Abstract
Background and Aims
Growth hormone (GH) not only supports hepatic metabolism but also protects against hepatocyte cell death. Hnf6 (or Oc1) belonging to the Onecut family of hepatocyte transcription factors known to regulate differentiated hepatic function, is a GH-responsive gene. We evaluate if GH mediates Hnf6 activity to attenuate hepatic apoptotic injury.
Methods
We used an animal model of hepatic apoptosis by bile duct ligation (BDL) with Hnf6 -/- (KO) mice in which hepatic Hnf6 was conditionally inactivated. GH was administered to adult wild type WT and KO mice for the 7 days of BDL to enhance Hnf6 expression. In vitro, primary hepatocytes derived from KO and WT liver were treated with LPS and hepatocyte apoptosis was assessed with and without GH treatment.
Results
In WT mice, GH treatment enhanced Hnf6 expression during BDL, inhibited Caspase -3, -8 and -9 responses and diminished hepatic apoptotic and fibrotic injury. GH-mediated upregulation of Hnf6 expression and parallel suppression of apoptosis and fibrosis in WT BDL liver were abrogated in KO mice. LPS activated apoptosis and suppressed Hnf6 expression in primary hepatocytes. GH/LPS co-treatment enhanced Hnf6 expression with corresponding attenuation of apoptosis in WT-derived hepatocytes, but not in KO hepatocytes. ChiP-on-ChiP and electromobility shift assays of KO and WT liver nuclear extracts identified Ciap1 (or Birc2) as an Hnf6-bound target gene. Ciap1 expression patterns closely follow Hnf6 expression in the liver and in hepatocytes.
Conclusion
GH broad protective actions on hepatocytes during liver injury are effected through Hnf6, with Hnf6 transcriptional activation of Ciap1 as an underlying molecular mediator.
Introduction
Growth Hormone (GH) regulates somatic growth, cell regeneration and metabolic activities. GH actions are mediated by Stat5 transcriptional activation of Igf-1 [1] and its downstream mediators, with the liver as a major site of Igf-1 production. GH-Stat5 pathway is also implicated in regulating the promoter function of Hepatocyte nuclear factor Hnf6 (Onecut; Oc-1) [2], a liver-enriched transcription factor critical to normal liver development, differentiation and function. Null mice with global disruption of Hnf6 have aberrant hepatoblast maturation, abnormal bile duct formation and die early of cholestasis [3]. In the differentiated liver, Hnf6 regulates hepatocyte target genes involved in metabolic [4] [5] [6] and regenerative functions [7] [8]. We previously treated mice with recombinant human GH to increase in vivo Hnf6 expression and found that GH treatment improved bile duct ligation (BDL) hepatic injury, enhanced cholesterol clearance and improved hepatocyte-specific cellular proliferation [8]. GH has also previously been shown to attenuate hepatocyte death. In GH receptor Ghr-deficient mice, cholic acid feeding worsens cholestasis and hepatocyte apoptosis [9]. Additionally, compared to mdr2 (multi-drug resistant transporter-2) null mutant mice, cholestasis, hepatic fibrosis and hepatocyte apoptosis were exacerbated in mdr2/Ghr or mdr2/Stat5 double null mice [9] [10] [11]. In these GH-resistant mice, Hnf6 expression was diminished, suggesting that increased susceptibility to hepatic apoptosis in the absence of GH function can be attributed to impaired Hnf6 hepatocyte-specific function, and that Hnf6 biological function is broader than the previously demonstrated Hnf6 regulation of hepatocyte proliferation and metabolic activities. We therefore test the hypothesis that GH apoptosis function during cholestatic liver injury is critically linked to Hnf6 function in hepatocytes. We used mice where Hnf6 was conditionally inactivated in the liver (Hnf6 -/-, referred to as KO mice) to evaluate if Hnf6 deficiency 1) worsens hepatic apoptotic injury; 2) impairs GH pro-survival effects in an in vivo model of hepatocyte apoptosis by BDL cholestatic injury; and 3) suppresses Hnf6 hepatic target genes underlying GH-mediated function.
Methods
Materials
Human recombinant GH was obtained from the NIDDK National Hormone and Peptide Program. Mouse monoclonal Hnf6; rabbit polyclonal antibodies against mouse procaspase-3, -8, -9 and cleaved caspases -3, -8, -9, α-Sma, and Igf1 were from Santa Cruz Biotechnology, CA; mouse anti-b-actin, rabbit anti-phospho-Stat5 A/B antibodies were from Sigma-Aldrich; Ciap1, pStat5, rabbit anti-mouse horseradish peroxidase, and goat anti-mouse horseradish peroxidase were from Cell Signaling Technology.
Hnf6 conditional null mice Hnf6 -/- (KO)
Mice homozygous for LoxP-containing Hnf6 allele (Hnf6flox/flox) [12] were interbred with transgenic mice expressing Cre-recombinase under the control of the albumin enhancer/promoter to generate Hnf6flox/flox; albumin-Cre animals. Albumin-Cre allowed Cre-mediated liver-specific recombination and inactivation of both floxed Hnf6 alleles to generate the KO mice.
Animal procedures
The animal study protocol was approved by and conducted in accordance with the Institutional Animal Care and Use Committee (IACUC) at the University of Illinois College of Medicine. Six to eight weeks F6 generation mice received care according to the IACUC guidelines. Following bile duct ligation (BDL n = 8–10, Sham n = 4), PBS or human recombinant GH was delivered at 5 ug/h by subcutaneous Alzet miniosmotic pumps for 1 week [8]. Because male mice were used in our previous work, as well as in Ghr and Stat5 null models, and because of the higher sensitivity of male mice to GH-responsive hepatic target genes [13,14], male mice were used.
Immunostaining
Paraffin-embedded liver tissues underwent TUNEL staining using TdT-FragELTM DNA fragmentation kit from Calbiochem (#QIA33), or α-Sma immunostaining [15]. The strength of α-Sma staining was quantified by Image J analysis program. The percentage of TUNEL-positive hepatocytes in 30 random microscopic fields for 1000 hepatocytes/mouse was counted.
Western blot assays
In three independent experiments, liver total or nuclear protein extract immune complexes [15] were detected with horseradish-conjugated secondary antibody (Fisher) followed by chemiluminescence (ECL + plus, Amersham Biosciences, Inc.).
Chromatin Immunoprecipitation (ChIP) Assays
Briefly [7], liver (n = 3/group) was homogenized, fixed in 1% Formaldehyde; and fragmented DNA samples from sonicated crude nuclear extracts were immunoprecipitated without antiserum, with rabbit or Hnf6 antiserum. Ciap1 forward and reverse primers were 5’-GAGCCTGGTGGTAGTGTGGT-3’ (-619/-599) and 5’-CCATGAGTGGGCTGATTTCT-3’ (-81/-61), Xiap forward and reverse primers were 5’-CCCAGATCCACCCACCTAAC-3’ and 5’-AACGAGCCTCAACCTCAGTC-3’ respectively.
ChiP on ChIP
Hnf6-immunoprecipitated DNA samples (with non-specific IgG as control) from three wild type and three KO liver were hybridized to Affymetrix Mouse Promoter 1.0R array containing over 25,500 mouse promoter regions. Hnf6-bound genes were grouped into functional categories using the biological module–centric algorithm DAVID Bioinformatics Resources 6.7, provided by National Institute of Allergy and Infectious Diseases (NIAID), NIH (https://david.ncifcrf.gov/home.jsp). Genes with Enrichment Scores ≥ 1.25 (p ≤ 0.05) were further investigated by hierarchical cluster analysis using MAT (model-based analysis of tiling-array) scores. The data have been deposited at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80498 in MIAME-compliant standard format. (S1 Table and S1 Fig)
Electrophoretic mobility shift assay (EMSA)
Crude nuclear extracts from WT and KO liver or in vitro-expressed Hnf6 proteins [16] were prepared with labeled double-stranded oligonucleotides 1, 2 and 3 bearing Hnf6-binding sites (Table 1). Protein-DNA complexes were separated from unbound DNA probes using 6% native polyacrylamide gels containing 0.5x Tris borate/EDTA in three independent experiments.
