Abstract
Colorectal cancer (CRC) is the fourth-ranked cause of cancer-related deaths worldwide. Despite recent advances in CRC management, distant recurrence (DR) remains the major cause of mortality in patients with preoperative chemotherapy and radiotherapy, underscoring a need to precisely identify novel gene signatures for predicting the risk of systemic relapse. Herein, we integrated two independent CRC gene expression datasets: the GSE71222 dataset, including 26 patients who developed DR and 126 patients who did not develop DR, and the GSE21510 dataset, including 23 patients who developed DR and 76 patients who did not develop DR. Our data revealed 37 common upregulated genes (fold change (FC) ≥ 1.5, P < 0.05) and three common downregulated genes (FC ≤ 1.5, P < 0.05) between DR and non-recurrent patients from the two datasets. We subsequently validated the upregulated gene panel in the Cancer Genome Atlas CRC datasets (379 patients), which identified a five-gene signature (S100A2, VIP, HOXC6, DACT1, KIF26B) associated with poor overall survival (OS, log-rank test P-value: 1.19 × 10−4) and poor disease-free survival (DFS, log-rank test P-value: 0.002). In a Cox proportional hazards multiple regression model, the five-gene signature and tumor stage retained their significance as independent prognostic factors for CRC DFS and OS. Therefore, our data identified a novel DR gene expression signature associated with worse prognosis in CRC.
Introduction
Colorectal cancer (CRC) is one of the most prevalent types of cancers and is currently ranked as the fourth leading cause of cancer-related deaths globally, and the third leading cause of death in the United States in both men and women [1, 2]. The 5-year survival rate for CRC patients with a localized tumor is approximately 90%, which declines to 70% for patients with regional disease, and to 12% for patients with metastatic disease [2]. Multiple molecular alterations occur during CRC development and progression. Therefore, the identification of clinical and pathological parameters that can accurately predict the prognosis of patients with CRC has been a daunting task. Some of the factors to consider for predicting the risk of systemic relapse include the differentiation status of the tumor, depth of tumor invasion, and vascular and perineural invasion [3, 4]. Over the past several years, numerous molecular signatures have been identified for CRC prognosis [5–7]. However, one major problem with many of the established molecular signatures for CRC relapse is the lack of validation across different groups and platforms. Therefore, large-scale analysis of multiple gene expression datasets might lead to the identification of more representative gene expression signatures associated with CRC relapse. Herein, we integrated three independent CRC gene expression datasets retrospectively, which led to the identification of a novel five-gene signature associated with CRC systemic relapse.
Materials and Methods
Patient information and data analysis
The current study was conducted on three different CRC cohorts: (1) the National Center for Biotechnology Information Gene Expression Omnibus (GEO) GSE71222 dataset, which included 26 patients who developed distant recurrence (DR) and 126 patients who did not develop DR; (2) the GSE21510 dataset, which included 23 patients who developed DR and 76 patients who did not develop DR; and (3) The Cancer Genome Atlas (TCGA) CRC dataset, which included a total of 379 CRC patients. Interrogation of the TCGA dataset was conducted as previously described [8–10]. The relationship of gene expression patterns with patient survival in the TCGA database was queried using the cBioportal database with the formula GENE: EXP > 0, where GENE represents a query gene. The clinical characteristics for the TCGA dataset are shown in Table 1. The clinical characteristics for the GSE71222 and GSE21510 datasets have been described previously [11, 12].
Table 1. The Cancer Genome Atlas CRC dataset patient and tumor characteristics.
| N = 379 | % | |
|---|---|---|
| Age, years | ||
| Median age | 66 | |
| Range | 31–90 | |
| Gender | ||
| Male | 206 | 54.4 |
| Female | 168 | 44.3 |
| Unknown | 5 | 1.3 |
| Overall survival, months | ||
| Median | 22.04 | |
| Range | 0–147.9 | |
| Disease-free survival, months | ||
| Median | 20.27 | |
| Range | 0–147.9 | |
| Stage | ||
| I | 56 | 14.8 |
| II | 135 | 35.6 |
| III | 112 | 29.6 |
| IV | 52 | 13.7 |
| NA | 24 | 6.3 |
Microarray data analysis
The GSE71222 and GSE21510 raw gene expression datasets were retrieved from the GEO and were imported into GeneSpring 13.0 software (Agilent Technologies, Palo Alto, CA, USA). Raw data were subsequently normalized using the percentile shift, and a 1.5 fold-change (FC) cutoff and P < 0.05 were used to determine significantly changed transcripts between groups [13].
