Abstract
Human immunodeficiency virus type 1 subtype B and C proteases were manipulated to contain 90M, 88D, or 89L, and their in vitro biological properties were studied. We showed that D30N has significantly more impact in subtype C than in subtype B counterparts, accounting for the reported low prevalence of this mutation in patients failing nelfinavir-based regimens.
Drugs designed to inhibit reverse transcriptase and protease (PR) of human immunodeficiency virus type 1 (HIV-1) have been implemented in the last decade, and their combination regimens have improved the treatment of infected individuals (13, 17). However, not all patients respond to highly active antiretroviral treatment and eventually develop drug resistance through the accumulation of several patterns of drug resistance mutations. Such mutations have been extensively characterized for subtype B isolates (9, 18; see the updated list in the Los Alamos National Laboratory HIV sequence database at www.hiv.lanl.gov), and their biological relevance and phenotypic impact in non-B subtypes need to be clarified.
The mutation D30N is a unique amino acid substitution found in the PR from isolates of patients failing nelfinavir (NFV)-containing regimens. Recently, Cane et al. (3) studied 58 HIV-1 subtype C-infected individuals treated with several combinations of PR inhibitor (PI)-containing regimens and found the D30N mutation in only one patient, together with the N88D mutation, although NFV was widely used in this patient group. This phenomenon was also observed in patients infected with CRF01(A/E) failing a PI-containing regimen in Japan, where the mutations D30N, A71V, and N88D were found exclusively in subtype B-infected individuals (2). Previous studies conducted with HIV-1 subtype B-infected patients indicated, however, that around 50% of patients receiving NFV fail therapy due to the development of the D30N mutation, whereas the remaining accumulate an alternative mutation pattern containing L90M as the primary mutation (14). This study explores this uneven distribution of D30N and L90M PR mutations between B and C subtype strains and shows the different in vitro behaviors of B and C isolates carrying these mutations.
Clone pNL4-3 (1) was used as the prototypic subtype B strain, and pNL43-C6 (7) was used as the subtype C prototypic clone. Clone C6 is a modified pNL4-3 infectious clone where the six amino acid differences between the subtype C and B consensus sequences (I15V, M36I, R41K, H69K, L89M, and I93L) were introduced through site-directed mutagenesis (7). Prototypic pNL43 and C6 PR genes were cloned in the pCR4 TOPO vector (Invitrogen) by following the manufacturer's protocol. Clones were then subjected to site-directed mutagenesis in order to modify the target codons by a PCR mutagenesis procedure described by the manufacturer (QuikChange kit; Stratagene). Using this technique, we constructed the clones CD30N, CD30N/N88D, CD30N/M89L, CL90M, BD30N, BD30N/N88D, and BL90M. Primer sequences used in those mutageneses are available upon request. Prototypic and mutant PR clones were inserted in a PR-deleted HXB2 backbone by a recombinant virus assay technology as previously described (8, 11). Cotransfections were always conducted with the same amounts of PR fragment and vector DNA (5 and 1 μg, respectively) in three independent experiments. BL90M and BD30N/N88D generated detectable recombinant viruses 6 days posttransfection (pt), 1 day after Bwt, whereas BD30N and BD30N/N88D were capable of generating detectable viral particles only after 10 days pt. In contrast, CD30N, CD30N/N88D, and CL90M viruses appeared in the cell supernatant at the same time as the parental clone, Cwt. This kinetics of appearance was reproducible in all three experiments.
RNA from viruses was extracted from transfection cell culture supernatants and reverse transcribed into cDNAs, and the products were used in PCRs targeting the HIV-1 PR gene as previously described (7). PR genes were then resequenced to confirm mutations. To check for possible adaptation of PR cleavage sites with a particular PR genotype (4, 5, 16), the sites located in the gag region were amplified with primers P24INF (5′-GTCCAAAATGCGAACCCAGATTGTAA-3′) and PNL43R (5′-CGCTGCCAAAGAGTGATCTGAGGGAA-3′). The gag fragment had 528 nucleotides and contained CA/p2, p2/NC, NC/p1, p1/TFP, and p1/p6 cleavage sites. PCR fragments were sequenced in both directions in an ABI 310 automated sequencer (Applied Biosystems, Foster City, Calif.), with the same primers used in the amplifications. The recovered virus from subtype B transfections did not accumulate any secondary mutation in either genomic region. In contrast, CD30N (but not Cwt, CD30N/N88D, or CL90M) accumulated a new mutation solely in the PR at position 83 (N83T), encoded by a G-to-T transversion at the first nucleotide of the 83rd PR codon. Of note, the mutation was observed when this experiment was repeated three times. To explore the reason for lack of recovery of C virus carrying solely the D30N mutation, the polymorphism L89M found in the subtype C consensus was reverted by site-directed mutagenesis. This specific position was chosen due to its nearness to positions 88 and 30. This subtype C signature was reverted in the CD30N clone, generating CD30N/M89L. This new construct was transfected, generating a productive infection in MT4 cells comparable to that for the mutated C clones. The viruses present in these two culture supernatants were isolated and sequenced, and no secondary mutations (including N83T) could be found.