Table 1. Primer DNA sequences of real time PCR genes.
| Name | Forward 5'-3' | Reverse 5'-3' |
|---|---|---|
| Ciap1 | TCAGGTGATGTGGAGCTCAG | GCATACATCCCTGCACACAC |
| Ciap2 | TCCCTGTCATCTCACCATGA | TGTCTAGCATCAGGCCACAG |
| Xiap | TTGGAACATGGACATCCTCA | TGCCCCTTCTCATCCAATAG |
| Survivin | GAGTGAGTCCCAGCTTCCAG | TGATTCCCAGAGATCGTTCC |
| Livin | ACGGTCAAAAGGAAGGGACT | CAGGCTGGGTCTCTCTTCTG |
| Naip | GCCAGGTACCATGAAGAGGA | AATTCACATTTGGGGAACCA |
| Apollon | CTGCGGGGTTGTGATTTACT | GCAGAGAGCATCCAACACAA |
| Bcl-2 | CACGCTGGGAGAACAGGGTA | GGATGTACTTCATCACTATCTCCCG |
| Bad | AGGATCGCTGTGTCCCTTTA | GCAGTCCAGAACAGGAGAGG |
| Bcl-x | CCTTCAGGCCTCTCTCTCCT | CCAGCAGCTCCTCACACATA |
| Bak-1 | CCAACATTGCATGGTGCTAC | AGGAGTGTTGGGAACACAGG |
| Caspase-3 | AGCTTGGAACGGTACGCTAA | CGTACCAGAGCGAGATGACA |
| Caspase-8 | CCTAGACTGCAACCGAGAGG | GCAGGCTCAAGTCATCTTCC |
| Caspase-9 | TGCCCTTGCCTCTGAGTAGT | AACAAAGAAACGCCCACAAC |
| Ctgf | CAAAGCAGCTGCAAATACCA | GGCCAAATGTGTCTTCCAGT |
| Tgfb2r | CGGAAATTCCCAGCTTCTGG | TTTGGTAGTGTTCAGCGAGC |
Forward primer and reverse primer sequences shown with position upstream of the transcription start site.
Real-Time PCR
Total liver RNAs were extracted using RNA-STAT-60 (Tel-Test "B" Inc. Friendswood, TX). cDNA were amplified and analyzed in triplicate using a MyiQ Single Color Real-Time PCR Detection System (Biorad, Hercules, CA). Primer sequences are listed in Table 1. Hnf6, Hnf1b, Hnf4a, Oc2, Foxa2, C/epb primer sequences have been reported previously [6,15,17]. Relative gene expression was calculated using the mathematical delta delta method by PE Applied Biosystems. Levels were normalized to Cyclophilin and reported relative to group with the lowest expression level.
Primary hepatocyte culture
Primary hepatocytes from 3–5 male KO or WT liver were isolated after collagenase perfusion, purified by Percoll centrifugation, plated at 5.5 x 105 cells/well and cultured in DMEM supplemented with 100 nM dexamethasone, 100 nM insulin, 100 U/ml penicillin G, 100 mg/ml streptomycin, and 10% bovine calf serum at 37°C in 5% CO2. After three hours, LPS (1 ug/mL) and/or GH (1 ug/mL) were added for 32 hours before cell harvest.
Statistical Analysis
Data are expressed as mean ± S.D.. Intergroup differences were evaluated by ANOVA for repeated measures. A p value of ≤ 0.05 is considered to be significant. All statistical analyses were performed with the software SPSS.
Results
1. GH diminishes BDL apoptosis in WT but not KO liver
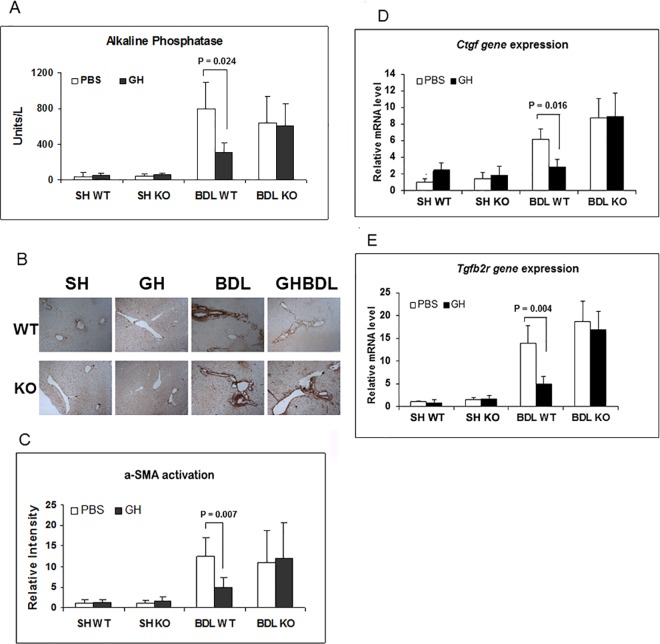
Previously, we reported that GH infusion during BDL increases hepatic Hnf6 and Igf1 expression, with corresponding diminished cholestatic liver injury, enhanced hepatocyte proliferation and improved hepatic metabolic function [8]. Common features in hepatic cholestatic injury are hepatocyte cell death and hepatic fibrosis, with apoptosis driving the fibrogenesis response (19, 20). Experimental models of cholestasis in GH-resistant mice provide strong evidence for GH role in protecting against hepatocyte death [9,10,18–21]. To evaluate if Hnf6 directly regulates hepatocyte apoptosis and if Hnf6 mediates GH function in apoptosis, BDL was performed in Hnf6 -/- (referred to as KO) mice with conditional inactivation of hepatic Hnf6 in hepatocytes and biliary epithelial cells. As previously reported [22], at baseline, KO adult mice have normal liver biochemistries and no intrahepatic abnormality. Except for a 4-fold elevation (p = 0.02) in Hnf1b, a transcription factor regulated by Hnf6 [3], the expression of Hnf6 paralogs Oc-2 and Oc-3 [23,24] and major hepatocyte nuclear factors such as Hnf1a, Hnf4a, Foxa2, and C/ebp are unaffected in KO mice (data not shown). Following 7 days of BDL injury, relative to Sham liver, PBS-treated BDL WT show enhanced hepatocyte apoptosis by TUNEL assay (Fig 1A and 1B), increased expression of Caspase-3, -8 and -9 by real-time PCR (Fig 1C and 1E), and increased cleaved caspase protein expression by western blot (Fig 1F). With GH treatment, TUNEL hepatocyte apoptosis is reduced (Fig 1A and 1B), as is Caspase-3, -8, -9 gene and protein expression in WT BDL liver (Fig 1C and 1F). This is paralleled by attenuated cholestasis with lower serum alkaline phosphatase [8] (Fig 2A), diminished α-smooth muscle actin (α-Sma) marker for fibrosis (Fig 2B and 2C) and profibrotic genes Ctgf, Tgfb2R (Fig 2D and 2E) in GH-treated BDL WT liver relative to PBS BDL liver. In the absence of GH, KO BDL and WT BDL hepatocytes show similar degree of apoptotic injury by TUNEL assay (Fig 1A and 1B); and in whole liver, similar real-time PCR of hepatic Caspase-3, -8 and -9 (Fig 1C–1E) and western blot for cleaved caspase protein expression (Fig 1F; with the exception of cleaved caspase-8). The extent of hepatic cholestasis (Fig 2A) and fibrosis (Fig 2B–2E) is also similar between WT BDL and KO BDL liver in the absence of GH. In KO mice, previously demonstrated GH-associated reduction of hepatocyte TUNEL labeling, and of hepatic Caspase -3, -8, -9 expression in WT BDL liver is lost (Fig 1C–1E). In fact, we observed increased caspase-3 expression, minimal to no reduction of caspase-8, and -9 proteins (Fig 1F), undiminished cholestatic injury, no differences in Ctfg, Tgfb2R expression or α-Sma staining relative to PBS-treated KO BDL liver (Fig 2). The data suggest that Hnf6 is essential to GH-mediated reduction of BDL hepatocyte apoptosis, hepatic cholestasis and fibrosis.
