Abstract
Fusidic acid-resistant epidemic Staphylococcus aureus strains causing impetigo bullosa have been reported in Scandinavia. We show that these strains form part of a European epidemic clonotype that carries the fusB determinant. In contrast, resistance to fusidic acid in a collection of nonepidemic strains resulted primarily from mutations in fusA.
Fusidic acid inhibits bacterial protein synthesis by interfering with dissociation of elongation factor G (EF-G) from the ribosome (15). It is frequently employed as a topical agent for treatment of superficial staphylococcal skin infections, including impetigo and atopic dermatitis (2, 5). Limited data suggest that resistance to fusidic acid in clinical isolates of Staphylococcus aureus arises from mutations in the gene encoding EF-G (fusA) (1, 8) or by acquisition of a plasmid determinant (fusB) that encodes a poorly characterized resistance mechanism (3, 9, 12).
Recently, fusidic acid-resistant epidemic strains of S. aureus causing impetigo bullosa were reported in Sweden and Norway (11, 16). In this paper we demonstrate that resistant strains reported in these Scandinavian outbreaks are clonally related and exist in other European countries. Furthermore, we found that this clonotype carries the fusB resistance determinant. In contrast, fusidic acid resistance arising in nonepidemic strains was attributed, in several cases, to mutations in fusA.
Clinical S. aureus isolates in which the genetic basis of fusidic acid resistance was established are listed in Table 1. In addition to representatives of epidemic fusidic acid-resistant strains from Scandinavia and Ireland, resistant strains collected during a comparative phase III study (FCF0001 INT) and from a dermatology unit in Harrogate, United Kingdom (strain designations with an H prefix) (13), were examined. Detection of fusA mutations in these strains was performed by PCR amplification and sequencing, as previously described (10). The fusB determinant was detected by Southern hybridization (14) with the Alkphos Direct kit (Amersham Biosciences, Amersham, United Kingdom) using the entire fusB gene (A. J. O'Neill and I. Chopra, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1064, 2002) as a probe. Further isolates were screened by PCR amplification of a 292-bp fragment of the fusB gene using oligonucleotide primers FB(iii) (5′-ATTCAATCGGAAACCTATAATGATA) and FB(iv) (5′-TTATATATTTCCGATTTGATGCAAG). Using an annealing temperature of 60°C, it was established that these primers generated an amplicon only from strains known to carry fusB (data not shown).
TABLE 1.
Mechanisms of fusidic acid resistance in clinical isolates of S. aureusa
| Strain | Country of origin | Reference or source | Fusidic acid MIC (μg/ml)b | Mechanism of fusidic acid resistance
|
||
|---|---|---|---|---|---|---|
| fusB | Polymorphism(s) in EF-G | Other (undefined) | ||||
| CS944 | Denmark | FCF0001 INT | 0.047 | − | − | |
| CS734 | Finland | FCF0001 INT | 0.094 | − | − | |
| CS675 | Belgium | FCF0001 INT | 0.125 | − | − | |
| CS823 | Spain | FCF0001 INT | 0.125 | − | − | |
| CS840 | Denmark | FCF0001 INT | 0.125 | − | − | |
| CS726 | Germany | FCF0001 INT | 0.125 | − | − | |
| CS689 | Spain | FCF0001 INT | 0.19 | − | − | |
| CS749 | Belgium | FCF0001 INT | 0.19 | − | V607I | |
| CS1121 | Denmark | FCF0001 INT | 0.47 | − | − | |
| CS1116 | Belgium | FCF0001 INT | 1.5 | − | H457Y (mixed population) | |
| CS992 | Denmark | FCF0001 INT | 2 | − | − | + |
| CS642 | Germany | FCF0001 INT | 2 | + | − | |
| CS730 | Finland | FCF0001 INT | 3 | − | − | + |
| CS808 | France | FCF0001 INT | 3 | − | − | + |
| CS866 | Denmark | FCF0001 INT | 3 | − | − | + |
| CS957 | Denmark | FCF0001 INT | 3 | − | G451V | |
| CS18 | Norway | 16 | 3 | + | − | |
| CS607 | Denmark | FCF0001 INT | 4 | + | − | |
| CS11 | Norway | 16 | 4 | + | − | |
| CS21 | Ireland | LEO Pharma Ireland | 4 | + | − | |
| CS6 | Sweden | 11 | 4 | + | − | |
| CS9 | Sweden | 11 | 4 | + | − | |
| H18 | UK | 13 | 4 | + | − | |
| H19 | UK | 13 | 4 | + | − | |
| H44 | UK | 13 | 4 | + | − | |
| H45 | UK | 13 | 4 | + | − | |
| H47 | UK | 13 | 4 | + | − | |
| H49 | UK | 13 | 4 | + | − | |
| H83 | UK | 13 | 4 | + | − | |
| H78 | UK | 13 | 6 | + | − | |
| H17 | UK | 13 | 8 | + | − | |
| CS767 | Belgium | FCF0001 INT | 12 | − | G452S | |
| CS858 | Spain | FCF0001 INT | 48 | − | H457Y | |
| CS688 | Spain | FCF0001 INT | 64 | − | H457Y | |
| CS697 | Belgium | FCF0001 INT | 64 | − | H457Y | |
| CS735 | Germany | FCF0001 INT | 96 | − | H457Y | |
| CS741 | Finland | FCF0001 INT | 96 | − | A71T, P404L, L461S | |
| CS1145 | Denmark | FCF0001 INT | >256 | − | A67T, H457Y | |
| CS854 | Denmark | FCF0001 INT | >256 | − | A67T, H457Y | |
| CS872 | Denmark | FCF0001 INT | >256 | − | L461K | |
| CS874 | Denmark | FCF0001 INT | >256 | − | L461K | |
Representatives of the epidemic clonotype are in bold.
