Abstract
A rapid, continuous method for noninvasively monitoring the effectiveness of several antibacterial agents in real time by using a model of wound infection was developed. This study was divided into three steps: (i) construction of a plasmid to transform Escherichia coli into a bioluminescent variant, (ii) study of the bioluminescent E. coli in vitro as a function of temperature and pH, and (iii) determination of the MIC and the minimal bactericidal concentration of sulfamethoxazole-trimethoprim (SMX-TMP). Finally, the efficacy of SMX-TMP was monitored in vivo in a cutaneous wound model (hairless rat) infected with this bioluminescent bacterium by using a bioluminescence imaging system. E. coli was transformed by electroporation with a shuttle vector (pRB474) containing the firefly (Photinus pyralis) luciferase gene, resulting in a bioluminescent phenotype. It was found that pH 5.0 was optimal for incorporation of the susbstrate d-luciferin for the luciferase reaction. In vitro, when the agar dilution method, standard turbidity assays, and the bioluminescence imaging system were used, E. coli(pRB474) proved to be susceptible to SMX-TMP. In vivo, at 4 h, SMX-TMP treatment was already efficient compared to no treatment (P = 0.034). At 48 h, no bioluminescence was detected in the wound, demonstrating the susceptibility of E. coli to SMX-TMP. In conclusion, this study points out the advantage of using bioluminescence imaging to evaluate the effects of antibiotics for the treatment of acute infections in vivo in a nondestructive and noninvasive manner.
The rates of multiantibiotic resistance among bacteria that infect wounds and burns are constantly on the rise (4, 12). Consequently, rapid control of wound infections and monitoring of therapeutic strategies by optical techniques have recently been proposed. The method of optically monitoring bacterial numbers and viability in real time in living animals by use of genetically engineered bacteria that emit luminescence, together with ultrasensitive photon-counting cameras, has been demonstrated with several models (7, 11, 16, 17, 20, 21, 33).
By this technique, quantification of the luminescence images can determine in real time the extent of infection in living animals and can thereby provide both temporal and spatial information about the labeled bacteria and their metabolic activities. Similarly, antibiotic effects can be detected directly, nondestructively, and noninvasively.
The study described here aimed to evaluate the effects of an antibiotic in situ by using bioluminescent bacteria. The study was divided into three steps. The first consisted of constructing a plasmid to transform Escherichia coli into a bioluminescent variant. The second step consisted of studying bioluminescent E. coli in vitro as a function of temperature and pH. The third step consisted of determining the MIC and the minimal bactericidal concentration (MBC) of an antibiotic, sulfamethoxazole-trimethoprim (SMX-TMP), for this bioluminescent strain. Finally, the efficacy of SMX-TMP was monitored in vivo in a model of cutaneous wound infection with this bioluminescent bacterium.
MATERIALS AND METHODS
Bacterial strain.
A relatively nonpathogenic strain of E. coli which lacks the virulence factors necessary to cause invasive infection was used (39). E. coli TOP10F′, a facultative anaerobe (reference strain, C615-00) in the normal intestinal flora of humans and animals (5), was obtained from Invitrogen SARL (Cergy Pontoise, France) and was routinely grown at 37°C. Furthermore, TOP10F′ is a recombination-negative strain designed for the stable replication of high-copy-number plasmids (18).
Plasmid construction.
A bioluminescent strain of E. coli was constructed by transformation with plasmid pRB474. Plasmid pRB474 is an expression vector that contains the firefly (Photinus pyralis) luciferase gene. This enzyme uses d-luciferin, O2, and ATP as substrates and produces AMP, oxyluciferin, inorganic pyrophosphate (PPi), water, and light (547 to 617 nm) (3, 9):
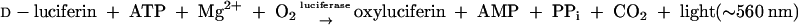 |
The luc gene of vector pSPluc (Table 1) was amplified by PCR. Reactions were carried out in 50-μl volumes containing 5 μl of 10× PCR buffer (supplied with Taq DNA polymerase), 50 pmol of each oligonucleotide primer, each deoxynucleotide triphosphate (dATP, dCTP, dGTP, and dTTP) at a concentration of 0.2 mM, 1 U of Taq DNA polymerase, and 10 ng of plasmid DNA containing firefly luciferase (pSPluc).
TABLE 1.
Bacterial strain, plasmids, and primers used in the study
| Strain, plasmid, or primer | Genotype or sequence | Source or reference |
|---|---|---|
| E. coli TOP10F′ | Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen (NO-C615-00) |
| Plasmids | ||
| pSP-luc | Ampr Ori colE1 | Promega (E1781) |
| pRB474 | Ampr Cmr Ori− Ori+ | 6 |
| Primers | ||
| LucF (5′-3′) | AGG AAG GAT CCA GAG CAG ATT GTA CTG AGA Ga | This study |
| LucR (5′-3′) | AAT GCA GGT TAA CCT GGC TTA TCG AAA TT |
Underlined nucleotides show the position of the BamHI restriction site.