Fig 1. TUNEL assay and Caspase expression in WT and KO liver.
(A) Representative TUNEL-labeled liver micrographs from Sham (SH) WT or KO mice without (WT, KO) or with GH treatment (WTGH and KOGH); and Bile duct ligation (BDL) WT or KO mice without (WTBDL, KOBDL) or with GH treatment (GHWTBDL and GHKOBDL) (n = 8–10/group). The arrows identify TUNEL + hepatocytes. (B) Bar graph of % TUNEL (+) hepatocytes/100x high power field from WT or KO SH or BDL liver treated with PBS (□) or GH (■) and significant p value. (C-E) Caspase-3 (C), -8 (D) and -9 (E) in WT or KO liver treated with PBS (□) or GH (■) and significant p values. Gene levels were calculated relative to SH WT liver. (F) Representative western blots of Procaspase-8, -9, -3, cleaved proteins and b-Actin in SH or BDL WT and KO liver and the corresponding bar graphs of caspase staining intensity in BDL samples with significant p values.
Fig 2. Hepatic cholestasis and fibrosis in WT and KO mice.
(A) Serum Alkaline Phosphatase levels (a marker of cholestasis) in SH or BDL WT and KO mice treated with PBS (□) or GH (■) and significant p values. (B) Representative micrographs of α-Sma immunostaining of SH or BDL WT and KO liver. (C) Bar graph of α-Sma immunostaining of SH or BDL WT and KO liver treated with PBS (□) or GH (■) with intensity relative to SH WT and significant p value. (D) Ctgf expression in SH or BDL WT and KO mice treated with PBS (□) or GH (■) with levels relative to Sham WT and significant p values. (E) Tgfb2R expression in SH or BDL WT and KO mice treated with PBS (□) or GH (■) with levels relative to SH WT and significant p values.
2. GH reduction of BDL apoptosis is Hnf6-dependent
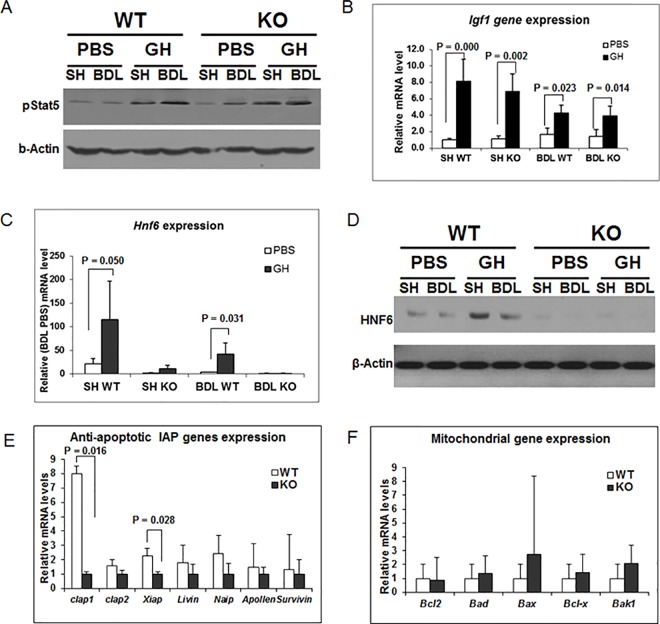
Since GH transcriptionally activates Hnf6 and Igf1, we assessed their contribution to GH protective effects. Following GH infusion in WT mice, GH signaling is enhanced as shown by higher hepatic phosphorylated Stat5 protein in GH-Sham relative to PBS-Sham liver; and similarly, in GH-BDL relative to PSB-BDL liver (Fig 3A). Correspondingly, GH-responsive Igf1 (Fig 3B), and Hnf6 gene (Fig 3C) and protein expression (Fig 3D) increase in GH-treated Sham WT relative to PBS-treated WT Sham; and similarly, in GH-BDL relative to PSB-BDL liver. Previous publication indeed showed that GH-induced increase of Hnf6 expression is hepatocyte specific with increased Hnf6 localization to hepatocyte nuclei [8]. While the Stat5 protein and Igf1 response is intact in GH-treated KO Sham and BDL liver, relative to PBS-treated Sham and BDL KO liver respectively, as expected, Hnf6 gene and protein (Fig 3C and 3D) expression is absent in KO liver. These data show that despite intact Stat5 pathways and Igf1 expression in KO mice, the loss of Hnf6 impairs GH improvement of BDL apoptosis and fibrosis in KO liver, suggesting that GH-mediated protective response is mediated through Hnf6, independent of Igf1.
Fig 3. Downstream signaling response to GH treatment.
(A) Representative western blot of phosphorylated Stat5 and b-Actin of liver nuclear proteins from Sham (SH) or BDL WT and KO liver treated with PBS or GH. (B) Igf1 expression in WT SH and KO SH as well as WT BDL and KO BDL liver treated with PBS (□) or GH (■), with levels relative to SH KO PBS liver and significant p values. (C) Hnf6 expression in SH or BDL WT and KO liver treated with PBS (□) or GH (■), with levels relative to SH KO liver and significant p values. (D) Representative western blot of Hnf6 nuclear protein extracts from SH or BDL WT and KO liver treated with PBS or GH. (E) Baseline expression levels of Iap (Ciap1, Ciap2, Xiap, Livin, Naip, Apollen and Survivin) family of genes from WT and KO liver with levels shown relative to KO liver and significant p values. (F) Baseline expression levels of Bcl2 (Bcl2, Bad, Bax, Bcl-x, Bak1) families of genes from WT and KO liver with levels shown relative to KO liver and significant p values.
