Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) strains (n = 455) collected in 2001 from 100 Belgian hospitals were characterized by molecular typing and by resistance gene distribution to macrolides-lincosamides-streptogramins and to aminoglycoside antibiotics. Rapid diversification of MRSA clones, compared with results of previous surveys, was evidenced by the broad geographic distribution of seven major clones belonging to the pandemic MRSA clonal complexes 5, 8, 22, 30, and 45 by multilocus sequence typing.
In Belgium, the proportion of methicillin-resistant strains isolated from patients with Staphylococcus aureus bacteremia has risen from 23% in 1999 to 28% in 2002 (6). Since 1995, the methicillin-resistant S. aureus (MRSA) reference laboratory has organized epidemiological surveillance to monitor the evolution of genotypes and of antimicrobial resistance profiles of MRSA strains isolated from hospitalized patients in Belgium (3). During the last two surveys, performed in 1995 and 1997, molecular typing of MRSA strains showed the prominence of three major epidemic pulsed-field gel electrophoresis (PFGE) types, A1, B2, and C3, together comprising nearly 90% of isolates (3). The proportion of gentamicin-susceptible strains rose from 22 to 48%, in parallel with an increasing prevalence of type B2 strains. In this report, we describe the distribution of clonal types and resistance genes to aminoglycoside antibiotics and macrolides-lincosamides-streptogramins (MLS) in MRSA strains from a national survey conducted in 2001.
From January to December 2001, 455 nonduplicate, clinical MRSA isolates were referred from 100 acute-care hospitals (51% of Belgian sites) to the reference laboratory. In the reference laboratory, identification was confirmed by coagulase testing, oxacillin agar screening, and PCR for the detection of 16S rRNA and mecA and nuc genes (14). MICs for 17 antimicrobial agents were determined by the agar dilution method (15). NCCLS breakpoints were used for MIC interpretation for all drugs except fusidic acid and mupirocin, which were interpreted according to the Committee for Antimicrobial Testing of the French Society of Microbiology and British Society for Antimicrobial Chemotherapy, respectively (1, 2). Erythromycin and clindamycin were tested by the disk diffusion method to differentiate the MLS resistance phenotype as inducible or constitutive. The aminoglycoside-modifying enzymes (AME) encoded by aac(6′)-aph(2"), ant(4′), and aph(3′), the ribosomal methylases encoded by erm(A) and erm(C), the macrolide efflux pumps encoded by msr(A) and msr(B), and the streptogramin A inactivating enzyme encoded by vat(B) were tested by PCR (4, 22). MRSA strains with decreased susceptibility to mupirocin (MIC > 8 μg/ml) were tested by PCR for the mupA gene coding for the isoleucyl-tRNA synthetase 2 (3). All strains were genotyped by SmaI macrorestriction analysis of genomic DNA resolved by PFGE, and patterns were classified as previously described (3, 5). Epidemic MRSA type strains were defined as type strains recovered in more than five hospitals. Multilocus sequence typing (MLST) was performed on selected MRSA strains (n = 26) belonging to the major epidemic PFGE types (7). The staphylococcal cassette chromosome mec (SCCmec) type was determined for representative MRSA strains from each PFGE type by multiplex PCR (17).
