Abstract
Three mutants of the extended-spectrum β-lactamase TEM-60, the P51L, K104E, and S164R mutants, were constructed by site-directed mutagenesis. The kinetic parameters of the mutated enzymes and interactions of inhibitors were significantly different from those of TEM-60, revealing that the L51P mutation plays an important role in enzyme activity and stability in the TEM-60 background.
The activity of oxyimino-cephalosporins and monobactams against gram-negative bacterial pathogens has been impaired by the emergence and dissemination of extended-spectrum β-lactamases (ESBLs) (1, 5). In the family Enterobacteriaceae, the majority of ESBLs are TEM- or SHV-type derivatives that have extended their substrate specificity after amino acid substitutions at some key positions (1). A large number of natural TEM- and SHV-type variants with ESBL activity have been described (http://www.lahey.org/studies/webt.htm). TEM-60 is a TEM-2 derivative originally detected in a clinical isolate of Providencia stuartii (3). TEM-60 differs from TEM-2 by three amino acid substitutions (L51P, E104K, and R164S). Two of the three amino acid substitutions (E104K and R164S) are common among TEM-type ESBLs and well-known for their role in extension of substrate specificity (6), while one (L51P) is unique to this TEM-type variant (http://www.lahey.org/studies/webt.htm). The goal of this study was to investigate the role of the mutations present in TEM-60 by a site-directed mutagenesis approach.
Construction and characterization of TEM-60 mutants.
Three TEM-60 mutants, the P51L, K104E, and S164R mutants, were generated by site-directed mutagenesis using the overlap extension method (12). Briefly, each mutation was introduced into a PCR amplicon using mutagenic primers in combination with primers TEM/F and TEM/R (Table 1) to generate two partially overlapping DNA fragments, which were subsequently used in an overlap extension reaction coupled to amplification of the entire coding sequence with the TEM/F and TEM/R primers. The resulting amplicons were cloned in plasmid pBC-SK (Stratagene, Inc., La Jolla, Calif.), using the BamHI and EcoRI restriction sites present in the TEM primers, to obtain plasmids pMUT1 (TEM-60 P51L), pMUT-2 (TEM-60 K104E), and pMUT-3 (TEM60 S164R). Plasmid pVR-1 (3) was used as the template for site-directed mutagenesis experiments. Escherichia coli strain HB101 (11) was used as the host for recombinant plasmids. The authenticity of cloned mutant genes was verified by sequencing both strands.
TABLE 1.
Oligonucleotide primers used for site-directed mutagenesis
| Primer | Sequencea | Positionsb | Codon change | Primer used in combination | Amplicon size (bp) |
|---|---|---|---|---|---|
| Mutagenic primers | |||||
| Mutagenic for P51L | |||||
| P51L/F | 5′-GAGCTGGATCTCAACAGCGGTAAG-3′ | 135-159 | CCT→CTC | ||
| P51L/R | 5′-CTTACCGCTGTTGAGATCCAGCTC-3′ | 159-135 | |||
| Mutagenic for K104E | |||||
| K104E/F | 5′-GAATGACTTGGTTGAGTACTCACCAG-3′ | 290-316 | AAG→GAG | ||
| K104E/R | 5′-CTGGTGAGTACTCAACCAAGTCATTC-3′ | 316-290 | |||
| Mutagenic for R164S | |||||
| R164S/F | 5′-CGCCTTGATAGATGGGAACCGGA-3′ | 474-497 | AGT→AGA | ||
| R164S/R | 5′-TCCGGTTCCCATCTATCAAGGCG-3′ | 497-474 | |||
| Amplification primers | |||||
| TEM/F | 5′-CCCGGATCCATGAGTATTCAACATTTCCGTGCT-3′ | P51L/R | 159 | ||
| K104E/R | 316 | ||||
| R164S/R | 497 | ||||
| TEM/R | 861 | ||||
| TEM/R | 5′-CCCGAATTCTTACCAATGCTTAATCAGTGAGGCA-3′ | P51L/F | 727 | ||
| K104E/F | 572 | ||||
| R164S/F | 388 |
The mutated positions are underlined.
Positions according to the numbering of the blaTEM-60 coding sequence.
The BamHI and EcoRI restriction sites introduced immediately before the start and stop codons (in boldface type) of the blaTEM-60 coding sequence to facilitate cloning are underlined.