3. Hnf6 binds inhibitor of Apoptosis Protein gene Ciap1 promoter
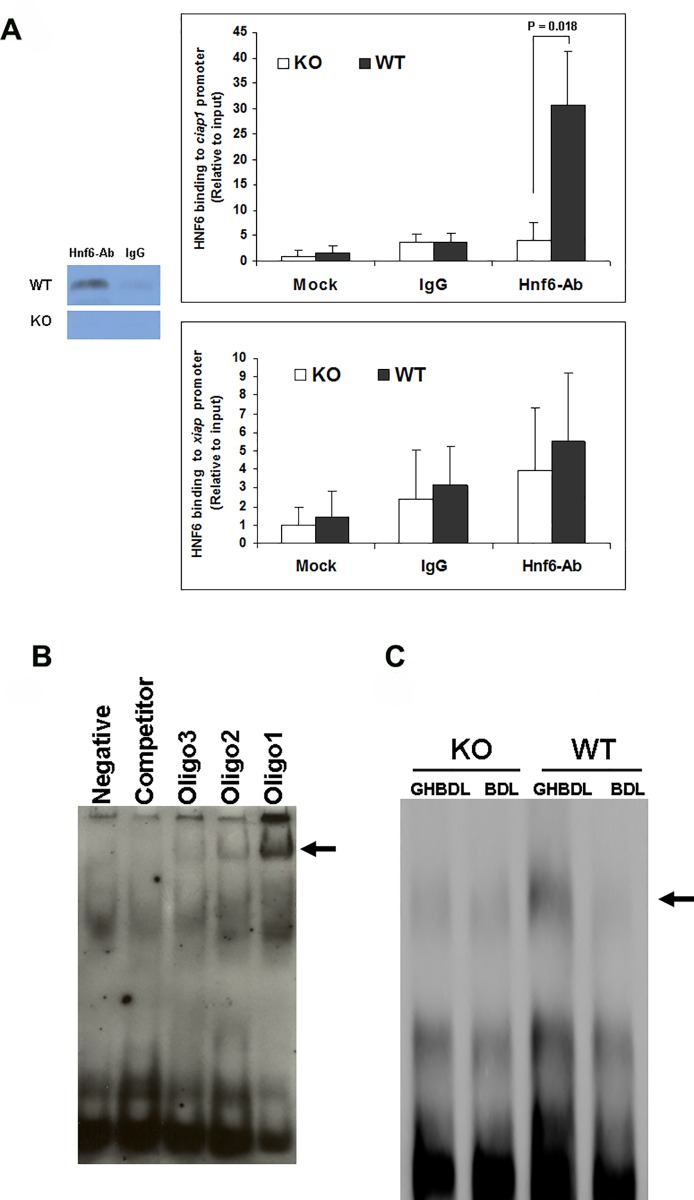
Genome-wide location analysis of Hnf6-bound genes by ChIP-on-ChIP assays of WT and KO liver (n = 3 each, S1 Table and S1 Fig) was performed to screen for potential Hnf6 target genes with antiapoptotic function. Hnf6 binding to Ciap1 (also known as Birc2, Baculoviral Iap repeat-containing protein 2, a member of the Iap Inhibitor of Apoptosis Protein families involved in TNFa- and TRAIL (tumor necrosis factor-related apoptosis-inducing ligand)-induced cell signaling for apoptosis [25] [26] [27]) is markedly reduced in KO liver. To confirm the ChIP-on-ChIP results, we performed real-time PCR of WT and KO liver for apoptosis-related gene profiles. Among members of the Iap families, notably Ciap1, Ciap2, Survivin, and Xiap (Fig 3E), and Bcl2 families (Fig 3F), only Ciap1 and Xiap are significantly downregulated in KO liver. ChIP assays for constitutive in vivo binding of Hnf6 nuclear protein to Ciap1 and Xiap promoters show that Ciap1 promoter sequences are amplified from Hnf6-immunoprecipitated but not mock-treated or irrelevant IgG-treated hepatic chromatin/DNA complex in WT liver (Fig 4A). KO liver have no Ciap1 promoter amplification, showing that baseline Hnf6 binding to Ciap1 promoter in WT liver is lost in KO liver. No difference between IgG- vs Hnf6-bound Xiap complex is found in WT or KO liver. Furthermore, anti-Hnf6 antibody-precipitated Xiap1 promoter amplification is similar to mock- or IgG-treated liver, consistent with the lack of constitutive binding of Hnf6 to Xiap1 promoter.
Fig 4. Ciap1 promoter occupancy by Hnf6.
(A) Ciap1- and Xiap-precipitated promoter fragments bound by mock, control IgG or anti-Hnf6 antibody were amplified by real time PCR and significant p value is shown. The gel insert shows precipitated anti-Hnf6 antibody treated Hnf6-DNA complex in WT vs KO nuclear extracts. Relative Ciap1 promoter amplification is graphed relative to the corresponding WT mock data. (B) Electromobility shift assay EMSA of in vitro synthesized Hnf6 protein in the absence (Negative) or in the presence of 3 Ciap1 promoter fragments with arrow showing the Oligonucleotide-Hnf6 protein precipitated complex. The Digoxin-labeled Oligonucleotide could be inhibited with DIG Oligo Competitor. (C) EMSA of nuclear extracts from KO and WT liver against Ciap1 promoter oligonucleotide 1, with the arrow showing the location of the DNA-protein complex.
Ciap1 promoter sequence analysis for Hnf6 consensus binding motifs [28] reveals three potential HNF6 binding sites (Table 2). Electromobility shift assays show that Ciap1 oligonucleotide 1 binding to in vitro synthesized Hnf6 protein is strongest (Fig 4B). These results further suggest that Ciap1 is likely a direct Hnf6-regulated target gene.
Table 2. Mouse Ciap1 promoter DNA sequences containing Hnf6 binding sites.
| Name | Position | Sequence |
|---|---|---|
| Oligo 1 | -539/-530 | ATATCgAcTT |
| Oligo 2 | -381/-372 | ATGTCCATTTG |
| Oligo 3 | +648/+657 | GTGTCAATTc |
| Hnf6 consensus sequence | DHWTCYATND |
Lower case letters represent nucleotides deviating from Hnf6 consensus binding sequence. Nucleotide abbreviations are as follows: D is not C, H is A or T, W is A or G, Y is A or C and N is A, T, C or G.
4. Ciap1 expression correlates with Hnf6 hepatic expression profile
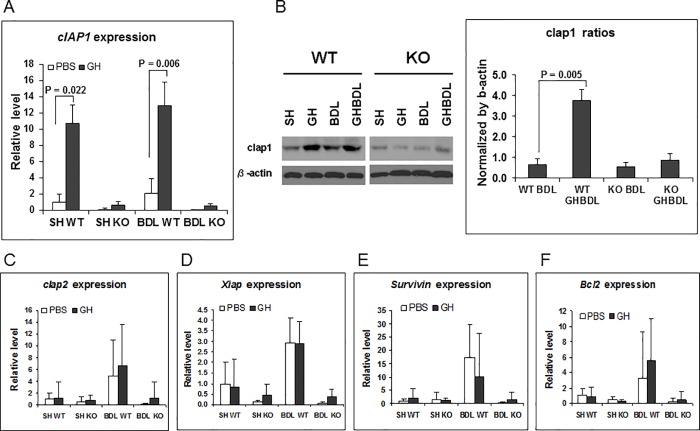
We next characterized the relationship between in vivo Hnf6 and hepatic expression of Iap during BDL. Previously, GH treatment reduces the extent of BDL-mediated downregulation of Hnf6 expression [8] (also Fig 3C and 3D). In the current study, GH treatment in Sham or BDL WT liver is associated with upregulation of hepatic Ciap1 (Fig 5A) relative to PBS Sham or BDL liver respectively, but not of Xiap, Survivin, or Ciap2 (Fig 5C–5E). In the KO liver, the loss of Hnf6 is associated with the loss of induction in the expression of all Iap family of genes during BDL, with or without GH treatment (Fig 5A). Of note, Bcl2 gene expression in both WT and KO liver is similar between PBS-treated BDL and GH-treated BDL samples (Fig 5F). Western blotting shows that paralleling Ciap1 expression pattern in WT liver, Ciap1 protein was higher in GH-treated Sham relative to PBS-Sham liver, and likewise, in GH-BDL liver relative to PBS-BDL liver (Fig 5B). In contrast, Ciap1 expression in KO liver is uniformly lower in Sham, PBS-BDL and GH-BDL relative to WT liver counterparts (Fig 5B). The data showing tight correlations between Hnf6 and Ciap1 patterns of expression are consistent with Ciap1 as an Hnf6-regulated target gene.
Fig 5. Iap gene and protein gene expression in GH-treated BDL liver.
(A) Hepatic Ciap1 expression in SH or BDL WT and BDL liver with PBS (□) or GH (■) treatment, with levels relative to SH WT liver and significant p values. (B) Representative western blot gel of SH or BDL WT and KO liver, and graph of quantitated signal from GH-treated WT and KO BDL liver with significant p value. (C-F) Ciap2, Xiap, Survivin, Bcl2 expression in SH or BDL WT and KO liver with PBS (□) or GH (■) treatment, with levels relative to SH WT liver.
5. Hnf6 binds to Ciap1 promoter in BDL liver
Electromobility shift assay of liver nuclear extracts against Ciap1 promoter oligonucleotide 1 (Fig 4C) shows that in vivo, Hnf6-Ciap1 signal complex is stronger in GH-treated BDL WT liver relative to PBS-BDL WT liver, reflecting enhanced Hnf6 expression and Hnf6 binding to the Ciap1 promoter in GH-treated samples. KO BDL liver shows poor HNF6-Ciap1 signal complex, even with GH treatment. These data are in further support of Hnf6 binding and regulation of the Ciap1 promoter as a target gene during the hepatic adaptive response to injury.