Amplification of each gene was achieved with 35 cycles at 94°C for 1 min and 50°C for 1 min, followed by a final extension step at 72°C for 1 min. The PCR was carried out in a Geneamp PCR system 9700 automated thermocycler (Perkin-Elmer Applied Biosystems, Foster City, Calif.). PCR-amplified luc DNA was purified prior to the cloning procedures by passing the complete reaction volumes through spin columns (PCR purification kit; Qiagen SA, Courtaboeuf, France).
The PCR-amplified luc gene and shuttle vector pRB474 were digested with BamHI and EcoRI after 2 h at 37°C. Then, the complementary ends of the restriction enzyme-digested PCR-amplified luc gene and pRB474 vector DNA were ligated by using the enzyme DNA ligase.
Transformation of E. coli to a bioluminescent phenotype.
E. coli cells were transformed with shuttle vector pRB474 by electroporation, as follows. The cells were cultivated in Luria broth to an optical density at 600 nm (OD600) of 0.6, centrifuged at 4,000 × g at 4°C for 15 min, washed twice with ice-cold buffer (10 mM HEPES buffer [pH 7.0] with 15% glycerol), and suspended in 5% sucrose containing 15% glycerol. The electroporation was performed in Equi-Bio (Eurogentec) electroporation cuvettes with a 0.2-cm distance between electrodes with 2 μl of plasmid (10 ng) and 50 μl of ice-cold electrocompetent cells. A single electric pulse of 5 ms with settings of 2.5 kV, 25 μF, and 200 Ω was given; and the cells were then rapidly removed from the electroporation apparatus (Bio-Rad) and suspended in 1,000 μl of SOC (20 g of Bacto Tryptone, 5 g of Bacto Yeast Extract, 0.5 g of NaCl, glucose to a concentration of 20 mM, and an MgCl2-MgSO4 mixture at a concentration of 10 mM each). After 1 h the cells were plated onto agar plates containing 50 μg of ampicillin per ml and were grown for 1 day at 35°C. Bioluminescent colonies were selected by using a luminometer (Lumat LB 9501; Berthold France S.A, Thoiry, France).
Bioluminescence imaging system.
The light emission of the bioluminescent bacteria was detected both in vitro and in vivo with a bioluminescence imaging system. This system included an ultra-high-sensitivity video camera (model C2400-25; Hamamatsu Photonics KK, Massy, France) developed for imaging under extremely low light levels. It was fitted with a stereoscopic zoom microscope (model SMZ; Nikon, Tokyo, Japan) and mounted in a specimen chamber. The camera used a two-stage multichannel plate image intensifier capable of detecting under low light levels images down to photon levels. In the photon-counting mode, an image of the emitted light was captured by using an integration time of 5 min at a 90% setting on the image-intensifier control module. The bioluminescence intensity of the same analysis area was calculated in vitro (surface area, 12 mm2, corresponding to 55,326 pixels) and in vivo (surface area, 18 mm2, corresponding to 85,098 pixels) with Argus software (Hamamatsu Photonics KK). This analysis area was kept constant for all wounds at all time points. For each measurement, the background signal was subtracted from the bioluminescent signal.
Influence of pH and temperature on E. coli bioluminescence. (i) pH.
The effect of pH was studied by adding a constant amount (500 μM) of d-luciferin to 0.1 M phosphate-citrate buffer adjusted to different pHs, ranging from 4 to 8, in 96-well plates in vitro. The bioluminescence reaction was started by adding 100 μl of substrate buffer to a 100-μl sample of growth-phase E. coli cells, and light emission was measured immediately with the imaging system.
(ii) Temperature.
Substrate buffer (50 μl; pH 5) was added in 100 μl of Luria broth containing the bioluminescent bacteria in the growth phase (OD600, 0.6) in 96-well plates in vitro. These plates were incubated for 6 h at two different temperatures (20 and 30°C). The bioluminescence emitted by these bacteria was measured every hour.
Animals.
All animal experiments conformed to the Ministère de l'Agriculture et de la Forêt Resolution on the use of animals in research and were approved by the Subcommittee on Research Animal Care of the Lille Medical University (protocol 2003-35).