Each mutant enzyme was purified from a culture of the corresponding E. coli strain grown overnight at 37°C in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.). Enzymes were extracted from bacterial cells suspended in 100 mM Tris-HCl (pH 8.0) by sonic disruption and purified by three chromatography steps: an anion-exchange chromatography on a Q-Sepharose FF column (Amersham Biosciences, Milan, Italy) equilibrated with 100 mM Tris-HCl (pH 8.0) and eluted with a linear NaCl gradient in the same buffer; a size-exclusion chromatography on a Superdex-200 column (XK 16/100; Amersham Biosciences) equilibrated and eluted with 20 mM sodium phosphate buffer (pH 7.0) containing 0.15 M NaCl; and a fast chromatofocusing on a MonoP HR 5/20 column (Amersham Biosciences) equilibrated with 25 mM Bis-Tris buffer (pH 7.1) and eluted with 25 ml of 10-fold-diluted Polybuffer 74 in the pH range of 7 to 4. During purification, β-lactamase activity was monitored by hydrolysis of 100 μM nitrocefin as described previously (3). The purity of the enzyme preparations was >95%, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7) (data not shown). The pIs of the purified TEM-60 P51L, K104E, and S164R mutants, determined by analytical isoelectric focusing (3), were 6.0, 5.4, and 6.1, respectively, in agreement with theoretical values. Steady-state kinetic parameters (Km and kcat) with β-lactam substrates were determined under initial rate conditions as described previously (9). Inhibition by β-iodopenicillanate (β-IP) and tazobactam was investigated using nitrocefin (200 to 300 μM) as the reporter substrate as described previously (3).
The kinetic parameters of the mutant enzymes were determined with several β-lactam substrates. Compared to TEM-60, the P51L mutant lost activity against cefotaxime and exhibited a reduced catalytic efficiency with penicillins and aztreonam; this was mostly related to an increase in the Km values. The K104E mutant lost activity against oxyimino-cephalosporins, aztreonam, and ampicillin. The S164R mutant lost activity against ceftazidime and exhibited increased Km values with the other β-lactam substrates. This resulted in a remarkable reduction of catalytic efficiency with cefotaxime and aztreonam, while with cephaloridine, ampicillin, and penicillin G, the decreased affinity was balanced out by an increase in the turnover rates, with minor changes in the kcat/Km ratios (Table 2).
TABLE 2.
Kinetic parameters determined with the purified β-lactamase TEM-60 mutants
| Substrate | TEM-60
|
TEM-60 P51L
|
TEM-60 K104E (TEM-7)
|
TEM-60 S164R (TEM-18)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kmb (μM) | kcat (s−1) | kcat/Km (μM−1 · s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 · s−1) | Kmb (μM) | kcat (s−1) | kcat/Km (μM−1 · s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 · s−1) | |
| Nitrocefin | 20 | 5 | 0.25 | 180 | 8 | 0.044 | 41 (NAc) | 176 (NA) | 4.3 (NA) | 65 (NA) | 600 (NA) | 9.2 (NA) |
| Cephaloridine | 28 | 15 | 0.54 | 18 | 22 | 1.2 | 70 (87) | 127 (26) | 1.8 (0.30) | 156 (321) | 590 (675) | 3.8 (2.1) |
| Cefotaxime | 31 | 1.5 | 0.048 | >1,000d | <0.01e | <10−5 | >1,000d (100) | <0.01e (1.5) | <10−5 (0.015) | 210 (1,975) | 0.45 (158) | 0.002 (0.080) |
| Ceftazidime | 59 | 12 | 0.20 | 80 | 60 | 0.75 | >1,000d (1,000) | <0.01e (9.0) | <10−5 (0.009) | >1,000d (2,875) | <0.01e (23) | <10−5 (0.008) |
| Aztreonam | 55 | 9 | 0.16 | 660 | 17 | 0.026 | >1,000d (1,333) | <0.01e (4.0) | <10−5 (0.003) | 1,660 (785) | 150 (11) | 0.090 (0.014) |
| Ampicillin | 9 | 18 | 2.0 | 450 | 3.2 | 0.007 | 4.2 (16) | <0.01e (18) | <10−5 (1.1) | 335 (35) | 146 (920) | 0.44 (26) |
| Penicillin G | 9 | 26 | 2.9 | 575 | 23 | 0.040 | 1.7 (3.1) | 33 (40) | 19 (13) | 15 (20) | 200 (1,400) | 13 (69) |
Km and kcat values are the means of three measurements. The standard deviation was always lower than 5%. Data for TEM-60 are from reference 3. Data for TEM-7 (10) and for TEM-18 (10) are shown in parentheses in comparison to those of the K104E and S164R mutants.
Km values lower than 10 μM were determined as Ki as described previously (4), using nitrocefin (200 to 300 μM) as the reporter substrate.
NA, not available.
Evaluated upon exposure of the enzyme to a substrate concentration of up to 1 mM, using nitrocefin (200 μM) as the reporter substrate.
Found using an enzyme concentration of 25 nM and a substrate concentration of 1 mM and recording the absorbance for 30 min.
β-IP was unable to inhibit the TEM-60 P51L mutant and with the K104E and S164R mutants exhibited lower acylation efficiencies than the acylation efficiency of TEM-60. Tazobactam, which with TEM-60 acts as a transient inactivator with a measurable turnover rate (k when a substrate concentration of 2 mM was used [k+2], 4.2 × 10−4 s−1), behaved as a competitive inhibitor for all mutants (Table 3).
TABLE 3.