The resistance breakpoint for fusidic acid recommended by the British Society for Antimicrobial Chemotherapy is 2 μg/ml (6).
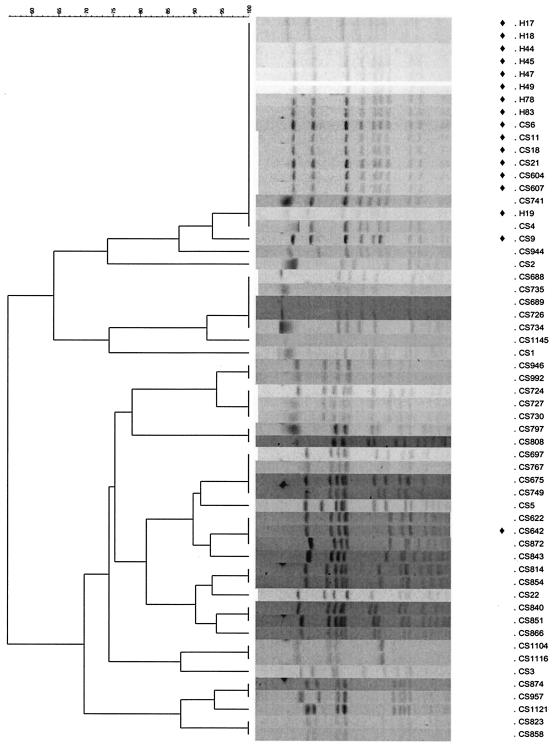
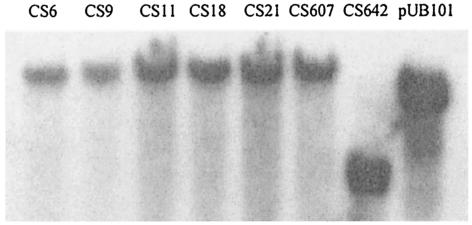
Pulsed-field gel electrophoresis analysis (7) of epidemic Fusr strains from Norway, Sweden, Ireland, and the United Kingdom indicated that they constitute a single clonotype (Fig. 1). The clone exhibited low-level resistance to fusidic acid (MIC, ∼4 μg/ml) (Table 1) and carried fusB. The fusB gene was detected in total DNA preparations from these strains (Fig. 2) but not in plasmid DNA preparations, indicating a chromosomal location for this resistance determinant.
FIG. 1.
Dendrogram and individual PFGE profiles showing relationships between S. aureus strains employed in this study. The cluster representing the epidemic European clonotype is at the top of the figure. Strains carrying the fusB determinant are indicated by the diamonds.
FIG. 2.
Detection of fusB in total DNA from clinical fusidic acid-resistant S. aureus strains. Southern hybridization with EcoRI-digested total DNA derived from six representatives of the epidemic clonotype (CS6 to CS607) and a distinct strain (CS642). Plasmid pUB101 is the positive control.
The fusidic acid MIC for the epidemic strains (Table 1) was ∼4-fold lower than that associated with strains carrying fusB on the archetypal fusidic acid resistance plasmid, pUB101 (MIC of 16 μg of fusidic acid/ml) (3). This is probably due to differences in resistance gene dosage, since chromosomal carriage likely involves a single copy of fusB, while pUB101 is typically present at 11 to 14 copies/cell (4).
Other Fusr strains belonging to this clonotype from dermatology patients in Harrogate, United Kingdom (13), were tested for the presence of fusB by PCR. A 292-bp amplicon was generated from strains belonging to the epidemic clonotype (Fig. 1; strains with the prefix H), indicating the presence of fusB (data not shown).
The genetic basis of fusidic acid resistance was also examined in nonepidemic isolates recovered from patients undergoing topical fusidic acid treatment for atopic dermatitis. In instances where fusidic acid resistance arose during treatment, both initial (fusidic acid-susceptible) and final (fusidic acid-resistant) isolates were examined.