6. GH enhances Ciap1 and attenuates apoptosis in primary hepatocytes
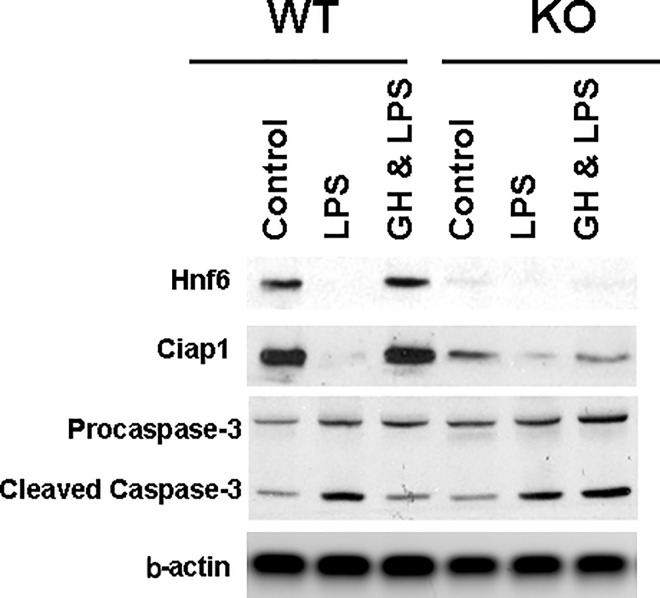
IAP proteins suppress TNFa– αnd TRAIL-induced cell death by inhibiting the activity of initiator and effector caspases [25] [26] [27]. Ciap1 knockdowns with either targeted SiRNA or antagonist to Ciap using endogenous or pharmacologic SMAC (Second Mitochondrial Activator of Caspases) increase stress-mediated apoptosis, while Ciap1 overexpression inhibits cell death in various cell-derived tissues [29] [30] [31] [32]. In primary rat hepatocytes, transfection with adenoviral vector expressing human CIAP1 suppresses LPS/cytokine mediated caspase-3 activation [33]. Conversely, hepatocyte cell lines as well as primary mouse hepatocytes treated either with ShRNA targeting Ciap1 or with SMAC mimetics become sensitized to apoptosis [34] [35]. To further evaluate Hnf6/Ciap1 transcriptional link with hepatocyte-specific apoptosis, primary hepatocytes from KO and WT liver were exposed in vitro to high dose LPS. Western blot (Fig 6) shows, as expected, severe suppression of Hnf6 along with Ciap1 expression in KO hepatocytes. LPS treatment is associated with diminished Hnf6 expression in WT hepatocytes as an acute phase response (an observation of Hnf6 downregulation in acute injury during BDL [15], and in ethanol- or carbon tetrachloride-treated liver (data not shown)). GH treatment however restores Hnf6 expression in LPS hepatocytes, with Ciap1 expression pattern paralleling that of Hnf6 in LPS and GH-LPS samples. In WT hepatocytes, suppression of Hnf6/Ciap1 at high LPS dose is associated with enhanced cleaved caspase-3, while GH-dependent increased Hnf6/Ciap1 expression correlates with suppression of LPS-stimulated apoptosis. In spite of Hnf6 loss and parallel reduction in Ciap1 expression, KO hepatocytes respond to LPS with similar ability as in WT hepatocytes to induce caspase-3. In these KO hepatocytes however, GH effect on enhancing Hnf6 and Ciap1 is impaired. This is associated with compromised ability of GH to attenuate LPS-induced apoptosis. In summary, the stress response in KO mice is characterized by the loss of GH-mediated attenuation of apoptotic injury in LPS-treated primary hepatocytes because of suppressed Hnf6- and Ciap1 expression.
Fig 6. Apoptosis in LPS-treated WT and KO primary hepatocytes.
Representative western blot of Hnf6, Ciap1, Procaspase-3, Cleaved Caspase-3, and b-Actin expression of total proteins from primary hepatocytes in WT and KO mice (n = 3–5) after treatment for 32 hours with 1 ug/ml of LPS, with or without GH cotreatment.
Discussion
Hepatocyte apoptosis is a universal finding in all forms of liver injury, including cholestasis. We previously demonstrated that Hnf6 mediates hepatocyte survival functions such as hepatocyte cellular proliferation, hepatic glucose and cholesterol metabolism through Hnf6 transcriptional activation of its target genes [4–7,36]. We herein evaluated the role of Hnf6 in hepatocyte-specific apoptosis in the previous model of BDL by using mice with conditional inactivation of Hnf6 in the liver. Null mice do not exhibit more severe hepatocyte apoptotic BDL injury, demonstrating that Hnf6 is not essential to protection against hepatic apoptosis. Hepatic Igf1 during BDL injury is unimpaired in KO mice. KO mice thus may have compensated for Hnf6 loss through, as previously described, Igf-1 pro-survival effects on cell proliferation [37,38] and anti-apoptotic activity [39–41]. The loss of Hnf6 function in KO mice may be further counterbalanced by preserved expression of Oc2 and Oc3 paralogs, Foxa2, and notably by compensatory increases in Hnf1b. Hnf1b and Foxa2 have been demonstrated to have prosurvival [42] and antiapoptotic [43] function in hepatocytes, respectively. These transcription factors may cooperate with Igf1 to further alleviate KO liver susceptibility to hepatic cell death.
Prior cholestatic injury models in Ghr, Ghr/Mdr2 or Stat5/Mdr2 null mice have implicated GH-Stat5-Igf1 pathway in GH-mediated cell growth, survival, metabolism, and particularly protection against hepatocyte apoptosis [9] [10]. GH can also modulate hepatic gene function in an Igf1-independent manner through GH-Stat5 regulation of Hnf6 promoter. We previously demonstrated that GH treatment during BDL in mice attenuates injury-related downregulation of Hnf6 expression. In these animals, enhanced Hnf6 expression is associated with improved hepatocyte proliferation, cholestasis, and cholesterol clearance. This protective response is linked to GH-Stat5 induction of Hnf6 and Hnf6 Cyclin D1 and Cyp7a1 target genes [8]. Our current data show that with GH treatment, broad reduction in hepatic apoptosis (extrinsic and intrinsic pathways) and in cholestasis is observed in WT BDL mice. GH protective function is however abrogated in KO BDL mice, despite intact Stat5 and Igf1 response. Hnf6 is therefore necessary for GH broad protection against apoptosis and cholestasis during BDL injury, independently of Igf1 [8].
Albumin-Cre-mediated liver-specific inactivation of Hnf6 affects both hepatocytes and cholangiocytes in our KO liver. While the contribution of Hnf6 inactivation in cholangiocytes cannot be excluded, the demonstrated GH-Hnf6 link to hepatic apoptosis is likely hepatocyte-dominant on the basis of hepatocyte-specific TUNEL immunostaining and primary hepatocyte data. Furthermore, hepatocytes constitute 85% of the liver cell mass while cholangiocytes only comprise 3–5% of the epithelial cell population. Interestingly, our previous work demonstrated that while Hnf6 has prosurvival effects on hepatocytes, Hnf6 impairs cholangiocyte proliferation [15]. While we do not observe baseline biliary duct abnormalities in the KO mice, it remains to be determined if the KO liver will respond during BDL injury to GH with an accentuated ductal proliferative reaction over biliary apoptosis.
The liver displays a sexual dimorphic response to GH [44], with Hnf6 participating in this sex-differential network to regulate many metabolic enzymes in a sex-specific fashion, such as CYP2C12, a member of cytochrome P450 family [45]. We have yet to determine if GH- and Hnf6-mediated responses to hepatocyte apoptosis are also sex-specific.