In many countries, nosocomial MRSA clones were previously coresistant to multiple classes of antimicrobials, like aminoglycosides, MLS, quinolones, and rifampin (9). In 1995, MRSA strains collected from Belgian hospitals were on average coresistant to seven antimicrobials (3). In 2001, in contrast, MRSA strains were, on average, coresistant to only three antimicrobials other than beta-lactams. The loss of coresistance was particularly pronounced with aminoglycosides and clindamycin. The proportion of resistance to MLS (Table 1) was comparable to that reported in recent European surveys (9, 21). All but one MLS-resistant strain contained either the erm(A) (34%) or the erm(C) (30%) gene or a combination of both erm genes (0.4%) (Table 2), as observed in previous studies in Europe (12, 20). The proportion of erm(C) genes in MLS-resistant strains was higher than that previously observed (4). The single isolate resistant to quinupristin-dalfopristin harbored the vat(B) gene associated with the erm(A) gene. Eighty-seven percent of strains harboring the erm(A) gene expressed the constitutive MLS phenotype. On the other hand, the erm(C) gene was present in 95% of the strains with the inducible MLS phenotype. Most of the strains were susceptible to aminoglycosides except to tobramycin. Among aminoglycoside-resistant strains, 40% of isolates carried the ant(4′) gene, 12% of isolates carried the aac(6′)-aph(2") gene, and 5% of isolates carried the aph(3′) gene (Table 2). One aminoglycoside-susceptible isolate carried the ant(4′) gene. Previous European and Japanese surveys conducted in the 1990s found a higher prevalence of AME genes, from 53 to 85% for the ant(4′) gene and from 62 to 100% for the aac(6′)-aph(2") gene, whereas the aph(3′) gene was found less frequently (11, 19, 23). The proportion of high-level resistance to mupirocin remained low (3.5%) but has increased significantly over the last years (P = 0.007). All strains with high-level resistance to mupirocin carried the mupA gene. More than 95% of strains were resistant to ciprofloxacin, whereas >90% of strains were susceptible to fusidic acid, minocycline, and trimethoprim-sulfamethoxazole. All isolates were susceptible to linezolid and glycopeptides. However, borderline glycopeptide-susceptible strains (with a vancomycin MIC of 4 μg/ml and a teicoplanin MIC of 8 μg/ml) suggest the presence of isolates with resistant subpopulations. These strains are being further characterized by population analysis (13).
TABLE 1.
Cumulative percentages of MRSA strains (n = 455) inhibited by increasing concentrations of 17 antimicrobial drugs and distribution by susceptibility categorya
| Drug | % of strains susceptible at MIC (μg/ml) of:
|
% of strains per susceptibility category
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | >128 | >1,024b | S | I | R | |
| OXA | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 3 | 9 | 42 | 68 | 82 | 100 | 0.1 | 0 | 99.9 | |
| VAN | 0 | 0 | 0 | 14 | 54 | 99 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | |
| TEC | 0 | 2 | 16 | 64 | 89 | 97 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | |
| ERY | 0 | 2 | 16 | 36 | 36 | 36 | 36 | 36 | 36 | 37 | 39 | 50 | 100 | 36.0 | 0 | 64.0 | |
| CLI | 13 | 33 | 58 | 62 | 62 | 62 | 62 | 62 | 62 | 62 | 62 | 62 | 100 | 62.2 | 0.2 | 37.6 | |
| Q-D | 1 | 6 | 31 | 88 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.8 | 0 | 0.2 | |
| CIP | 0 | 0 | 1 | 5 | 5 | 5 | 6 | 11 | 23 | 29 | 36 | 51 | 100 | 5.1 | 0.4 | 94.5 | |
| LZD | 0 | 0 | 0 | 2 | 12 | 53 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | |
| GEN | 1 | 10 | 74 | 88 | 88 | 89 | 89 | 89 | 89 | 90 | 92 | 98 | 100 | 88.6 | 0.2 | 11.2 | |
| AMK | 1 | 1 | 1 | 3 | 33 | 58 | 64 | 80 | 94 | 98 | 99 | 99 | 100 | 93.8 | 4.4 | 1.8 | |
| NET | 0 | 6 | 60 | 87 | 90 | 90 | 93 | 98 | 100 | 100 | 100 | 100 | 100 | 97.8 | 2.0 | 0.2 | |
| TOB | 0 | 17 | 49 | 51 | 53 | 53 | 53 | 55 | 60 | 65 | 69 | 78 | 100 | 53.4 | 1.3 | 45.3 | |
| MIN | 12 | 42 | 82 | 87 | 89 | 91 | 93 | 95 | 98 | 100 | 100 | 100 | 100 | 93.4 | 2.0 | 4.6 | |
| RIF | 93 | 93 | 93 | 93 | 94 | 96 | 96 | 97 | 97 | 97 | 97 | 97 | 100 | 94.5 | 2.0 | 3.5 | |
| SXT | 80 | 91 | 98 | 99 | 99 | 99 | 99 | 100 | 100 | 100 | 100 | 100 | 100 | 99.3 | 0 | 0.7 | |
| FUS | 42 | 60 | 75 | 92 | 93 | 94 | 95 | 98 | 99 | 99 | 100 | 100 | 100 | 93.6 | 5.7 | 0.7 | |
| MUP | 7 | 12 | 42 | 89 | 89 | 90 | 90 | 92 | 94 | 96 | 96 | 96 | 97 | 100 | 89.7 | 6.8 | 3.5 |
Abbreviations: OXA, oxacillin; VAN, vancomycin; TEC, teicoplanin; ERY, erythromycin; CLI, clindamycin; Q-D, quinupristin-dalfopristin; CIP, ciprofloxacin; LZD, linezolid; GEN, gentamicin; AMK, amikacin; NET, netilmicin; TOB, tobramycin; MIN, minocycline; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; FUS, fusidic acid; MUP, mupirocin.