Statistical analysis
Kaplan-Meier survival curve comparison was conducted using the log-rank test, and a P-value of ≤0.05 was considered statistically significant. The Cox proportional hazards multiple regression model was used to identify the independent prognostic factors and to correct the effect of potential confounding variables, such as gender (male vs female), age (> 65y vs < 65y), tumor stage (stage 3/4 vs stage 1/2), and of cancer type (colon adenocarcinoma vs rectal adenocarcinoma vs mucinous adenocarcinoma of the colon and rectum) on OS and DFS using MedCalc 16.8.4 (MedCalc, Mariakerke, Belgium). Pathway analyses were conducted using DAVID functional annotation and clustering bioinformatics tool, as described in our previous reports [14, 15]. Statistical analyses and graphing were performed using Graphpad Prism 6.0 software (Graphpad Software, San Diego, CA, USA).
Results
Generation of a gene expression panel associated with risk of DR
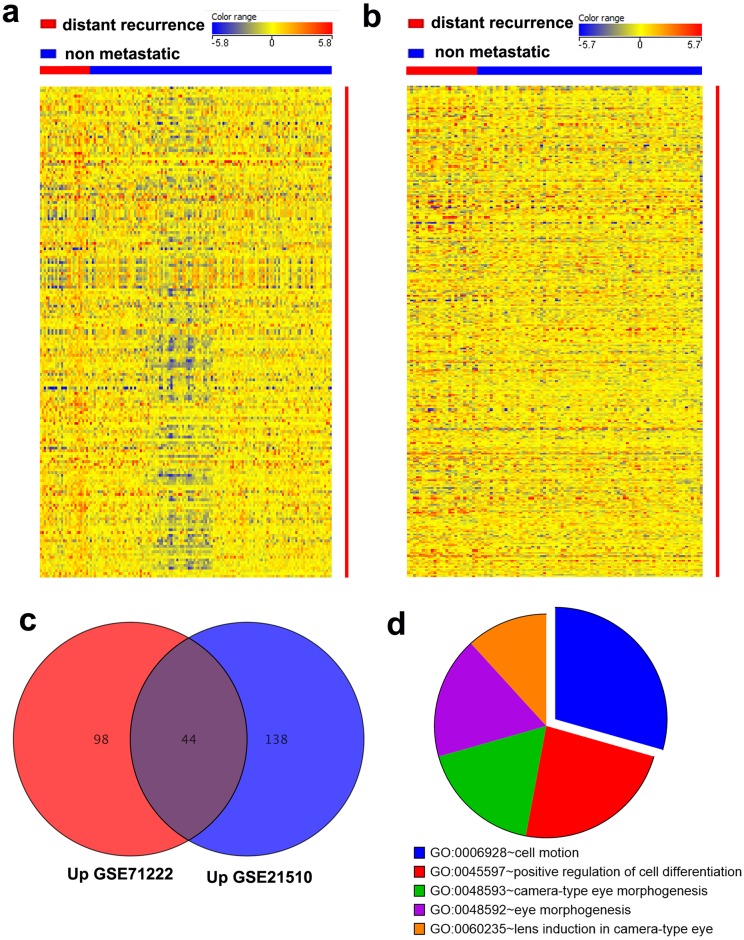
To devise a gene expression panel associated with CRC DR with high confidence, we analyzed two independent CRC gene expression datasets (GSE71222 and GSE21510) and identified the genes associated with patient recurrence. Analysis of the GSE71222 and GSE21510 datasets revealed 180 (1.5 FC, P < 0.05) and 317 (1.5 FC, P < 0.05) differentially expressed transcripts between DR and non-metastatic tumors, respectively (Fig 1a and 1b). To identify DR-related genes with high confidence, we crossed the differentially expressed genes from the two datasets that revealed 44 common upregulated transcripts, comprising 37 genes (Fig 1c, Table 2), and three common downregulated genes (Table 2). Pathway analysis performed on the common upregulated genes revealed enrichment in several cellular pathways, including cell motion and regulation of cell differentiation (Fig 1d).