With respect to cholestasis, bile transport gene promoters contain Hnf6 binding sites (data not shown), suggesting that Hnf6-targeted genes may be involved in bile uptake and excretion. Further evaluation of Hnf6 regulation of cholestatic genes is pending. Overall, our data suggest that previously described aggravated hepatic injury in Gh-deficient mice is likely from dual Igf1 and Hnf6 defective functions. The implication is that preservation of both Igf1 and Hnf6 axes is important to hepatic survival responses against apoptosis and cholestasis. Future evaluations of cholestatic injury in double null Igf1/Hnf6 against single Igf1 or Hnf6 null mice will shed further light on their relative biological importance.
Hepatocellular cell death is a driver and sensitizer for liver inflammation and fibrosis [46]. GH attenuation of apoptosis is therefore expected to improve BDL-fibrosis [8]. GH-dependent Igf1 is an important protective mechanism against fibrosis, since hepatic fibrosis is accelerated in Igf-deficient cholestatic mice [10], but attenuated following Igf1 replacement [21]. Despite a normal Igf1 response to GH during BDL, GH does not improve hepatic fibrosis in KO mice. The GH-Hnf6 regulated pathway, independently of Igf1, is therefore an alternative mechanism for GH protective effect in fibrosis. A potential mediator of this pathway is Hnf6-regulated Tgfb2r and Ctgf target genes. We previously demonstrated that in vivo, Hnf6-adenoviral vector suppresses Tgfb2r hepatic expression [8]. With GH treatment, increased Hnf6 and reduced BDL hepatic fibrosis are associated with diminished Tgfb2r gene and protein [8], suggesting that Hnf6 negatively regulates Tgfb2r. Furthermore, Tgfb2r signaling is increased in global Hnf6 null mice embryonic liver [47], while our ChIP-on ChIP data show over-amplified Tgfb2r promoter in KO relative to WT liver (data not shown). These data are consistent with Hnf6 as a transcriptional suppressor of Tgfb2r promoter function. Of note, pancreatic Ctgf expression was upregulated in mice with pancreas-specific Hnf6 inactivation [48], suggesting that HNF6 antifibrotic function may be mediated through its transcriptional inhibition of Tgfb2r and Ctgf. It remains to be seen if GH-dependent alleviation of fibrosis is secondary to suppression of apoptosis by GH/Igf1-, GH/Hnf6-dependent pathways, and/or through GH/Hnf6-regulated suppression of Hnf6 fibrogenic Tgfb2r or Ctgf target genes.
Among members of the IAP Inhibitor of Apoptosis Protein families modulating TNF-induced apoptosis and survival [25] [26] [27], Ciap1 (Bicr2) promoter expression is markedly reduced in KO liver. Consistent with Ciap1 as an Hnf6-transcriptionally regulated gene, ChiP assays reveal that Hnf6 constitutively binds Ciap1 promoter while baseline hepatic Ciap1 expression is severely diminished in KO mice. Electromobility shift assays additionally show that GH treatment of WT BDL mice increases Hnf6-Ciap1 nuclear complex signal over untreated WT BDL liver, with real time PCR and Hnf6 and Ciap1 protein expression recapitulating the gel shift data. All of these GH-enhancing effects are suppressed in Hnf6 KO mice. Together, the data are in support of Hnf6-regulated Ciap1 transcriptional activation as an underlying mechanism for GH antiapoptotic function during BDL.
During injury, Ciap1 inhibits apoptosis signaling through a Caspase-activation and -recruitment domain, and several Baculo Iap repeat domains to interact with TNF-receptor associated factors TRAF2/3 and caspase-3, -7, -9 [25] [26]. Despite lower baseline Ciap1 expression in KO primary hepatocytes, LPS stimulates caspase expression equally well in both WT and KO hepatocytes, in support of the complexity of LPS-induced apoptosis. These in vitro findings are compatible with our in vivo data showing that BDL KO liver do not exhibit worse apoptotic injury. GH restores Ciap1 and attenuates apoptosis in WT but not KO primary hepatocytes, consistent with Ciap1 as a mediator of GH hepatic antiapoptotic function in a hepatocyte-specific fashion through Hnf6-dependent pathways. These data recapitulate our in vivo whole liver findings that the protective effect of GH on apoptosis is lost in Hnf6- and Ciap1-deficient KO BDL mice. It is worth noting that Ciap is implicated in viral clearance in mouse model of hepatitis B infection [49], as well as in chemoresistance [32,50,51], providing the clinical rationale for IAP antiviral [52] and antitumor therapies [53]. It remains to be determined if Ciap1 link to the protective effect of GH on in vivo hepatocyte apoptosis can be explored as a potential application for liver support therapies.
Conclusion
Hnf6 is not necessary to protection against LPS-apoptotic injury in hepatocytes, or to hepatic protection against apoptosis in BDL cholestasis. Hnf6 is however is critical to GH prosurvival function during LPS-injury in primary hepatocytes, as well as in the cholestastic liver, to attenuate hepatocyte apoptosis independently of GH/Igf1 axis. This is likely mediated through Hnf6 transcriptional target gene Ciap1. The implication is that for patients with hepatic Igf1 dysfunction in clinical settings such as chronic liver diseases [54], therapeutic interventions using Hnf6 and/or Ciap1 gain-of-function approaches may diminish hepatocellular death, improve hepatocyte proliferation and enhance liver function.
Supporting Information
Cluster diagram of the Cluster Analysis showing Hnf6-bound genes. R2 measures Hierarchical cluster analysis using MAT (model-based analysis of tiling-array) scores with Ciap1 (or Birc2, denoted with *) having the highest MAT-score (664), and significant variance.
(DOC)
The table depicts Hnf6-bound genes with Enrichment Scores ≥ 1.25 (corresponding to a ≤ 0.05) and Group Enrichment Score ranking by biological significance based on overall EASE (Expression Analysis Systematic Explorer) scores (corresponding to the one-tailed Fisher exact probability of gene over-representation by functional class). Baculoviral IAP repeat-containing 2 (NM_007465, Birc2 or Ciap1) is shown in Group 4, with Enrichment Score of 1.59.
(DOC)
Acknowledgments
This work is dedicated to the late Robert Costa, for his mentorship and support.
Abbreviations
- α-Sma
α-smooth muscle actin
- BDL
Bile duct ligation
- Bicr2
Baculoviral Iap repeat-containing protein 2
- Ciap
Cellular inhibitor of apoptosis protein
- Ctgf
Connective tissue growth factor
- GH
Growth hormone
- Hnf
Hepatocyte nuclear factor
- IAP
inhibitor of apoptosis protein
- Igf
Insulin-like growth factor
- KO
Hnf6 conditional null mice
- LPS
Lipopolysaccharide
- Oc
Onecut
- SH
Sham
- Tgfb2r
Transforming growth factor b2 Receptor
- Xiap
X-linked inhibitor of apoptosis protein
Data Availability
The ChIP-on-ChIP data files are available from the://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80498.
Funding Statement
Grant support was provided by NIH R21 DK070784-02 to AH, and U01DK089540 and R01DK105689 to MG.