Male hairless rats (Charles River France, Les Oncins, France) weighing from 200 to 300 g were used for this study. A full-thickness, circular, 20-mm-diameter wound was created with surgical scissors on the backs of the rats while they were under general anesthesia (140 mg of ketamine per kg of body weight and 3 mg of chlorpromazine per kg). The position of this wound was 4 cm caudal to the ears, and the wound was placed on the midline. A Teflon chamber with a 2.2-cm diameter, similar to the chamber developed by Balazs et al. (2), was applied around the wound, glued into the edge of the skin with Histoacryl adhesive (B. Braun Surgical GmbH, Melsungen, Germany), and sutured (Ethicon 4-0 sutures). A sterile glass window (GF-C; Whatman) was placed inside the chamber to protect the wound against other infections.
Antibiotics.
SMX-TMP is a synthetic antibacterial combination product. TMP is 2,4-diamino-5-(3,4,5-trimethoxybenzyl) pyrimidine (molecular weight, 290.32). SMX is N1-(5-methyl-3-isoxazolyl)-sulfanilamide (molecular weight, 253.28). This drug combination has proved to be an effective therapeutic agent with broad-spectrum antibacterial activity against both gram-positive and gram-negative organisms. It is recognized as having excellent activity against staphylococci in the skin (13, 35, 40). This makes SMX-TMP a good choice for the treatment of skin infections or as a general antibiotic when the actual identity of the infecting organism is not known (10). SMX-TMP (Bactrim; Produits Roche, Neuilly sur Seine, France) was injected into the animals intravenously (dosage, 30 mg/kg).
MIC and MBC determinations.
The MIC of SMX-TMP for E. coli(pRB474) was determined by using (i) an automated system (model Vitek 2; bioMerieux Vitek, Marcy l'Etoile, France) and (ii) the agar dilution method by the procedure of the National Committee for Clinical Laboratory Standards (29). MICs and MBCs were also determined in vitro with the imaging system.
For MIC and MBC evaluations with the bioluminescence imaging system, 100 μl of Luria broth (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl per liter [pH 7.0]) containing E. coli(pRB474) in exponential growth phase (OD600, 0.6) was centrifuged, washed with phosphate-buffered saline, and resuspended in fresh Luria broth at the same density. The suspension was added to a series of SMX-TMP solutions with increasing concentrations (0.3, 0.6, 0.9, and 1.2 mg/liter) in 96-well plates and the plates were incubated for 4 h at 35°C. For each concentration, the bioluminescence was quantified with the imaging software.
Determination of SMX-TMP efficacy in vivo.
Wound infections were created in six animals. A suspension (50 μl of phosphate-buffered saline) containing 5 × 107 mid-log-phase bioluminescent E. coli cells (108 cells/ml) was inoculated into each wound. Thirty minutes later (the duration required for the bacteria to attach to the tissue), 50 μl of substrate solution (1 mM of d-luciferin in 0.1 M phosphate-citrate buffer [1]) was added to the wound.
Bacterial loading was controlled with the imaging system to confirm that it was equivalent in each wound. Three rats were treated with two intravenous injections of SMX-TMP (dosage, 30 mg/kg) at 12-h intervals (SMX-TMP-treated group). Three rats remained untreated (control [CTRL] group). Each wound was imaged at 0, 4, 8, 24, 36, and 48 h postinfection with the bioluminescent imaging system. Before each measurement, 50 μl of substrate solution (1 mM of d-luciferin in 0.1 M phosphate-citrate buffer) was added exogenously to the wound. The difference between the CTRL group and the SMX-TMP group was evaluated by the Student t test.
RESULTS
Control of E. coli bioluminescence.
The bioluminescence signal of E. coli measured with the luminometer was linearly proportional to the numbers of bacterial CFU (as determined by serial dilution and plating) from 103 to 107 organisms (data not shown).
Influence of pH and temperature on E. coli bioluminescence.
As expected, pH and temperature were shown to have significant effects on bioluminescent E. coli(pRB474). The slightly acid pH modifies the luminescence of E. coli(pRB474) in exponential growth phase. The light emission of these cells was maximum at pH 5.0.
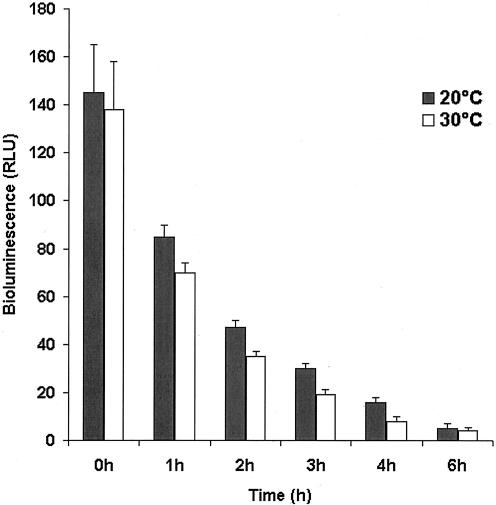
The bioluminescence of E. coli(pRB474) bacteria in growth phase is temperature dependent (Fig. 1). The results show that E. coli(pRB474) bacteria stored in Luria broth remain bioluminescent for at least 6 h after addition of d-luciferin. The light emission of bacteria incubated at 30°C (which corresponds to the rat's skin temperature) was only slightly reduced (5 to 10%) compared to that observed at 20°C (room temperature).
