyl group; Et, ethyl group; SPKS, PKS bound; SCoA, CoA bound.
The present work revealed a novel iterative type I PKS involved in NCZ biosynthesis in “S. carzinostaticus” var. F-41. Particularly noteworthy is the existence of the genes encoding a putative FADH2-dependent halogenase. Detailed functional analysis of these genes will lead to the discovery of the novel types of biosynthetic enzymes involved in aliphatic halometabolites.
Acknowledgments
We gratefully acknowledge Kayaku Co. Ltd. for the supply of “S. carzinostaticus” strains. We are indebted to Tomihisa Ohta, Kanazawa University, for the generous gift of NCZ-A. We thank Josef Altenbuchener, University of Stuttgart, for useful discussion. We are grateful to Heinz G. Floss, University of Washington, for kindly supplying plasmids. We thank David A. Hopwood for critical reading of the manuscript.
A part of this work was financially supported by a Grant-in-Aid for Scientific Research (S) (grant 15101007) from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Ahlert, J., E. Shepard, N. Lomovskaya, E. Zazopoulos, A. Staffa, B. O. Bachmann, K. Huang, L. Fonstein, A. Czisny, R. E. Whitwam, C. M. Farnet, and J. S. Thornson. 2002. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science 297:1173-1175. [DOI] [PubMed] [Google Scholar]
- 2.Bibb, M. J., P. R. Findlay, and M. W. Johnson. 1984. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30:157-166. [DOI] [PubMed] [Google Scholar]
- 3.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Bisang, C., P. F. Long, J. Cortes, J. Westcott, J. Crosby, A. L. Matharu, R. Cox, T. J. Simpson, J. Staunton, and P. F. Leadlay. 1999. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature 401:502-505. [DOI] [PubMed] [Google Scholar]
- 5.Butler, A. R., N. Bate, and E. Cundliffe. 1999. Impact of thioesterase activity on tylosin biosynthesis in Streptomyces fradiae. Chem. Biol. 6:287-292. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Z., P. Flatt, W. H. Gerwick, V.-A. Nguyen, C. L. Willis, and D. H. Sherman. 2002. The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene 296:235-247. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, Y.-Q., G.-L. Tang, and B. Shen. 2002. Identification and localization of the gene cluster encoding biosynthesis of the antitumor macrolactam leinamycin in Streptomyces atroolivaceus S-140. J. Bacteriol. 184:7013-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Y.-Q., G.-L. Tang, and B. Shen. 2003. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc. Natl. Acad. Sci. USA 100:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dairi, T., T. Nakano, K. Aisaka, R Katsumata, and M. Hasegawa. 1995. Cloning and nucleotide sequence of the gene responsible for chlorination of tetracycline. Biosci. Biotechnol. Biochem. 59:1099-1106. [DOI] [PubMed] [Google Scholar]
- 10.Denoya, C. D., R. W. Fedechko, E. W. Hafner, A. I. McArthur, M. R. Morgenstern, D. D. Skinner, K. Stutzman-Engwall, R. G. Wax, and W. C. Wernau. 1995. A second branched-chain α-keto acid dehydrogenase gene cluster (bkdFGH) from Streptomyces avermitilis: its relationship to avermectin biosynthesis and the construction of a bkdF mutant suitable for the production of novel antiparasitic avermectins. J. Bacteriol. 177:3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duitman, E. H., L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ullrich, T. Stein, F. Leenders, and J. Vater. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96:13294-13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edo, K., M. Mizugaki, Y. Koide, H. Seto, K. Furihata, N. Õtake, and N. Ishida. 1985. The structure of neocarzinostatin chromophore possessing a novel bycyclo[7,3,0]dodecadiyne system. Tetrahedron Lett. 26:331-334. [Google Scholar]
- 13.El-Sayed, A. K., J. Hothersall, S. M. Cooper, E. Stephens, T. J. Simpson, and C. M. Thomas. 2003. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem. Biol. 10:419-430. [DOI] [PubMed] [Google Scholar]
- 14.Funa, N., Y. Ohnishi, I. Fujii, M. Shibuya, Y. Ebizuka, and S. Horinouchi. 1999. A new pathway for polyketide synthesis in microorganisms. Nature 400:897-899. [DOI] [PubMed] [Google Scholar]