Fig 1. Genes associated with CRC distant recurrence (DR).
Heatmap depicting the expression levels of differentially expressed genes (1.5 fold changes and P ≤ 0.05) between DR and non-recurrent (NR) CRC patients from the GSE71222 (a) and GSE21510 (b) datasets. Each column represents an individual sample and each row represents a single transcript. The expression level of each mRNA in a single sample is depicted according to the color scale. (c) Venn diagram depicting the common upregulated genes between DR and NR CRC samples from the GSE71222 and GSE21510 datasets. (d) Pie chart illustrating the distribution of the top 5 pathway designations for the 44 common upregulated transcripts from (c). The pie size corresponds to the number of matched entities.
Table 2. Common recurrence-related genes in the GSE71222 and GSE21510 datasets.
| Gene Symbol | FC (GSE71222) | FC GSE21510 |
|---|---|---|
| Upregulated genes | ||
| LAMC2 | 1.51 | 1.60 |
| SERPINA3 | 1.58 | 2.33 |
| LPL | 1.74 | 2.42 |
| S100A2 | 1.79 | 2.12 |
| PROM1 | 1.99 | 2.33 |
| COL9A3 | 1.77 | 2.13 |
| SERPINB5 | 1.85 | 2.57 |
| TNFRSF11B | 2.08 | 2.28 |
| TCN1 | 2.15 | 2.73 |
| C4BPA | 1.60 | 2.12 |
| SLC14A1 | 1.50 | 1.80 |
| REG1B | 2.50 | 2.42 |
| VIP | 1.67 | 2.06 |
| HOXC6 | 1.75 | 2.50 |
| MSX2 | 1.56 | 1.63 |
| BMP4 | 1.50 | 1.60 |
| TNIK | 1.62 | 1.56 |
| PRUNE2 | 1.71 | 1.66 |
| KRT6B | 1.90 | 3.45 |
| NOV | 1.62 | 1.73 |
| TESC | 1.71 | 1.83 |
| DACT1 | 1.52 | 1.72 |
| BHLHE41 | 1.60 | 2.06 |
| ABHD2 | 1.59 | 1.58 |
| AMIGO2 | 1.90 | 1.87 |
| DCDC2 | 1.82 | 2.18 |
| CD109 | 1.67 | 1.86 |
| EPHA4 | 1.80 | 2.32 |
| PPP2R2C | 1.71 | 1.85 |
| SOX2 | 1.58 | 1.82 |
| EPHB1 | 1.84 | 2.03 |
| GPR155 | 1.72 | 1.72 |
| SBSPON | 1.86 | 1.93 |
| TMEM71 | 2.16 | 2.91 |
| KIF26B | 1.97 | 1.52 |
| C3ORF70 | 1.50 | 1.70 |
| CPA6 | 1.56 | 1.76 |
| Downregulated genes | ||
| PTPRD | -2.09 | -2.04 |
| PID1 | -1.52 | -1.54 |
| ELF5 | -1.67 | -1.62 |
Selected genes are based on a fold-change (FC) of 1.5 and P < 0.05 cut-off threshold.