FIG. 1.
Emission of light by E. coli cells as a function of storage time in Luria broth at two different temperatures (20 and 30°C). RLU, relative light units.
Study of SMX-TMP efficacy with the imaging system in vitro and in vivo. (i) Efficacy in vitro.
The antibiotic susceptibilities of E. coli(pRB474) were determined by standard turbidity assays. The result showed that any concentration equal to or slightly less than 20 μg of SMX-TMP per ml inhibited the growth of 5 × 105 CFU of E. coli(pRB474) per ml (MIC ≤ 20 μg/ml). The agar dilution method gave an MIC of 0.08 mg/liter.
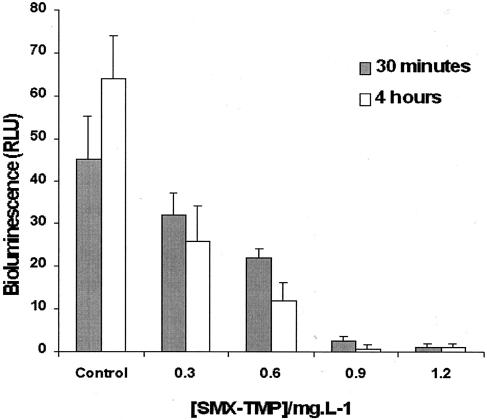
Increasing concentrations of SMX-TMP (0.3, 0.6, 0.9, and 1.2 mg/liter) in 100 μl of Luria broth containing 105 mid-log-phase bioluminescent E. coli cells were applied to determine the MICs and the MBCs with the imaging system. The viabilities of these bioluminescent cells were detected with the imaging system after 30 min and 4 h of incubation at 35°C (Fig. 2).
FIG. 2.
Effects of increasing concentrations of SMX-TMP on the light emission (relative light units) of E. coli detected with the bioluminescence imaging system. Each concentration of SMX-TMP (0.3, 0.6, 0.9, and 1.2 mg/liter) was incubated with the bioluminescent E. coli for 30 min (▪) and 4 h (□) at 35°C in Luria broth. The sample labeled “Control” is the untreated strain.
The results showed that the bioluminescence of E. coli(pRB474) remained almost stable in SMX-TMP at a concentration 300 mg/liter for 4 h of incubation at 35°C and that the bioluminescence corresponded to the MIC of SMX-TMP for 107 CFU of E. coli(pRB474) per ml. By using these data, it was also possible to determine that the MBC of SMX-TMP for bioluminescent E. coli(pRB474) corresponded to 900 mg/liter.
(ii) Efficacy in vivo of SMX-TMP in cutaneous wounds induced in live hairless rats infected with bioluminescent E. coli.
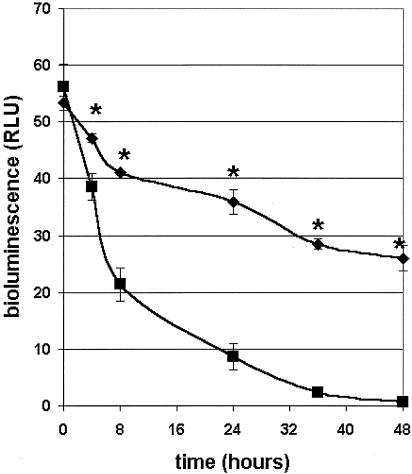
Two doses (15 mg/kg each) of SMX-TMP were injected into infected rats at a 12-h interval, giving a total dose of 30 mg/kg. The viabilities of the bioluminescent bacteria were detected with the imaging system immediately after infection and at 4, 8, 24, 36, and 48 h postinfection (Fig. 3 and 4).
FIG. 3.
In vivo monitoring of E. coli(pRB474) bioluminescence in the cutaneous wound by use of the bioluminescence imaging system. Each set of data represents the mean (±standard deviation |) number of relative light units (RLU) for three hairless rats either untreated (♦) or treated with SMX-TMP (▪; 30 mg/kg) and imaged for 5 min at 0, 4, 8, 24, 36, and 48 h postinfection. *, statistically different by the Student t test.