A chimeric virus containing the HXB2/wt pNL43 PR was also generated as a susceptible reference, and 50% inhibitory concentrations (IC50) for all mutated viruses were determined in phenotyping assays for all Food and Drug Administration-approved PIs, amprenavir, indinavir, lopinavir, ritonavir, saquinavir, and nelfinavir. Phenotyping results shown are the means of three independent assays using the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide)-based cell viability assay as previously described (8). All statistical methods to calculate the IC50 of the clones and isolates were performed with the Analyze-it statistics package, version 1.62, for Microsoft Excel and Sigmaplot software. Briefly, a Hill's three-parameter nonlinear regression was performed to obtain the sigmoid curve of viable cells and the IC50 value of each drug for each virus tested. Table 1 shows the mean IC50 values ± standard deviations (SD) for three independent replicates. The level of resistance to NFV induced by L90M is lower than that induced by D30N regardless of the subtype. However, the isolate BD30N had a higher level of resistance to NFV than CD30N/N83T. There was a noticeable hypersusceptibility to amprenavir in clones carrying N88D regardless of their subtype. The phenomenon was also observed when ritonavir was tested in CD30N/N88D and CD30N/N83T clones, but not in the BD30N/N88D counterpart. Similar to what has been shown in previous work (7) there is a noticeable hypersusceptibility to lopinavir in all subtype C clones.
TABLE 1.
Phenotypic analysis of B and C clones and reference HXB2\NL-43 PRa
| Clone | IC50 (μM) ± SDb (fold resistancec) of:
|
|||||
|---|---|---|---|---|---|---|
| APV | SQV | IDV | RTV | NFV | LPV | |
| HXB2\NL-43 | 78.75 ± 5.31 | 3.07 ± 0.81 | 8.5 ± 1.9 | 25.25 ± 3.9 | 41.75 ± 9 | 28 ± 2.1 |
| BD30N | 88.00 ± 6.61 (1.1) | 1.89 ± 1.01 (0.6) | 6.35 ± 0.58 (0.74) | 23.5 ± 5.2 (0.94) | 1,743 ± 87.69 (41.7) | 20.96 ± 7.5 (0.75) |
| BD30N/N88D | 36.25 ± 4.42 (0.46) | 4.02 ± 1.02 (1.3) | 8.55 ± 1.1 (1) | 12.75 ± 5.1 (0.51) | 520.2 ± 34 (12.4) | 31 ± 7 (1.1) |
| BL90M | 95.5 ± 10.72 (1.21) | 13.75 ± 3.5 (4.47) | 32 ± 3.2 (3.7) | 86.25 ± 9.6 (3.45) | 178 ± 6.8 (4.28) | 27 ± 4 (0.96) |
| CD30N/N83T | 27.75 ± 3.096 (0.35) | 1.72 ± 0.51 (0.56) | 8.27 ± 0.75 (0.9) | 2.55 ± 1.2 (0.102) | 1,447.5 ± 50 (34.6) | 2.27 ± 1 (0.08) |
| CD30N/N88D | 32.375 ± 4.151 (0.41) | 2.1 ± 0.2 (0.7) | 8.9 ± 1.5 (1.05) | 2.05 ± 0.12 (0.082) | 636.5 ± 21 (15.2) | 2.3 ± 0.5 (0.08) |
| CL90M | 122.75 ± 13.961 (1.5) | 14.25 ± 2.6 (4.6) | 42.2 ± 4.1 (4.97) | 92.5 ± 3.8 (3.7) | 241 ± 82 (5.7) | 3.15 ± 0.7 (0.11) |
PI abbreviations: APV, amprenavir; SQV, saquinavir; IDV, indinavir; RTV, ritonavir; LPV, lopinavir.
SD of six independent experiments.
Resistance compared to that for HXB2\NL-43 PR. Resistance values significantly different from that for the HXB2\NL-43 PR reference clone (t test with P < 0.05) are in boldface.