Kinetic parameters of TEM-60 mutants with β-lactamase inhibitorsa
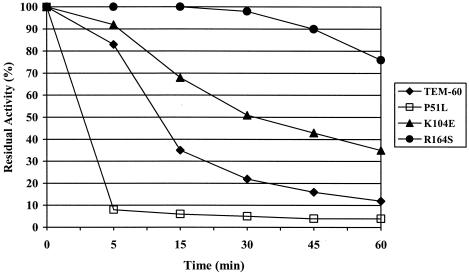
The thermal stability of the TEM-60 mutants was determined by measuring activity (at 30°C against 50 μM nitrocefin in 30 mM sodium phosphate buffer [pH 7.2]) after incubation at 42°C for up to 60 min. Residual activity was calculated relative to the activity of the corresponding enzyme after incubation at 30°C for the same time, which was set at 100%. Compared to TEM-60, the thermal stability of the P51L mutant was greatly decreased, while that of the two other mutants was increased (Fig. 1).
FIG. 1.
Thermal stability of TEM-60 mutants in comparison with that of the TEM-60 enzyme. Data are mean values of three measurements. The standard deviation was always lower than 10%. The experiments were performed as described in the text.
MICs of several β-lactams for the E. coli HB101 derivatives producing the different TEM-60 mutants were determined by a broth macrodilution assay (8). Overall, results of susceptibility testing were consistent with kinetic data (Table 4).
TABLE 4.
Patterns of β-lactam resistance mediated by different TEM-60 mutants produced in E. coli HB101a
| Antibiotic | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| HB101 (pVR-1) TEM-60 | HB101 (pMUT1) TEM-60 P51L | HB101 (pMUT2) TEM60 K104E | HB101 (pMUT3) TEM-60 R164S | HB101 | |
| Ampicillin | >64 | >64 | >64 | >64 | 2 |
| Cefotaxime | 4 | ≤0.5 | ≤0.5 | 1 | ≤0.5 |
| Ceftazidime | >64 | >64 | 1 | 1 | ≤0.5 |
| Aztreonam | >64 | 64 | 4 | 16 | ≤0.5 |
The in vitro susceptibilities of HB101(pVR1) producing TEM-60 and of HB101 are shown for comparison.
Concluding remarks.
The decreased stability of the TEM-60 P51L mutant and impaired activity with some substrates indicate that the L51P mutation, which is unique to TEM-60, plays an important role in the activity and stability of the enzyme. This finding is also consistent with the fact that, although the Q39K, E104K, and R164S substitutions are common among natural TEM-type variants, the combination of these three mutations has never been reported alone but always associated with other mutations, such as in TEM-8, TEM-24, TEM-46, and TEM-60 (http://www.lahey.org/studies/webt.htm). Interestingly, in a background such as that of TEM-7 or TEM-18, which could represent potential precursors of TEM-60, the L51P mutation was not neutral but appeared to be detrimental to the enzyme activity with some substrates, although it increased the enzyme stability. This fact might explain why the occurrence of this mutation among natural TEM-type derivatives is rare. In the TEM-1 structure, the L51 residue lies in the S1 β-sheet, only a few residues apart from the S2 β-sheet (3). The presence of a proline residue in this location, close to the R191, L194, and T195 residues (belonging to the H8 helix) and to the I260 residue (belonging to the S5 β sheet), could influence the hydrolytic properties of the enzyme by affecting stabilization of the helix and stability of the enzyme.
REFERENCES
- 1.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Meester, F., J. M. Frère, S. G. Waley, S. J. Cartwright, R. Virden, and F. Lindberg. 1986. 6-β-Iodopenicillanate as a probe for the classification of β-lactamases. Biochem. J. 239:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franceschini, N., M. Perilli, B. Segatore, D. Setacci, G. Amicosante, A. Mazzariol, and G. Cornaglia. 1998. Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived extended-spectrum β-lactamase, TEM-60. Antimicrob. Agents Chemother. 42:1459-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galleni, M., N. Franceschini, B. Quinting, L. Fattorini, G. Orefici, A. Oratore, J. M. Frère, and G. Amicosante. 1994. Use of the chromosomal class A β-lactamase of Mycobacterium fortuitum D316 to study potentially poor substrates and inhibitory β-lactam compounds. Antimicrob. Agents Chemother. 38:1608-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacoby, G. A. 1997. Extended-spectrum β-lactamases and other enzymes providing resistance to oxyimino-β-lactams. Infect. Dis. Clin. N. Am. 11:875-887. [DOI] [PubMed] [Google Scholar]
- 6.Knox, J. R. 1995. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob. Agents Chemother. 39:2593-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard. NCCLS document M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 9.Perilli, M., B. Segatore, M. R. De Massis, N. Franceschini, C. Bianchi, G. M. Rossolini, and G. Amicosante. 2002. Characterization of a new extended-spectrum β-lactamase (TEM-87) isolated in Proteus mirabilis during an Italian survey. Antimicrob. Agents Chemother. 46:925-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raquet, X., J. Lamotte-Brasseur, E. Fonzè, S. Goussard, P. Courvalin, and J. M. Frère. 1994. TEM β-lactamase mutants hydrolysing third-generation cephalosporins. J. Mol. Biol. 244:625-639. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Steffan, N. H., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using a polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 13.Vakulenko, S., and D. Golemi. 2002. Mutant TEM β-lactamase producing resistance to ceftazidime, ampicillins, and β-lactamase inhibitors. Antimicrob. Agents Chemother. 46:646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]