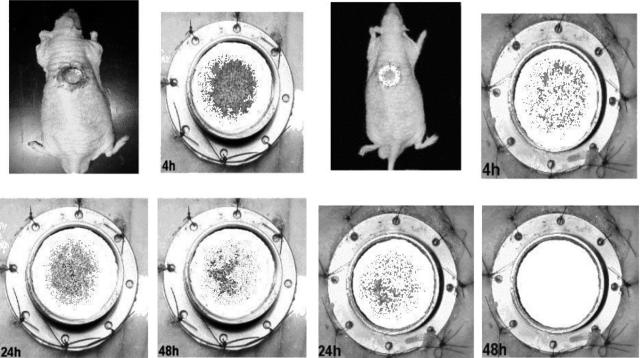
FIG. 4.
Monitoring of the effects of SMX-TMP on bioluminescent E. coli(pRB474) in a hairless rat. Bioluminescent bacteria were injected into the wound (5 × 107 CFU). Shown is one treated (SMX-TMP) E. coli(pRB474)-infected rat (right set of panels) and one untreated E. coli(pRB474)-infected rat (left set of panels) imaged dorsally for 5 min at 4, 24, and 48 h postinfection.
At time zero the values obtained for the untreated group (CTRL group) and the treated group (SMX-TMP group) were not significantly different (P = 0.278). At 4 h, SMX-TMP treatment was already efficient, since the values between the two groups were significantly different (P = 0.034). This observation was confirmed at 8 h (P = 0.0039), 24 h (P = 0.082), 36 h (P = 0.0011), and finally, 48 h (P = 0.0026). Bioluminescent bacteria were not detected at 48 h. Conversely, bioluminescent bacteria were still present in the untreated CTRL group, even though a 50% reduction in bioluminescence was observed at 48 h.
DISCUSSION
The firefly (P. pyralis) luciferase gene used in this study is a single polypeptide. Light production is started by the addition of the substrate, d-luciferin. Therefore, the metabolic stress caused by light production (and ATP consumption) takes place only at the luminescence measurement stage. Recently, other bioluminescent reporter systems, such as the bacterial luciferase operon luxCDABE, have also been used to provide a means of detecting bacterial viability (7, 8, 11, 33). The bacteria containing the luxCDABE operon continuously produce light and are under constant metabolic stress. In addition, cells must produce five polypeptides. These reasons may explain the difference in the light production power of the firefly luciferase operon and the bacterial luciferase operon (15).
In this study, rather than using pSPluc, a shuttle vector, pRB474, was constructed for use in E. coli. This choice was dictated by the stability of the pRB474 shuttle vector, which was tested over a period of 60 generations in E. coli and Bacillus subtilis. Furthermore, in the pRB474 shuttle vector, a vegetative B. subtilis promoter drives the transcription of genes that are inserted (6). Moreover, the pSPluc vector is a luciferase cassette vector containing the engineered firefly luciferase gene, which has been optimized for genetic reporter applications (34). The pSPluc vector is not itself intended for the expression of luciferase in gram-positive bacteria because it contains only one ColE1 replication origin for E. coli and none for gram-positive bacteria.
ATP is one of the substrates of the firefly luciferase reaction, and each cell contains a constant intracellular ATP pool which is effectively regulated. The light emission in vivo is therefore a very sensitive indicator of the intracellular state of the cells (37). Others (42) have shown earlier that the substrate for the luciferase reaction does not readily pass procaryotic or eucaryotic cell membranes at physiological pH but is efficiently incorporated under slightly acidic conditions in sodium citrate buffer, consistent with the results of Wood and DeLuca (44). The optimum pH for B. subtilis was shown to be 5.0 (23, 24). In our study, when we evaluated the penetration of d-luciferin into E. coli cells, pH 5.0 was found to be the optimum.
As an alternative approach, Luque-Ortega et al. (25) have optimized an easy luminescent method involving a technique that bypasses the poor permeation of d-luciferin at neutral pH by using d-luciferin (DMNPE [1-4,5-dimethoxy-2-mitrophenyl)ethyl ester]-luciferin) for Leishmania donovani promastigotes transfected with a firefly luciferase gene.
Cells kept in Luria broth at 30°C gave levels of light emission slightly lower than those of cells stored at room temperature (20°C). Analytically, this situation was probably due to a decrease in the cellular ATP content rather than the proteolytic degradation of the luciferase enzyme or cell death (14). In addition, these bioluminescent bacteria incubated at 30°C grow old and more quickly enter stationary growth phase than those preserved at room temperature (26, 41, 43).
Monitoring of the effects of antibiotics such as SMX-TMP in vitro and in animal model test systems could enhance our basic understanding of their actions and facilitate unique studies of disease in vivo.