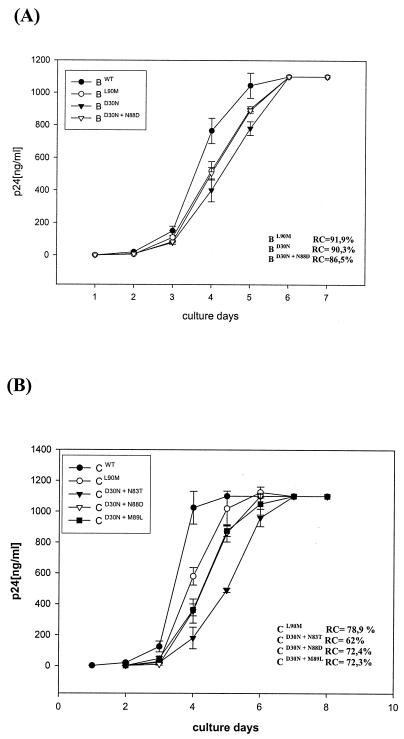
Viral replication capacities (RC) were evaluated by following the protocols previously described (6, 10). Briefly, 1,000 50% tissue culture infectious doses were used to infect 106 MT4 cells (multiplicity of infection [MOI] = 0.001). Cell culture supernatants were collected every day for nine consecutive days. The amount of virus produced in the cell supernatants was evaluated by determining p24 antigen content. For RC estimation, the time in days to reach a level of 5 × 105 pg of p24 antigen/ml in culture (replication constant K) was calculated by using Hill′s three-parameter nonlinear regression modeling. Relative RC were determined from ratios of resistant-strain K (Krs) to wild-type strain K (Kwt) and were expressed as percentages of the latter. For the evaluation of Kwt, each drug-sensitive clone from either subtype C or B was used as a reference for its respective mutated clones. The Kwt and Krs used in the determination of the relative RC of HIV isolates were the mean K values for the duplicate experiments with each virus. Figure 1 depicts the results of these experiments. We observed similar RC in subtype B clones carrying NFV resistance mutations (RC of 91.9 and 90.3% of that for Bwt for BL90M and BD30N/N88D, respectively), except for clone BD30N, which had a delayed-growth-curve profile (RC = 86.5%). Of note, all B clones were first detected in the cell supernatant at the same time (3 days postinfection). On the other hand, the RC for subtype C clones after infection corroborate the data obtained in the transfections. The clone with lowest RC, as evidenced by its delayed virus growth, was CD30N/N83T (RC of 62% of that for Cwt). The CL90M clone had an intermediate RC (78.9%), while the CD30N/M89L and CD30N/N88D clones both replicated with the same efficiency (RC = 72.3 and 72.4%, respectively). There was an evident reversion of the impairment of the D30N mutation in the subtype C PR when the M89L reversion mutation was introduced, similar to what was noticed with the CD30N/N88D clone (Fig. 1).
FIG. 1.
Virus growth kinetics after infection of MT4 cells (MOI = 0.001) with recombinant infectious virus of subtypes B (A) and C (B). The level of p24 was measured as described in the text. Results shown are means of six independent experiments, and bars represent SD.
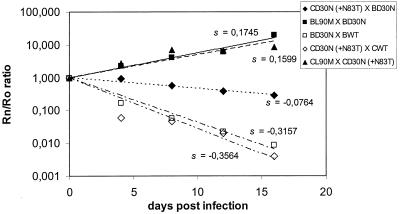
The relative viral fitness of two recombinant viruses was analyzed by following the methodology previously described by García-Lerma et al. (6). The viral lysate from two different viruses was used to infect 106 MT4 cells in exponential growth with the same MOI of 0.001 for each isolate. Every 4 days, 200 μl of viral supernatant was used to infect 106 fresh MT4 cells. Five passages were done, and aliquots were collected. The relative amount of each virus in the culture was evaluated by reverse transcriptase PCR, followed by a single dideoxynucleotide incorporation assay using the Big Dye kit (ABI, Alameda, Calif.) as previously described by García-Lerma et al. (6). Briefly, the methodology is based on the measurement of viral relative proportions based on the chromatogram peak areas of each variant, where each one is marked by extension with a dideoxynucleotide of a different fluorescent color. For each competition assay point we calculated the ratio of one virus to the other one (RN) by dividing the peak area for one virus by the peak area for the other one. The ratio at time zero (R0) was calculated at the conclusion of the mixture of the two competing viruses at the beginning of the assay (10). Relative fitness was determined by determining the adaptive coefficient s, which is the slope of the plotted curve. MT4 cells were also separately infected with each virus to assess possible spontaneous reversion of the mutated codons. The following competition experiments were conducted: clone Cwt versus CD30N, Bwt versus BD30N, CL90M versus CD30N, BL90M versus BD30N, and BD30N versus CD30N. Figure 2 shows the R0/RN curves for these experiments, where we could observe a lower fitness (relative adaptive coefficient s) of clone CD30N than of Cwt and CL90M. The same phenomenon was observed when subtype B clones were analyzed. However, the differences in coefficients for BD30N versus those for Bwt and to BL90M were lower than those for the equivalent comparisons between subtype C clones (Fig. 2).