Validation of the DR-associated gene panel in the TCGA CRC dataset
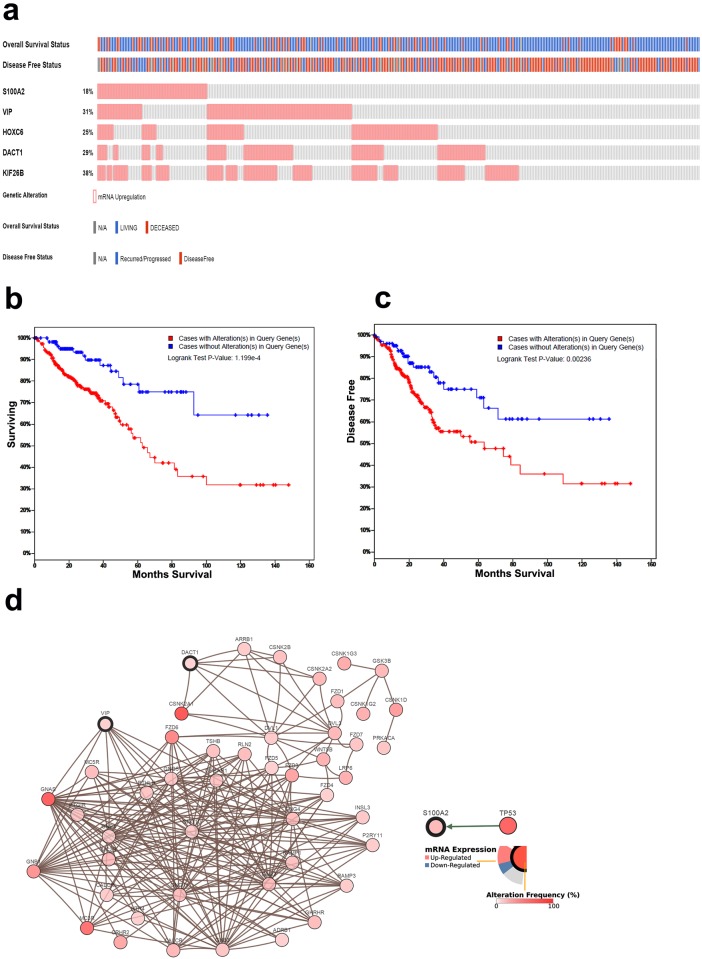
We subsequently focused on the potential role of the upregulated genes in CRC recurrence. Therefore, each of the 37 upregulated genes was further validated using the TCGA CRC dataset to determine their relationship to overall survival (OS) and disease-free survival (DFS). S100A2, VIP, HOXC6, DACT1, and KIF26B were significantly associated with OS (P≤0.01) and DFS (P≤0.05), while LAMC2, NOV, and AMIGO2 were only associated with DFS (P≤0.05). We subsequently focused on the five-gene panel that was associated with OS and DFS. The OncoPrint for this gene panel in the TCGA CRC dataset with the proportion of patients overexpressing each gene is presented in Fig 2a. Interestingly, the combination of this five-gene panel revealed a higher prognostic value, in which patients overexpressing at least one of the five genes showed a worse OS (log-rank test P-value: 1.19 × 10−4, Fig 2b) and worse DFS (log-rank test P-value: 0.002, Fig 2c) than those with lower expression of these genes. Data from the univariate analysis were subsequently put into the Cox proportional hazards multiple regression model to identify the independent factors for prognosis. The results showed that expression of the five-gene panel and tumor stage retained their significance as independent prognostic factors for CRC DFS and OS (p = 0.0023 and 0.0001 for DFS and p = 0.0086 and <0.0001 for OS, respectively), while age at diagnosis only correlated with OS, p = 0.0004 (Table 3). Network analysis of this five-gene signature revealed multiple network interactions in CRC, such as between VIP and GNG11, GNB3, GNG12, GNB2, GNG5, GNAS, GNG2, GNB4, GNG4, GNG10, and GNB1; between DACT1 and ARRB1, DVL1, CSNK2B, CSNK2A1, and CSNK2A2; and between S100A2 and TP53 (Fig 2d).
Fig 2. Validation of the distant recurrence (DR) gene panel in the TCGA dataset.
(a) OncoPrint of the DR five-gene signature in the TCGA CRC dataset. Alteration in the expression of different members of the five-gene signature (rows) in relation to each sample (columns). Relationships to overall and disease-free survival are also shown. CRC cases with upregulated expression of the DR signature showed worse overall (b) and disease-free (c) survival than cases with lower expression. (d) Network view of the VIP/DACT1/S100A2 neighborhood in CRC. VIP, DACT1, and S100A2 are seed genes (indicated with thick borders), and all other genes were identified as altered in CRC.