- 15.Gaisser, S., A. Trefzer, S. Stockert, A. Kirschning, and A. Bechthold. 1997. Cloning of an avilamycin biosynthetic gene cluster from Streptomyces viridochromogenes Tü57. J. Bacteriol. 179:6271-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaitatzis, N., B. Silakowski, B. Kunze, G. Nordsiek, H. Blöcker, G. Höfle, and R. Müller. 2002. The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by novel type of modular polyketide synthase. J. Biol. Chem. 277:13082-13090. [DOI] [PubMed] [Google Scholar]
- 17.Gribble, G. W. 2003. The diversity of naturally produced organohalogens. Chemosphere 52:289-297. [DOI] [PubMed] [Google Scholar]
- 18.Hammer, P. E., D. S. Hill, S. T. Lam, K.-H. van Pée, and J. M. Ligon. 1997. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 63:2147-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heathcote, M. L., J. Staunton, and P. F. Leadlay. 2001. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem. Biol. 8:207-220. [DOI] [PubMed] [Google Scholar]
- 20.Hohaus, K., A. Altmann, W. Burd, I. Fischer, P. E. Hammer, D. S. Hill, J. M. Ligon, and K.-H. van Pée. 1997. NADH-dependent halogenases are more likely to be involved in halometabolite biosynthesis than haloperoxidases. Angew. Chem. Int. Engl. Ed. 36:2012-2013. [Google Scholar]
- 21.Hopwood, D. A. 1997. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97:2465-2497. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda, H., T. Nonomiya, M. Usami, T. Ohta, and S. Õmura. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA 96:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishida, N., K. Miyazaki, K. M. Kumagai, and M. Rikimaru. 1965. Neocarzinostatin, an antitumor antibiotic of high molecular weight. J. Antibiot. 18:68-76. [PubMed] [Google Scholar]
- 24.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa, J., N. Tsuchizaki, M. Yoshida, D. Ishiyama, and K. Hotta. 2000. Colony PCR for detection of specific DNA sequences in actinomycetes. Actinomycetologica 14:1-5. [Google Scholar]
- 26.Kaneda, T. 1991. Iso- and anteoiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 55:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller, S., T. Wage, K. Hohaus, M. Hölzer, E. Eichhorn, and K.-H. van Pée. 2000. Purification and partial characterization of tryptophan 7-halogenase (PrnA) from Pseudomonas fluorescens. Angew. Chem. Int. Ed. 39:2300-2302. [DOI] [PubMed] [Google Scholar]
- 28.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical streptomyces genetics. The John Innes Foundation, Norwich, England.
- 29.Lau, J., D. E. Cane, and C. Khosla. 2000. Substrate specificity of the loading didomain of the erythromycin polyketide synthase. Biochemistry 39:10514-10520. [DOI] [PubMed] [Google Scholar]
- 30.Liu, W., S. D. Christenson, S. Standage, and B. Shen. 2002. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science 297:1170-1173. [DOI] [PubMed] [Google Scholar]
- 31.Lomoskaya, N. D., K. F. Chater, and N. M. Mkrtumian. 1980. Genetics and molecular biology of Streptomyces bacteriophages. Microbiol. Rev. 44:206-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDaniel, R., S. E. Khosla, D. A. Hopwood, and C. Khosla. 1993. Engineered biosynthesis of novel polyketides. Science 262:1546-1550. [DOI] [PubMed] [Google Scholar]
- 33.Nowak-Thompson, B., N. Chaney, J. S. Wing, S. J. Gould, and J. E. Loper. 1999. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J. Bacteriol. 181:2166-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nozoe, S., N. Ishii, G. Kusano, K. Kikuchi, and T. Ohta. 1992. Neocarzilins A and B, novel polyenones from Streptomyces carzinostaticus. Tetrahedron Lett. 33:7547-7550. [Google Scholar]
- 35.Nozoe, S., K. Kikuchi, N. Ishii, and T. Ohta. 1992. Synthesis of neocarzilin A: an absolute stereochemistry. Tetrahedron Lett. 33:7551-7552. [Google Scholar]
- 36.Onaka, H., S. Taniguchi, Y. Igarashi, and T. Furumai. 2003. Characterization of the biosynthetic gene cluster of rebeccamycin from Lechevaleria aerocolonigenes ATCC 39243. Biosci. Biotechnol. Biochem. 67:127-138. [DOI] [PubMed] [Google Scholar]