FIG. 2.
Fitness vectors and corresponding fitness values for diverse virus competitions: CD30N versus BD30N, BL90M versus BD30N, BD30N versus BWT, CD30N versus CWT, and CL90M versus CD30N. In each competing passage, the ratio between both viruses (RN) was compared to the ratio for the initial mixtures (R0). The value in each passage was used to derive the fitness vectors by linear regression. Details of the procedure are described in the text. The slopes of fitness vectors (s) for the HIV-1 clone at the right in each pair are indicated in the plotting area.
This work has explored the uneven distribution of D30N and L90M PR mutations in B and C subtype strains and showed in vitro evidence that D30N can drastically impair subtype C virus replication. This fact is corroborated by a lower RC for the CD30N clone than for CL90M and Cwt, and all C clones except Cwt showed a lower RC when matched with equivalent subtype B clones. RC from subtype B clones were not as drastically impacted by NFV-related mutations as those from subtype C clones. In addition, only CD30N virus carrying a spontaneous mutation (N83T) incorporated upon culture could be obtained. This mutation has been previously associated with PI treatment in B isolates (4, 19). However, the role of this substitution in PI resistance is largely unknown. We speculate that this mutation was selected to restore the fitness of the CD30N clone, and this might be a favorable in vitro genetic route, as we observed independently three times. Clone BD30N, however, did not accumulate secondary mutations in the PR gene.
Our study further explored the cause of D30N′s unusual behavior in a subtype C backbone, implicating PR polymorphism L89M in this phenotype. We speculate that the methionine found in C PR (but not in B) at this position disturbs the stability of this protease, which carries the D30N substitution, as this polymorphism involves the only amino acid that differs between C and B PR in PR C terminal and that spatially neighbors residues 90, 88, and 30. The reversion of L89M to M89L in subtype C PR isolates carrying D30N clearly recovered the viral RC of this variant, similarly to what happened with the addition of N88D (see Fig. 1), without incorporation of secondary mutations such as N83T or N88D.
We have showed that the mutation D30N confers a higher level of resistance to NFV (42-fold for BD30N and 34-fold for CD30N/N83T) than L90M (4.2-fold for BL90M and 5.7-fold for CL90M) regardless of the subtype analyzed (Table 1). This contrasts with the findings reported by Perrin et al. (15), who observed only a twofold difference in NFV resistance level between virus with the D30N and L90M mutations when included separately in subtype B clones. Of note, BD30N viruses have a level of resistance to NFV comparable to that of the CD30N virus. The inclusion of the N88D mutation in BD30N and CD30N clones reduced the IC50 of NFV and induced in the clones a hypersusceptibility to amprenavir (P < 0.001; t test) compared to that for the NL43 reference strain, as previously reported for subtype B (20). However, we could find a specific hypersusceptibility to ritonavir when CD30N/N83T and CD30N/N88D clones were tested. In this case, IC50s for these two clones were shown to be approximately 10 times lower than that for the reference strain (P < 0.003; t test). Although reaching higher NFV resistance levels in the phenotyping assay, the viruses of both subtypes carrying D30N substitutions were less fit than their counterparts carrying the NFV resistance mutation L90M. The virus CD30N/N83T was slightly less fit than its correlate BD30N, reflecting the importance of the emerging N83T substitution for the rescue of fitness of the CD30N clone. The fitness data, taken together with the RC results, explain the preferential route of NFV resistance in subtype C clinical isolates through L90M and not D30N after NFV failure (3). This pathway is less costly to viral fitness and does not require any compensatory mutation in that subtype. This might be a specific phenotype from subtypes C and CRF01_AE and should not be generalized to all non-B strains. Although the results shown here explain the low prevalence of D30N mutations in subtype C strains, we have to bear in mind that the subtype C isolates studied here are really hybrid viruses because they contain a subtype B backbone, and this can be a limitation to the data presented here. This knowledge, however, may influence future therapeutic strategies and interpretation of genotypic resistance in areas where these variants prevail.