References
- 1.Woelfle J, Chia DJ, Rotwein P (2003) Mechanisms of growth hormone (GH) action. Identification of conserved Stat5 binding sites that mediate GH-induced insulin-like growth factor-I gene activation. J Biol Chem 278: 51261–51266. 10.1074/jbc.M309486200 [DOI] [PubMed] [Google Scholar]
- 2.Lahuna O, Rastegar M, Maiter D, Thissen JP, Lemaigre FP, and Rousseau GG. (2000) Involvement of STAT5 (signal transducer and activator of transcription 5) and HNF-4 (hepatocyte nuclear factor 4) in the transcriptional control of the hnf6 gene by growth hormone. Mol Endocrinol 14: 285–294. 10.1210/mend.14.2.0423 [DOI] [PubMed] [Google Scholar]
- 3.Clotman F, Lannoy VJ, Reber M, Cereghini S, Cassiman D, Jackemin P, et al. (2002) The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development 129: 1819–1828. [DOI] [PubMed] [Google Scholar]
- 4.Costa RH, Kalinichenko VV, Holterman AX, Wang X (2003) Transcription factors in liver development, differentiation, and regeneration. Hepatology 38: 1331–1347. 10.1016/j.hep.2003.09.034 [DOI] [PubMed] [Google Scholar]
- 5.Lannoy VJ, Decaux JF, Pierreux CE, Lemaigre FP, Rousseau GG (2002) Liver glucokinase gene expression is controlled by the onecut transcription factor hepatocyte nuclear factor-6. Diabetologia 45: 1136–1141. 10.1007/s00125-002-0856-z [DOI] [PubMed] [Google Scholar]
- 6.Wang M, Tan Y, Costa RH, Holterman AX (2004) In vivo regulation of murine CYP7A1 by HNF-6: a novel mechanism for diminished CYP7A1 expression in biliary obstruction. Hepatology 40: 600–608. 10.1002/hep.20349 [DOI] [PubMed] [Google Scholar]
- 7.Tan Y, Yoshida Y, Hughes DE, Costa RH (2006) Increased expression of hepatocyte nuclear factor 6 stimulates hepatocyte proliferation during mouse liver regeneration. Gastroenterology 130: 1283–1300. 10.1053/j.gastro.2006.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M, Chen M, Zheng G, Dillard B, Tallarico M, Ortiz Z, et al. (2008) Transcriptional activation by growth hormone of HNF-6-regulated hepatic genes, a potential mechanism for improved liver repair during biliary injury in mice. Am J Physiol Gastrointest Liver Physiol 295: G357–366. 10.1152/ajpgi.00581.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stiedl P, McMahon R, Blaas L, Stanek V, Svinka J, Grabner B, et al. (2015) Growth hormone resistance exacerbates cholestasis-induced murine liver fibrosis. Hepatology 61: 613–626. 10.1002/hep.27408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaas L, Kornfeld JW, Schramek D, Musteanu M, Zollner G, Gumhold J, et al. (2010) Disruption of the growth hormone—signal transducer and activator of transcription 5—insulinlike growth factor 1 axis severely aggravates liver fibrosis in a mouse model of cholestasis. Hepatology 51: 1319–1326. 10.1002/hep.23469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fickert P, Fuchsbichler A, Wagner M, Zollner G, Kaser A, Tilg H, et al. (2004) Regurgitation of bile acids from leaky bile ducts causes sclerosing cholangitis in Mdr2 (Abcb4) knockout mice. Gastroenterology 127: 261–274. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Ables ET, Pope CF, Washington MK, Hipkens S, Means AL, et al. (2009) Multiple, temporal-specific roles for HNF6 in pancreatic endocrine and ductal differentiation. Mech Dev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waxman DJ, Holloway MG (2009) Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol 76: 215–228. 10.1124/mol.109.056705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lahuna O, Fernandez L, Karlsson H, Maiter D, Lemaigre FP, Rousseau GG, et al. (1997) Expression of hepatocyte nuclear factor 6 in rat liver is sex-dependent and regulated by growth hormone. Proc Natl Acad Sci U S A 94: 12309–12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holterman AX, Tan Y, Kim W, Yoo KW, Costa RH (2002) Diminished hepatic expression of the HNF-6 transcription factor during bile duct obstruction. Hepatology 35: 1392–1399. 10.1053/jhep.2002.33680 [DOI] [PubMed] [Google Scholar]
- 16.Rausa F, Samadani U, Ye H, Lim L, Fletcher CF, Jenkins NA, et al. (1997) The cut-homeodomain transcriptional activator HNF-6 is coexpressed with its target gene HNF-3 beta in the developing murine liver and pancreas. Dev Biol 192: 228–246. 10.1006/dbio.1997.8744 [DOI] [PubMed] [Google Scholar]
- 17.Glaser S, Wang M, Ueno Y, Venter J, Wang K, Chen H, et al. (2010) Differential transcriptional characteristics of small and large biliary epithelial cells derived from small and large bile ducts. Am J Physiol Gastrointest Liver Physiol 299: G769–777. 10.1152/ajpgi.00237.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castilla-Cortazar I, Garcia M, Muguerza B, Quiroga J, Perez R, Santidrian S, et al. (1997) Hepatoprotective effects of insulin-like growth factor I in rats with carbon tetrachloride-induced cirrhosis. Gastroenterology 113: 1682–1691. [DOI] [PubMed] [Google Scholar]
- 19.Held MA, Cosme-Blanco W, Difedele LM, Bonkowski EL, Menon RK and Denson LA. (2005) Alterations in growth hormone receptor abundance regulate growth hormone signaling in murine obstructive cholestasis. Am J Physiol Gastrointest Liver Physiol 288: G986–993. 10.1152/ajpgi.00287.2004 [DOI] [PubMed] [Google Scholar]
- 20.Mair M, Zollner G, Schneller D, Musteanu M, Fickert P, Gumhold J, et al. (2010) Signal transducer and activator of transcription 3 protects from liver injury and fibrosis in a mouse model of sclerosing cholangitis. Gastroenterology 138: 2499–2508. 10.1053/j.gastro.2010.02.049 [DOI] [PubMed] [Google Scholar]
- 21.Mirpuri E, Garcia-Trevijano ER, Castilla-Cortazar I, Berasain C, Quiroga J, Rodrizuez-Ortigoza C, et al. (2002) Altered liver gene expression in CCl4-cirrhotic rats is partially normalized by insulin-like growth factor-I. Int J Biochem Cell Biol 34: 242–252. [DOI] [PubMed] [Google Scholar]
- 22.Vanderpool C, Sparks EE, Huppert KA, Gannon M and Means AL. (2012) Genetic interactions between hepatocyte nuclear factor-6 and Notch signaling regulate mouse intrahepatic bile duct development in vivo. Hepatology 55: 233–243. 10.1002/hep.24631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margagliotti S, Clotman F, Pierreux CE, Beaudry JB, Jacquemin P, Rousseau GG, et al. (2007) The Onecut transcription factors HNF-6/OC-1 and OC-2 regulate early liver expansion by controlling hepatoblast migration. Dev Biol 311: 579–589. 10.1016/j.ydbio.2007.09.013 [DOI] [PubMed] [Google Scholar]
- 24.Jacquemin P, Lannoy VJ, Rousseau GG, Lemaigre FP (1999) OC-2, a novel mammalian member of the ONECUT class of homeodomain transcription factors whose function in liver partially overlaps with that of hepatocyte nuclear factor-6. J Biol Chem 274: 2665–2671. [DOI] [PubMed] [Google Scholar]
- 25.Silke J, Vucic D (2014) IAP family of cell death and signaling regulators. Methods Enzymol 545: 35–65. 10.1016/B978-0-12-801430-1.00002-0 [DOI] [PubMed] [Google Scholar]