Table 3. Multivariate analyses for the prognostic value of the 5-gene signature in TCGA CRC dataset.
| Parameters | Categories | DFS hazard ratio (95% CI) | P value | OS hazard ratio (95% CI) | P value |
|---|---|---|---|---|---|
| Five-gene expression | High vs Low | 1.95 (1.27 to 3.01) | 0.0023 | 1.84 (1.16 to 2.90) | 0.0086 |
| Age at diagnosis | <65 vs. >65 | 0.86 (0.56 to 1.33) | 0.5103 | 2.47 (1.50 to 4.09) | 0.0004 |
| Type | CA vs RA vs MA | 0.87 (0.67 to 1.12) | 0.2925 | 1.14 (0.86 to 1.51) | 0.3560 |
| Tumor stage | (3/4 vs 1/2) | 1.92 (1.37 to 2.69) | 0.0001 | 3.18 (1.96 to 5.16) | <0.0001 |
| Gender | M vs F | 1.35 (0.86 to 2.10) | 0.1857 | 1.04 (0.65 to 1.65) | 0.8614 |
CA: Colon Adenocarcinoma; RA: Rectal Adenocarcinoma; MA: Mucinous Adenocarcinoma of the Colon and Rectum
Discussion
In the current study, we retrospectively derived and validated a gene expression signature associated with the risk of systemic relapse in patients with CRC. Analysis of the GSE71222 and GSE21510 datasets identified 37 upregulated and three downregulated genes associated with DR in CRC. Interestingly, several of the identified genes (LAMC2, LPL, SERPINB5, TCN1, VIP, MSX2, PRUNE2, KRT6B, TESC, EPHA4, GPR155, KIF26B, C3ORF70, and PID1) were also found to be differentially expressed in our previous global mRNA expression profiling of CRC compared to adjacent normal mucosa, suggesting a plausible role of these genes in driving CRC in addition to DR [16]. Concordant with our data, Takahashi and colleagues [11] reported a worse prognosis in CRC patients overexpressing Traf2- and Nck-interacting kinase (TNIK). Higher expression of MSX2 was found to be associated with metastasis in different types of human cancers [17]. PROM1, also known as CD133, was among the 37 upregulated genes in both datasets. Interestingly, PROM1 has previously been reported as a cancer stem cell marker in CRC [18, 19]. Similarly, two of the identified genes in the current study (SLC14A1 and KIF26B) were identified in an intestinal stem cell signature previously reported to be associated with poor clinical outcome in CRC [20]. Therefore, it is possible that patients with an enriched CSC phenotype are more likely to develop DR. We subsequently validated this gene signature in the TCGA CRC dataset, which includes 379 patients. Our analysis narrowed down the CRC recurrence signature to five genes (S100A2, VIP, HOXC6, DACT1, and KIF26B) whose expression was associated with poor OS (log-rank test P-value: 1.19 × 10−4) and DFS (log-rank test P-value: 0.002), which was further confirmed in a multivariate analysis. Therefore, we here present a novel gene expression signature for predicting the risk of systemic relapse in CRC. Concordant with our data, overexpression of S100A2 has been associated with poor clinical outcome in colorectal [21] and oral [22] cancers. The HOXC6 gene is frequently upregulated in prostate cancer, although no association with patient relapse was observed [23]. DACT1 was recently shown to promote CRC tumorigenicity and invasion via stabilization of β-catenin [24]. Concordantly, overexpression of DACT1 was observed during the transition of ductal carcinoma in situ to invasive ductal carcinoma in breast cancer [25].
Conclusion
Herein, we integrated multiple gene expression datasets and devised a novel five-gene signature as an independent predictor of CRC DR. This signature adds to the current prognostic value of tumor staging. Before this five-gene-signature can be utilized in the clinic; however, additional validations are required
Acknowledgments
We would like to thank the Deanship of Scientific Research, King Saud University, Riyadh, Saudi Arabia for their support.
Data Availability
Data are available form the NCBI Gene Expression Omnibus (GEO) under accession numbers: GSE71222 and GSE21510.