The study of antimicrobial activity by use of the bioluminescence imaging system was performed both in vitro and in vivo with SMX-TMP and demonstrated that decreases in viable cell numbers are associated with similar changes in bioluminescence. Since one of the substrates is ATP, an essential energy source of each living cell, any disturbance in the intracellular ATP levels is directly reflected in the light emission levels measured from the reporter cells. Therefore, minor events that occur before the bacteria are killed are detected as decreases in light emission. The combination of SMX and TMP inhibits E. coli(pRB474) folic acid synthesis. This inhibition is called the “sequential blockade” and produces death of the bacterium (13, 35).
The d-luciferin substrate for P. pyralis luciferase and TMP-SMX are amphipathic molecules (i.e., hydrophilic and hydrophobic areas are present within the same molecule) (19, 32). The d-luciferin substrate and SMX-TMP antibiotic must permeate the outer membrane on the way into the periplasmic space and the cell, which means that that the outer membrane has special sieving properties. The active components involved in the molecular sieving of the outer membrane are porins (27, 28). SMX-TMP treatment leads to a reduction in ATP levels in E. coli. Consequently, a decrease in ATP levels in the cell leads to a reduction in the levels of light emission and inhibits the reaction of d-luciferin luciferase.
In in vitro studies, E. coli was proved to be susceptible to SMX-TMP, as determined by three methods: agar dilution, standard turbidity assays, and bioluminescence imaging. The MICs of TMP-SMX for E. coli(pRB474) were shown to be close when they were determined by either the agar dilution method or bioluminescence imaging (0.08 and 0.3 mg/liter, respectively), confirming that the bioluminescence imaging technique is a powerful tool.
The evaluation of antibiotic activity in vivo by use of the bioluminescence imaging system has clearly shown that SMX-TMP is efficient for the treatment of cutaneous wound infections caused by E. coli(pRB474). The activity of SMX-TMP was already detected at 4 h compared to the findings for the CTRL group. Complete clearance of the infection was obtained at 48 h. Antibiotic efficacy is usually affected by environmental conditions, such as the local production of enzymes, purulent and fibrinous exudates, and pH changes, which are known to affect the actions of drugs (36). Consequently, this observation confirms that TMP administered with SMX has a high degree of efficacy (30, 31). This study shows that fewer animals are required to reliably detect the action of an antimicrobial therapy, in accordance with previous studies (22, 33, 38).
This study demonstrates that bioluminescent E. coli could serve as a biosensor of antibacterial activity for both in vitro and in vivo studies. The use of bioluminescent imaging strategies to reveal the real-time effects of potential therapeutic agents on gram-negative bacterial infections in animal models of cutaneous wounds would greatly accelerate the analysis of compounds under development. The spatial and temporal differences in the bioluminescent signals between untreated and treated animals could be used to assess the effects of compounds on specific biological processes in vivo.
Acknowledgments
We thank Guy Dhelin for excellent technical assistance and Jean Claude D'Halluin (INSERM U524, Institut de Recherches sur le Cancer 2) and René Courcol (Laboratoire de Bacteriologie-Hygiene, Hôpital A. Calmette, CHRU de Lille) for free access to their laboratories and helpful suggestions. We are grateful to Reinhold Brückner (Mikrobielle Genetik, Universität Tubingen, Tubingen, Germany) for having kindly provided plasmid pRB474.