- 37.Paitan, Y., G. Alon, E. Orr, E. Z. Ron, and E. Rosenberg. 1999. The first gene in the biosynthesis of the polyketide antibiotic TA of Myxococcus xanthus codes for a unique PKS module coupled to a peptide synthetase. J. Mol. Biol. 286:465-474. [DOI] [PubMed] [Google Scholar]
- 38.Pelzer, S., R. Süβmuth, D. Heckmann, J. Recktenwald, P. Huber, G. Jung, and W. Wohlleben. 1999. Identification and analysis of the balhimycin biosynthetic gene cluster and its use for manipulating glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908. Antimicrob. Agents Chemother. 43:1565-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereda, A., R. G. Summers, D. L. Stassi, X. A. Ruan, and L. Katz. 1998. The loading domain of the erythromycin polyketide synthase is not essential for erythromycin biosynthesis in Saccharopolyspora erythraea. Microbiology 144:543-553. [DOI] [PubMed] [Google Scholar]
- 40.Piel, J. 2002. A polyketide synthase-peptide synthase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 99:14002-14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piraee, M., and L. C. Vining. 2002. Use of degenerate primers and touchdown PCR to amplify a halogenase gene fragment from Streptomyces venezuelae ISP5230. J. Ind. Microbiol. Biotechnol. 29:1-5. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez, C., I. A. Butovich, A. F. Brana, J. Rohr, C. Mendez, and J. A. Salas. 2002. The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarbazole derivatives. Chem. Biol. 9:519-531. [DOI] [PubMed] [Google Scholar]
- 43.Scrutton, N. S., A. Berry, and R. N. Perham. 1990. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 343:38-43. [DOI] [PubMed] [Google Scholar]
- 44.Shima, J., A. Hesketh, S. Okamoto, S. Kawamoto, and K. Ochi. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 178:7276-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silakowski, B., G. Nordsiek, B. Kunze, H. Blöcker, and R. Müller. 2001. Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: the myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sga15. Chem. Biol. 8:59-64. [DOI] [PubMed] [Google Scholar]
- 46.Sitachitta, N., B. L. Márquez, R. T. Williamson, J. Rossi, M. A. Roberts, W. H. Gerwick, V.-A. Nguyen, and C. L. Willis. 2000. Biosynthetic pathway and origin of the chlorinated methyl group in barbamide and dechlorobarbamide, metabolites from marine cyanobacterium Lyngbya majuscula. Tetrahedron 56:9103-9113. [Google Scholar]
- 47.Skinner, D. D., M. R. Morgenstern, R. W. Fedechko, and C. D. Denoya. 1995. Cloning and sequencing of a cluster of genes encoding branched-chain alpha-keto acid dehydrogenase from Streptomyces avermitilis and the production of a functional E1 [αβ] component in Escherichia coli. J. Bacteriol. 177:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smokvina, T., P. Mazodier, F. Boccard, C. J. Thompson, and M. Guérieneau. 1990. Construction of a series of pSAM2-based integrative vectors for use in actinomycetes. Gene 94:53-59. [DOI] [PubMed] [Google Scholar]
- 49.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taguchi, T., K. Itou, Y. Ebizuka, F. Malpartida, D. A. Hopwood, C. M. Surti, K. I. Booker-Milburn, G. R. Stephenson, and K. Ichinose. 2000. Chemical characterisation of disruptants of the Streptomyces coelicolor A3(2) actVI genes involved in actinorhodin biosynthesis. J. Antibiot. 53:144-152. [DOI] [PubMed] [Google Scholar]
- 51.Tang, L., S. Shah, L. Chung, J. Carney, L. Katz, C. Khosla, and B. Julien. 2000. Cloning and heterologous expression of the epothilone gene cluster. Science 287:640-642. [DOI] [PubMed] [Google Scholar]
- 52.Van Wageningen, A. M. A., P. N. Kirkpatrick, D. H. Williams, B. R. Harris, J. K. Kershaw, N. J. Lennard, M. Jones, S. J. M. Jones, and P. J. Solenberg. 1998. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 5:155-162. [DOI] [PubMed] [Google Scholar]
- 53.Xue, Y., L. Zhao, H.-W. Liu, and D. H. Sherman. 1998. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc. Natl. Acad. Sci. USA 95:12111-12116. [DOI] [PMC free article] [PubMed] [Google Scholar]