>1,024 column contains data only for mupirocin; no other strains were tested at that concentration.
TABLE 2.
Distribution of MLS resistance and AME genes among MRSA strains by PFGE type
| Gene(s) | No. of genes detected per PFGE type
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| A1 (n = 14) | A20 (n = 60) | A21 (n = 19) | B2 (n = 215) | G10 (n = 15) | L1 (n = 13) | C3 (n = 10) | Other (n = 109) | Total (n = 450) | |
| MLS resistance gene(s) | |||||||||
| erm(A) | 13 | 59 | 5 | 3 | 14 | 1 | 2 | 56 | 153 |
| erm(C) | 1 | 0 | 1 | 105 | 1 | 12 | 4 | 11 | 135 |
| erm(A) + erm(C) | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 2 |
| erm(A) + vat(B) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| No gene detected | 0 | 1 | 13 | 106 | 0 | 0 | 4 | 40 | 164 |
| AME gene(s) | |||||||||
| aac(6′)-aph(2") | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 7 | 11 |
| aac(6′)-aph(2") + ant(4′) | 9 | 2 | 0 | 1 | 0 | 0 | 0 | 12 | 24 |
| aac(6′)-aph(2") + aph (3′) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20 | 20 |
| ant(4′) | 1 | 51 | 17 | 30 | 14 | 1 | 7 | 37 | 158 |
| aph(3′) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| No gene detected | 2 | 7 | 2 | 184 | 1 | 12 | 1 | 31 | 240 |
The SCCmec types could be determined for 56 (93%) MRSA strains tested by multiplex PCR and were found to be the following: type IV (n = 34), type I (n = 12), type II (n = 8), and type III (n = 2). The genotyping of 455 MRSA strains showed 124 PFGE patterns categorized into 23 groups and 57 types. Group A strains (n = 116) were subdivided into 13 types. Types A20 (n = 60), A21 (n = 19), and A1 (n = 14) represented 79% of group A strains (Table 3). Type A1 strains belonged to the sequence type 247 (ST247)-SCCmec I clone. Type A20 and A21 strains belonged to the same clonal complex, CC8, but had a different sequence type (ST8) and carried the SCCmec IV type. Group B strains (n = 233) were clustered into nine types. The major type, B2, represented 92% of strains (n = 215) and belonged to the ST45-SCCmec IV clone. Group C strains (n = 37) belonged to the ST5-SCCmec IV clone and included nine PFGE types, of which C3 (n = 10) was the most frequently found. Group G strains (n = 18), subdivided into two types, had the same ST (ST5) but carried the SCCmec II type. Group L strains (n = 13), all of which shared a single type (L1), were identical by MLST and SCCmec type analysis to the “UK-epidemic MRSA (EMRSA) 15 clone” (ST22-MRSA-IV) (8). Seventy-six percent of the MRSA strains were distributed into seven major epidemic types: B2, A20, A21, G10, A1, L1, and C3 (Table 2), belonging to five clonal complexes causing nosocomial outbreaks worldwide (5, 8, 18).
TABLE 3.