REFERENCES
- 1.Albrich, W. C., M. Angstwurm, L. Bader, and R. Gartner. 1999. Drug resistance in intensive care units. Infection 27(Suppl. 2):S19-S23. [DOI] [PubMed] [Google Scholar]
- 2.Balazs, L., J. Okolicany, M. Ferrebee, B. Tolley, and G. Tigyi. 2001. Topical application of the phospholipid growth factor lysophosphatidic acid promotes wound healing in vivo. Am. J. Physiol. Reg. Integr. Comp. Physiol. 280:R466-R472. [DOI] [PubMed] [Google Scholar]
- 3.Biggley, W. H., J. E. Lloyd, and H. H. Seliger. 1967. The spectral distribution of firefly light. II. J. Gen Physiol. 50:1681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blondeau, J. M., D. Vaughan, et al. 2000. In vitro activity of 19 antimicrobial agents against 3513 nosocomial pathogens collected from 48 Canadian medical centres. Int. J. Antimicrob. Agents 15:213-219. [DOI] [PubMed] [Google Scholar]
- 5.Bonten, M., E. Stobberingh, J. Philips, and A. Houben. 1990. High prevalence of antibiotic resistant Escherichia coli in faecal samples of students in the south-east of The Netherlands. J. Antimicrob. Chemother. 26:585-592. [DOI] [PubMed] [Google Scholar]
- 6.Bruckner, R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187-192. [DOI] [PubMed] [Google Scholar]
- 7.Contag, C. H., P. R. Contag, J. I. Mullins, S. D. Spilman, D. K. Stevenson, and D. A. Benaron. 1995. Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol. 18:593-603. [DOI] [PubMed] [Google Scholar]
- 8.Contag, C. H., S. D. Spilman, P. R. Contag, M. Oshiro, B. Eames, P. Dennery, D. K. Stevenson, and D. A. Benaron. 1997. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem. Photobiol. 66:523-531. [DOI] [PubMed] [Google Scholar]
- 9.de Wet, J. R., K. V. Wood, D. R. Helinski, and M. DeLuca. 1985. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc. Natl. Acad. Sci. USA 82:7870-7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein, M. E., M. Amodio-Groton, and N. S. Sadick. 1997. Antimicrobial agents for the dermatologist. II. Macrolides, fluoroquinolones, rifamycins, tetracyclines, trimethoprim-sulfamethoxazole, and clindamycin. J. Am. Acad. Dermatol. 37:365-381. [DOI] [PubMed] [Google Scholar]
- 11.Francis, K. P., D. Joh, C. Bellinger-Kawahara, M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 68:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fung, H. B., J. Y. Chang, and S. Kuczynski. 2003. A practical guide to the treatment of complicated skin and soft tissue infections. Drugs 63:1459-1480. [DOI] [PubMed] [Google Scholar]
- 13.Gleckman, R., N. Blagg, and D. W. Joubert. 1981. Trimethoprim: mechanisms of action, antimicrobial activity, bacterial resistance, pharmacokinetics, adverse reactions, and therapeutic indications. Pharmacotherapy 1:14-20. [DOI] [PubMed] [Google Scholar]
- 14.Greger, J. E., and A. D. Eisenberg. 1985. Adenosine 5′-triphosphate content of Streptococcus mutans GS-5 during starvation in a buffered salt medium. Caries Res. 19:314-319. [DOI] [PubMed] [Google Scholar]
- 15.Hakkila, K., M. Maksimow, M. Karp, and M. Virta. 2002. Reporter genes lucFF, luxCDABE, gfp, and dsred have different characteristics in whole-cell bacterial sensors. Anal. Biochem. 301:235-242. [DOI] [PubMed] [Google Scholar]
- 16.Hamblin, M. R., D. A. O'Donnell, N. Murthy, C. H. Contag, and T. Hasan. 2002. Rapid control of wound infections by targeted photodynamic therapy monitored by in vivo bioluminescence imaging. Photochem. Photobiol. 75:51-57. [DOI] [PubMed] [Google Scholar]
- 17.Hamblin, M. R., T. Zahra, C. H. Contag, A. T. McManus, and T. Hasan. 2003. Optical monitoring and treatment of potentially lethal wound infections in vivo. J. Infect. Dis. 187:1717-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Hollis, R. P., K. Killham, and L. A. Glover. 2000. Design and application of a biosensor for monitoring toxicity of compounds to eukaryotes. Appl. Environ. Microbiol. 66:1676-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadurugamuwa, J. L., L. Sin, E. Albert, J. Yu, K. Francis, M. DeBoer, M. Rubin, C. Bellinger-Kawahara, T. R. Parr, Jr., and P. R. Contag. 2003. Direct continuous method for monitoring biofilm infection in a mouse model. Infect. Immun. 71:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadurugamuwa, J. L., L. V. Sin, J. Yu, K. P. Francis, R. Kimura, T. Purchio, and P. R. Contag. 2003. Rapid direct method for monitoring antibiotics in a mouse model of bacterial biofilm infection. Antimicrob. Agents Chemother. 47:3130-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuklin, N. A., G. D. Pancari, T. W. Tobery, L. Cope, J. Jackson, C. Gill, K. Overbye, K. P. Francis, J. Yu, D. Montgomery, A. S. Anderson, W. McClements, and K. U. Jansen. 2003. Real-time monitoring of bacterial infection in vivo: development of bioluminescent staphylococcal foreign-body and deep-thigh-wound mouse infection models. Antimicrob. Agents Chemother. 47:2740-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lampinen, J. 1995. Development of recombinant bacterial strains for cytotoxicity testing. Ann. Univ. Turkuensis 232:498-504. [Google Scholar]