Acknowledgments
We acknowledge the NIH AIDS Research and Reference Reagent Program for providing the antiretroviral compounds used in this work. We are also indebted to Charles Boucher for providing us with the pHXB-ΔPro plasmid.
This work was supported by the AIDS/STD National Program, Brazilian Ministry of Health, the State Science Foundation of Rio de Janeiro (grant E-26/151.970/00), and the Brazilian Council for Scientific and Technologic Development (grant 462394/00-0).
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin.1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ariyoshi, K., M. Matsuda, H. Miura, S. Tateishi, K. Yamada, and W. Sugiura. 2003. Patterns of point mutations associated with antiretroviral drug treatment failure in CRF01_AE (subtype E) infection differ from subtype B infection. J. Acquir. Immune Defic. Syndr. 33:336-342. [DOI] [PubMed] [Google Scholar]
- 3.Cane, P. A., A. de Ruiter, P. Rice, M. Wiselka, R. Fox, and D. Pillay. 2001. Resistance-associated mutations in the human immunodeficiency virus type 1 subtype c protease gene from treated and untreated patients in the United Kingdom. J. Clin. Microbiol. 39:2652-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cote, H. C., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallego, O., C. de Mendoza, A. Corral, and V. Soriano. 2003. Changes in the human immunodeficiency virus p7-p1-p6 gag gene in drug-naive and pretreated patients. J. Clin. Microbiol. 41:1245-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García-Lerma, G., S. Nidtha, K. Blumoff, H. Weinstock, and W. Heneine. 2001. Increased ability for selection of zidovudine resistance in a distinct class of wild-type HIV-1 from drug-naïve persons. Proc. Natl. Acad. Sci. USA 98:13907-13912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez, L. M. F., R. M. Brindeiro, M. Tarin, A. Calazans, M. A. Soares, S. Cassol, et al. 2003. The subtype C of human immunodeficiency virus type 1 protease presents an in vitro hypersusceptibility to protease inhibitor lopinavir. Antimicrob. Agents Chemother. 47:2117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hertogs, K., M.-P. De Bethune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, et al. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 10.Holland, J. J., J. C. de la Torre, D. K. Clarke, and E. Duarte. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maschera, B., E. Furfine, and E. D. Blair. 1995. Analysis of resistance to human immunodeficiency virus type 1 protease inhibitors by using matched bacterial expression and proviral infection vectors. J. Virol. 69:5431-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicastri, E., L. Sarmati, G. d′Ettorre, L. Palmisano, S. G. Parisi, I. Uccella, A. Rianda, E. Concia, V. Vullo, S. Vella, and M. Andreoni. 2003. Replication capacity, biological phenotype, and drug resistance of HIV strains isolated from patients failing antiretroviral therapy. J. Med. Virol. 69:1-6. [DOI] [PubMed] [Google Scholar]
- 13.Palella, F. J., Jr., K. M. Delaney, and A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 14.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrin, V., and F. Mammano. 2003. Parameters driving the selection of nelfinavir-resistant human immunodeficiency virus type 1 variants. J. Virol. 77:10172-10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters, S., M. Muñoz, S. Yerly, V. Sanchez-Merino, C. Lopez-Galindez, L. Perrin, B. Larder, D. Cmarko, S. Falkan, P. Meylan, and A. Telenti. 2001. Resistance to nucleoside analog reverse transcriptase inhibitors mediated by human immunodeficiency virus type 1 p6 protein. J. Virol. 75:9644-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puchhammer-Stockl, E., C. Kunz, E. Faatz, P. Kasper, F. X. Heinz, et al. 1999. Introduction of HIV-1 subtypes C, E and A into Austria. Clin. Diagn. Virol. 9:25-28. [DOI] [PubMed] [Google Scholar]
- 18.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 2000. Mutations in retroviral genes associated with drug resistance: 2000-2001 update. Int. Antivir. News 5:65-91. [Google Scholar]
- 19.Wu, T. D., C. A. Schiffer, M. J. Gonzales, J. Taylor, R. Kantor, S. Chou, D. Israelski, A. R. Zolopa, W. J. Fessel, and R. W. Shafer. 2003. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 77:4836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziermann, R., K. Limoli, K. Das, E. Arnold, C. J. Petropoulos, and N. T. Parkin. 2000. A mutation in human immunodeficiency virus type 1 protease, N88S, that causes in vitro hypersensitivity to amprenavir. J. Virol. 74:4414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]