- 26.Budhidarmo R, Day CL (2015) IAPs: Modular regulators of cell signalling. Semin Cell Dev Biol 39: 80–90. 10.1016/j.semcdb.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 27.Lemke J, von Karstedt S, Zinngrebe J, Walczak H (2014) Getting TRAIL back on track for cancer therapy. Cell Death Differ 21: 1350–1364. 10.1038/cdd.2014.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, et al. (2000) Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol 20: 4445–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi Y, Xia P (2012) Cellular inhibitor of apoptosis protein-1 (cIAP1) plays a critical role in beta-cell survival under endoplasmic reticulum stress: promoting ubiquitination and degradation of C/EBP homologous protein (CHOP). J Biol Chem 287: 32236–32245. 10.1074/jbc.M112.362160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, et al. (2007) IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 131: 682–693. 10.1016/j.cell.2007.10.037 [DOI] [PubMed] [Google Scholar]
- 31.Zhao D, Sun Y, Wei X, Liang H, Zhao L, Dong X, et al. (2015) cIAP1 attenuates shear stress-induced hBMSC apoptosis for tissue-engineered blood vessels through the inhibition of the mitochondrial apoptosis pathway. Life Sci 137: 81–88. 10.1016/j.lfs.2015.07.011 [DOI] [PubMed] [Google Scholar]
- 32.Jung SA, Park YM, Hong SW, Moon JH, Shin JS, Lee HR, et al. (2015) Cellular inhibitor of apoptosis protein 1 (cIAP1) stability contributes to YM155 resistance in human gastric cancer cells. J Biol Chem 290: 9974–9985. 10.1074/jbc.M114.600874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoemaker MH, Ros JE, Homan M, Trautwein C, Liston P, Poelstra K, et al. (2002) Cytokine regulation of pro- and anti-apoptotic genes in rat hepatocytes: NF-kappaB-regulated inhibitor of apoptosis protein 2 (cIAP2) prevents apoptosis. J Hepatol 36: 742–750. [DOI] [PubMed] [Google Scholar]
- 34.Akazawa Y, Guicciardi ME, Cazanave SC, Bronk SF, Werneburg NW, Kakisaka K, et al. (2013) Degradation of cIAPs contributes to hepatocyte lipoapoptosis. Am J Physiol Gastrointest Liver Physiol 305: G611–619. 10.1152/ajpgi.00111.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guicciardi ME, Werneburg NW, Bronk SF, Franke A, Yagita H, Thomas G, et al. (2014) Cellular inhibitor of apoptosis (cIAP)-mediated ubiquitination of phosphofurin acidic cluster sorting protein 2 (PACS-2) negatively regulates tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) cytotoxicity. PLoS One 9: e92124 10.1371/journal.pone.0092124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan Y, Adami G, Costa RH (2002) Maintaining HNF6 expression prevents AdHNF3beta-mediated decrease in hepatic levels of Glut-2 and glycogen. Hepatology 35: 790–798. 10.1053/jhep.2002.32482 [DOI] [PubMed] [Google Scholar]
- 37.Baserga R, Hongo A, Rubini M, Prisco M, Valentinis B (1997) The IGF-I receptor in cell growth, transformation and apoptosis. Biochim Biophys Acta 1332: F105–126. [DOI] [PubMed] [Google Scholar]
- 38.Gatto M, Drudi-Metalli V, Torrice A, Alpini G, Cantafora A, Biotta I, et al. (2008) Insulin-like growth factor-1 isoforms in rat hepatocytes and cholangiocytes and their involvement in protection against cholestatic injury. Lab Invest 88: 986–994. 10.1038/labinvest.2008.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pi-Chieh Wang K, Lee LM, Lin TJ, Sheen-Chen SM, Lin JW, Chiu WT, et al. (2011) Gene transfer of IGF1 attenuates hepatocellular apoptosis after bile duct ligation. J Surg Res 167: 237–244. 10.1016/j.jss.2009.07.051 [DOI] [PubMed] [Google Scholar]
- 40.Drudi Metalli V, Mancino MG, Mancino A, Torrice A, Gatto M, et al. (2007) Bile salts regulate proliferation and apoptosis of liver cells by modulating the IGF1 system. Dig Liver Dis 39: 654–662. 10.1016/j.dld.2007.03.008 [DOI] [PubMed] [Google Scholar]
- 41.Onori P, Alvaro D, Floreani AR, Mancino MG, Franchitto A, Guido M, et al. (2007) Activation of the IGF1 system characterizes cholangiocyte survival during progression of primary biliary cirrhosis. J Histochem Cytochem 55: 327–334. 10.1369/jhc.6R7125.2006 [DOI] [PubMed] [Google Scholar]
- 42.De Vas MG, Kopp JL, Heliot C, Sander M, Cereghini S, Haumaitre C. (2015) Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Development 142: 871–882. 10.1242/dev.110759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Brems JJ, Gamelli RL, Holterman AX (2013) Foxa2 may modulate hepatic apoptosis through the cIAP1 pathway. Cell Signal 25: 867–874. 10.1016/j.cellsig.2012.12.012 [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Laz EV, Waxman DJ (2012) Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol Cell Biol 32: 880–896. 10.1128/MCB.06312-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delesque-Touchard N, Park SH, Waxman DJ (2000) Synergistic action of hepatocyte nuclear factors 3 and 6 on CYP2C12 gene expression and suppression by growth hormone-activated STAT5b. Proposed model for female specific expression of CYP2C12 in adult rat liver. J Biol Chem 275: 34173–34182. 10.1074/jbc.M004027200 [DOI] [PubMed] [Google Scholar]
- 46.Koyama Y, Brenner DA (2015) New therapies for hepatic fibrosis. Clin Res Hepatol Gastroenterol 39 Suppl 1: S75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, et al. (2005) Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev 19: 1849–1854. 10.1101/gad.340305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Ables ET, Pope CF, Washington MK, Hipkens S, Means AL, et al. (2009) Multiple, temporal-specific roles for HNF6 in pancreatic endocrine and ductal differentiation. Mech Dev 126: 958–973. 10.1016/j.mod.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebert G, Preston S, Allison C, Cooney J, Toe JG, Stutz MD, et al. (2015) Cellular inhibitor of apoptosis proteins prevent clearance of hepatitis B virus. Proc Natl Acad Sci U S A 112: 5797–5802. 10.1073/pnas.1502390112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arnt CR, Chiorean MV, Heldebrant MP, Gores GJ, Kaufmann SH (2002) Synthetic Smac/DIABLO peptides enhance the effects of chemotherapeutic agents by binding XIAP and cIAP1 in situ. J Biol Chem 277: 44236–44243. 10.1074/jbc.M207578200 [DOI] [PubMed] [Google Scholar]
- 51.Bockbrader KM, Tan M, Sun Y (2005) A small molecule Smac-mimic compound induces apoptosis and sensitizes TRAIL- and etoposide-induced apoptosis in breast cancer cells. Oncogene 24: 7381–7388. 10.1038/sj.onc.1208888 [DOI] [PubMed] [Google Scholar]
- 52.Lucifora J, Trepo C (2015) Hepatitis: After HCV cure, HBV cure? Nat Rev Gastroenterol Hepatol 12: 376–378. 10.1038/nrgastro.2015.103 [DOI] [PubMed] [Google Scholar]
- 53.Saleem M, Qadir MI, Perveen N, Ahmad B, Saleem U, Irshad T, et al. (2013) Inhibitors of apoptotic proteins: new targets for anticancer therapy. Chem Biol Drug Des 82: 243–251. 10.1111/cbdd.12176 [DOI] [PubMed] [Google Scholar]
- 54.Bonefeld K, Moller S (2011) Insulin-like growth factor-I and the liver. Liver Int 31: 911–919. 10.1111/j.1478-3231.2010.02428.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cluster diagram of the Cluster Analysis showing Hnf6-bound genes. R2 measures Hierarchical cluster analysis using MAT (model-based analysis of tiling-array) scores with Ciap1 (or Birc2, denoted with *) having the highest MAT-score (664), and significant variance.
(DOC)
The table depicts Hnf6-bound genes with Enrichment Scores ≥ 1.25 (corresponding to a ≤ 0.05) and Group Enrichment Score ranking by biological significance based on overall EASE (Expression Analysis Systematic Explorer) scores (corresponding to the one-tailed Fisher exact probability of gene over-representation by functional class). Baculoviral IAP repeat-containing 2 (NM_007465, Birc2 or Ciap1) is shown in Group 4, with Enrichment Score of 1.59.
(DOC)
Data Availability Statement
The ChIP-on-ChIP data files are available from the://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80498.