- 24.Lampinen, J., M. Virta, and M. Karp. 1995. Comparaison of gram positive and gram negative bacterial strains cloned with different types of luciferase genes in bioluminescence cytotoxity tests. Environ. Toxicol. Water Qual. 10:157-166. [Google Scholar]
- 25.Luque-Ortega, J. R., O. M. Rivero-Lezcano, S. L. Croft, and L. Rivas. 2001. In vivo monitoring of intracellular ATP levels in Leishmania donovani promastigotes as a rapid method to screen drugs targeting bioenergetic metabolism. Antimicrob. Agents Chemother. 45:1121-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marincs, F. 2000. On-line monitoring of growth of Escherichia coli in batch cultures by bioluminescence. Appl. Microbiol. Biotechnol. 53:536-541. [DOI] [PubMed] [Google Scholar]
- 27.Nakae, T. 1976. Identification of the outer membrane protein of E. coli that produces transmembrane channels in reconstituted vesicle membranes. Biochem. Biophys. Res. Commun. 71:877-884. [DOI] [PubMed] [Google Scholar]
- 28.Nakae, T. 1976. Outer membrane of Salmonella. Isolation of protein complex that produces transmembrane channels. J. Biol. Chem. 251:2176-2178. [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 30.Nowak, A., M. Kadykow, and A. Klimowicz. 1983. Penetration of trimethoprim and sulfamethoxazole into skin blister fluid. Eur. J. Clin. Pharmacol. 25:825-827. [DOI] [PubMed] [Google Scholar]
- 31.Pien, F. D., S. Shrum, J. M. Swenson, B. C. Hill, C. Thornsberry, and J. J. Farmer III. 1985. Colonization of human wounds by Escherichia vulneris and Escherichia hermannii. J. Clin. Microbiol. 22:283-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitts, C., J. Yin, D. Bowern, C. Maxwell, and W. Southerland. 2002. Interaction energy analysis of nonclassical antifolates with Pnemocystis carinii dihydrofolate reductase. Int. J. Mol. Sci. 3:1188-1202. [Google Scholar]
- 33.Rocchetta, H. L., C. J. Boylan, J. W. Foley, P. W. Iversen, D. L. LeTourneau, C. L. McMillian, P. R. Contag, D. E. Jenkins, and T. R. Parr, Jr. 2001. Validation of a noninvasive, real-time imaging technology using bioluminescent Escherichia coli in the neutropenic mouse thigh model of infection. Antimicrob. Agents Chemother. 45:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherf, B. 1994. Firefly luciferase engineered for improved genetic reporting. Promega Notes 14. Promega Corp., Madison, Wis.
- 35.Smilack, J. D. 1999. Trimethoprim-sulfamethoxazole. Mayo Clin. Proc. 74:730-734. [DOI] [PubMed] [Google Scholar]
- 36.Spurlock, S. L., and E. A. Hanie. 1989. Antibiotics in the treatment of wounds. Vet. Clin. N. Am. Equine Pract. 5:465-482. [DOI] [PubMed] [Google Scholar]
- 37.Stanley, P. E. 1989. A review of bioluminescent ATP techniques in rapid microbiology. J. Biolumin. Chemilumin. 4:375-380. [DOI] [PubMed] [Google Scholar]
- 38.Tenhami, M., K. Hakkila, and M. Karp. 2001. Measurement of effects of antibiotics in bioluminescent Staphylococcus aureus RN4220. Antimicrob. Agents Chemother. 45:3456-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theilman, N. M., and R. L. Guerrant. 1999. Escherichia coli, p. 188-200. In V. L. Yu, T. C. Merigan, Jr., and S. L. Barriere (ed.), Antimicrobial therapy and vaccines. The Williams & Wilkins Company, Baltimore, Md.
- 40.Thornton, V. B., J. A. Davis, M. B. St. Clair, and M. N. Cole. 2003. Inoculation of Staphylococcus xylosus in SJL/J mice to determine pathogenicity. Contemp. Top. Lab. Anim. Sci. 42:49-52. [PubMed] [Google Scholar]
- 41.Unge, A., R. Tombolini, L. Molbak, and J. K. Jansson. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl. Environ. Microbiol. 65:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virta, M., J. Lampinen, and M. Karp. 1995. A luminescence-based mercury biosensor. Anal. Chem. 34:667-669. [Google Scholar]
- 43.Waterfield, N. R., R. W. Le Page, P. W. Wilson, and J. M. Wells. 1995. The isolation of lactococcal promoters and their use in investigating bacterial luciferase synthesis in Lactococcus lactis. Gene 165:9-15. [DOI] [PubMed] [Google Scholar]
- 44.Wood, K. V., and M. DeLuca. 1987. Photographic detection of luminescence in Escherichia coli containing the gene for firefly luciferase. Anal. Biochem. 161:501-507. [DOI] [PubMed] [Google Scholar]