One nonepidemic strain (CS642) carried fusB (Fig. 2) and exhibited low-level resistance (Table 1). As for the epidemic strains, fusB appeared to be chromosomally located in strain CS642. Higher-level fusidic acid resistance (MIC > 12 μg/ml) in the nonepidemic strains resulted exclusively from nucleotide substitutions in fusA (Table 1). In addition, two low-level Fusr strains (CS1116 and CS957) were also resistant as a result of mutation in EF-G (Table 1). All mutations identified in EF-G save one (A71T) have previously been shown to confer fusidic acid resistance or to participate in compensatory adaptation to the costs of fusidic acid resistance (8, 10). Since an alternative Fusr substitution at the A71 locus (A71V) has been described (8), the A71T mutation discussed above is likely to confer resistance to fusidic acid. Several mutations (P404L, G451V, and G452S) previously identified only in Fusr strains derived in vitro (8, 10) were detected here for the first time in clinical S. aureus isolates (Table 1).
In four nonepidemic strains exhibiting relatively low-level resistance (CS992, CS730, CS808, and CS866), neither fusB nor polymorphic variations in fusA were detected. This suggests that while mutations in fusA and possession of fusB are common routes to fusidic acid resistance in clinical isolates, other mechanisms may also exist.
In conclusion, we have demonstrated that several fusidic acid-resistant clinical isolates of S. aureus recovered from patients with impetigo bullosa in European countries represent a clonal epidemic strain carrying the fusB determinant on the chromosome. Mutations in fusA were identified in nonepidemic fusidic acid-resistant strains, but such mutations were not identified in the clonal epidemic strain. The factors favoring dissemination of the epidemic clonal strain have yet to be identified.
Acknowledgments
This work was supported by a research grant to I. Chopra from LEO Pharma, Ballerup, Denmark.
We thank J. Ravenscroft, Y. Tveten, and G. Kahlmeter for provision of strains and A. M. Moita for technical assistance.
REFERENCES
- 1.Besier, S., A. Ludwig, V. Brade, and T. A. Wichelhaus. 2003. Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol. Microbiol. 47:463-469. [DOI] [PubMed] [Google Scholar]
- 2.Brown, E. M., and P. Thomas. 2002. Fusidic acid resistance in Staphylococcus aureus isolates. Lancet 359:803. [DOI] [PubMed] [Google Scholar]
- 3.Chopra, I. 1976. Mechanisms of resistance to fusidic acid in Staphylococcus aureus. J. Gen. Microbiol. 96:229-238. [DOI] [PubMed] [Google Scholar]
- 4.Chopra, I., P. M. Bennett, and R. W. Lacey. 1973. A variety of staphylococcal plasmids present as multiple copies. J. Gen. Microbiol. 79:343-345. [DOI] [PubMed] [Google Scholar]
- 5.Greenwood, D. 2003. Fusidanes, p. 297-299. In R. G. Finch (ed.), Antibiotic & chemotherapy, 8th ed. Churchill Livingstone, London, United Kingdom.
- 6.MacGowan, A. P., and R. Wise. 2001. Establishing MIC breakpoints and the interpretation of in vitro susceptibility tests. J. Antimicrob. Chemother. 48(Suppl. 1):17-28. [DOI] [PubMed] [Google Scholar]
- 7.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagaev, I., J. Bjorkman, D. I. Andersson, and D. Hughes. 2001. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 40:433-439. [DOI] [PubMed] [Google Scholar]
- 9.O'Brien, F. G., C. Price, W. B. Grubb, and J. E. Gustafson. 2002. Genetic characterization of the fusidic acid and cadmium resistance determinants of Staphylococcus aureus plasmid pUB101. J. Antimicrob. Chemother. 50:313-321. [DOI] [PubMed] [Google Scholar]
- 10.O'Neill, A. J., J. Bostock, A. Morais Moita, and I. Chopra. 2002. Antimicrobial activity and mechanisms of resistance to cephalosporin P1, an antibiotic related to fusidic acid. J. Antimicrob. Chemother. 50:839-848. [DOI] [PubMed] [Google Scholar]
- 11.Osterlund, A., T. Eden, B. Olsson-Liljequist, S. Haeggman, and G. Kahlmeter. 2002. Clonal spread among Swedish children of a Staphylococcus aureus strain resistant to fusidic acid. Scand. J. Infect. Dis. 34:729-734. [DOI] [PubMed] [Google Scholar]
- 12.Projan, S. J. 2000. Antibiotic resistance in the staphylococci, p. 463-470. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 13.Ravenscroft, J. C., A. Layton, and M. Barnham. 2000. Observations on high levels of fusidic acid resistant Staphylococcus aureus in Harrogate, North Yorkshire, UK. Clin. Exp. Dermatol. 25:327-330. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook, J., and D. W. Russell. 2001. Molecular cloning; a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Tanaka, N., T. Kinoshita, and H. Masukawa. 1968. Mechanism of protein synthesis inhibition by fusidic acid and related antibiotics. Biochem. Biophys. Res. Commun. 30:278-283. [DOI] [PubMed] [Google Scholar]
- 16.Tveten, Y., A. Jenkins, and B. E. Kristiansen. 2002. A fusidic acid-resistant clone of Staphylococcus aureus associated with impetigo bullosa is spreading in Norway. J. Antimicrob. Chemother. 50:873-876. [DOI] [PubMed] [Google Scholar]