Funding Statement
This work was supported by the Deanship of Scientific Research, King Saud University, Riyadh, Saudi Arabia. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clinics in colon and rectal surgery. 2009;22(4):191–7. 10.1055/s-0029-1242458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(2):104–17. [DOI] [PubMed] [Google Scholar]
- 3.Tsai HL, Cheng KI, Lu CY, Kuo CH, Ma CJ, Wu JY, et al. Prognostic significance of depth of invasion, vascular invasion and numbers of lymph node retrievals in combination for patients with stage II colorectal cancer undergoing radical resection. Journal of surgical oncology. 2008;97(5):383–7. 10.1002/jso.20942 [DOI] [PubMed] [Google Scholar]
- 4.Knijn N, Mogk SC, Teerenstra S, Simmer F, Nagtegaal ID. Perineural Invasion is a Strong Prognostic Factor in Colorectal Cancer: A Systematic Review. The American journal of surgical pathology. 2016;40(1):103–12. 10.1097/PAS.0000000000000518 [DOI] [PubMed] [Google Scholar]
- 5.Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. The New England journal of medicine. 2001;344(16):1196–206. 10.1056/NEJM200104193441603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salazar R, Roepman P, Capella G, Moreno V, Simon I, Dreezen C, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(1):17–24. [DOI] [PubMed] [Google Scholar]
- 7.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS medicine. 2013;10(5):e1001453 10.1371/journal.pmed.1001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6(269):pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alajez NM. Significance of BMI1 and FSCN1 expression in colorectal cancer. Saudi J Gastroenterol. 2016;22(4):288–93. 10.4103/1319-3767.187602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi H, Ishikawa T, Ishiguro M, Okazaki S, Mogushi K, Kobayashi H, et al. Prognostic significance of Traf2- and Nck- interacting kinase (TNIK) in colorectal cancer. BMC cancer. 2015;15:794 10.1186/s12885-015-1783-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukamoto S, Ishikawa T, Iida S, Ishiguro M, Mogushi K, Mizushima H, et al. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17(8):2444–50. [DOI] [PubMed] [Google Scholar]
- 13.Al-Toub M, Vishnubalaji R, Hamam R, Kassem M, Aldahmash A, Alajez NM. CDH1 and IL1-beta expression dictates FAK and MAPKK-dependent cross-talk between cancer cells and human mesenchymal stem cells. Stem cell research & therapy. 2015;6(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alajez NM, Shi W, Hui AB, Bruce J, Lenarduzzi M, Ito E, et al. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell death & disease. 2010;1:e85. Epub 2011/03/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-toub M, Almusa A, Almajed M, Al-Nbaheen M, Kassem M, Aldahmash A, et al. Pleiotropic effects of cancer cells' secreted factors on human stromal (mesenchymal) stem cells. Stem cell research & therapy. 2013;4(5):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vishnubalaji R, Hamam R, Abdulla MH, Mohammed MA, Kassem M, Al-Obeed O, et al. Genome-wide mRNA and miRNA expression profiling reveal multiple regulatory networks in colorectal cancer. Cell death & disease. 2015;6:e1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. 10.1038/ng1060 [DOI] [PubMed] [Google Scholar]
- 18.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–10. 10.1038/nature05372 [DOI] [PubMed] [Google Scholar]
- 19.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–5. 10.1038/nature05384 [DOI] [PubMed] [Google Scholar]
- 20.Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell stem cell. 2011;8(5):511–24. 10.1016/j.stem.2011.02.020 [DOI] [PubMed] [Google Scholar]
- 21.Masuda T, Ishikawa T, Mogushi K, Okazaki S, Ishiguro M, Iida S, et al. Overexpression of the S100A2 protein as a prognostic marker for patients with stage II and III colorectal cancer. International journal of oncology. 2016;48(3):975–82. 10.3892/ijo.2016.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar M, Srivastava G, Kaur J, Assi J, Alyass A, Leong I, et al. Prognostic significance of cytoplasmic S100A2 overexpression in oral cancer patients. Journal of translational medicine. 2015;13:8 10.1186/s12967-014-0369-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamid AR, Hoogland AM, Smit F, Jannink S, van Rijt-van de Westerlo C, Jansen CF, et al. The role of HOXC6 in prostate cancer development. The Prostate. 2015;75(16):1868–76. 10.1002/pros.23065 [DOI] [PubMed] [Google Scholar]
- 24.Yuan G, Wang C, Ma C, Chen N, Tian Q, Zhang T, et al. Oncogenic function of DACT1 in colon cancer through the regulation of beta-catenin. PloS one. 2012;7(3):e34004 10.1371/journal.pone.0034004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer research. 2006;66(10):5278–86. 10.1158/0008-5472.CAN-05-4610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available form the NCBI Gene Expression Omnibus (GEO) under accession numbers: GSE71222 and GSE21510.