Distribution of MRSA strains (n = 455) by PFGE, MLST, and SCCmec types
| PFGE group | PFGE type | No. of isolates | No. of hospitals | MLST allelic profile | Sequence type | Clonal complex | SCCmec type |
|---|---|---|---|---|---|---|---|
| A | A1 | 14 | 12 | 3-3-1-12-4-4-16 | 247 | 8 | I |
| A20 | 60 | 32 | 3-3-1-1-4-4-3 | 8 | 8 | IV | |
| A21 | 19 | 13 | 3-3-1-1-4-4-3 | 8 | 8 | IV | |
| Other | 23 | 18 | |||||
| B | B2 | 215 | 80 | 10-14-8-6-10-3-2 | 45 | 45 | IV |
| Other | 18 | 16 | |||||
| C | C3 | 10 | 6 | 1-4-1-4-12-1-10 | 5 | 5 | IV |
| Other | 27 | 21 | |||||
| G | G10 | 15 | 12 | 1-4-1-4-12-1-10 | 5 | 5 | II |
| Other | 3 | 3 | |||||
| L | L1 | 13 | 6 | 7-6-1-5-8-8-6 | 22 | 22 | IV |
| Other | 38 | 27 |
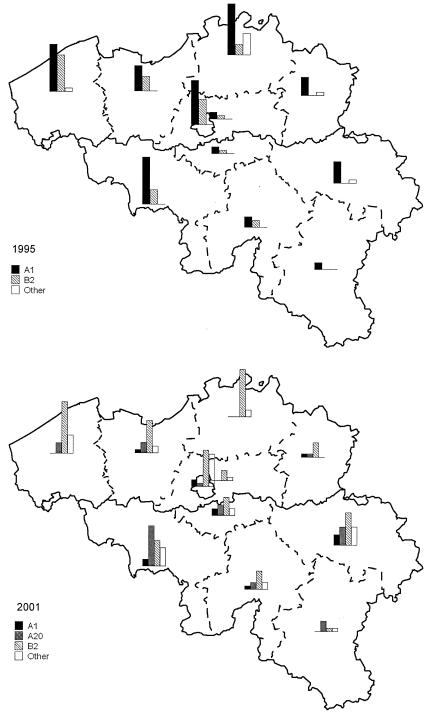
In the late 1980s, type A1 strains were associated with hospital outbreaks in Belgium and spread to the whole country in the early 1990s (Fig. 1) (3, 5). In 2001, type A1 strains had been progressively supplanted by other epidemic clones and were present only in the southern part of the country (3). Type B2 strains emerged in 1992 in the western part of the country and spread thereafter to the neighboring provinces to become predominant in 2001 (3, 5). Type A20 strains were mainly recovered in the southern part of the country, whereas type A21 strains were predominantly disseminated in the north. G10 strains were recovered only from hospitals in Brussels and Walloon. L1 and C3 strains were found with similar low frequencies in the three regions.
FIG. 1.
Evolution between 1995 and 2001 of the geographical distribution by province in Belgium of epidemic MRSA strains of PFGE types A1, A20, B2, and others.
The majority of type A1 strains harbored ermA and the combination of aac(6′)-aph(2") and ant(4′) genes (Table 2). Most of the type A20 and A21 strains, carrying predominantly only ant(4′), were more susceptible to aminoglycosides. However, most type A20 strains contained erm(A), conferring resistance to MLS. The majority of B2 strains were susceptible to aminoglycosides (86%), although some isolates harbored the AME genes. For the MLS resistance genes, type B2 strains contained either no MLS resistance gene (49%) or erm(C) (49%). G10 strains harbored the ant(4′) (93%) and erm(A) (93%) genes, whereas 92% of L1 strains showed only the erm(C) gene and no resistance gene to aminoglycosides. The distribution of resistance genes was heterogeneous in type C3 strains. The mupA gene was recovered in MRSA type A20, C3, and B2 strains.
The proportion of MRSA infections acquired after 48 h of hospitalization in acute-care hospitals decreased significantly, from 85% in 1995 to 69% in 2001 (P < 0.001) (3). This phenomenon probably reflects the constitution of an increasing reservoir of chronic MRSA carriers. Recently, Hoefnagels-Schuermans et al. have reported an MRSA strain carriage prevalence of 4.9% in residents of 17 Belgian nursing homes (10). They observed that 98% of MRSA isolates belonged to the three major Belgian epidemic types that were widely disseminated in acute-care hospitals at that time.
In summary, data from consecutive national surveys showed a recent diversification of MRSA clones disseminated in Belgian hospitals, with the broad diffusion of seven major clones belonging to the five pandemic MRSA lineages causing nosocomial infections worldwide. Of note, there was a marked expansion in the number of epidemic strains carrying the type IV SCCmec cassette that was previously thought to characterize community-acquired MRSA strains (16). The changes in the prevalence of epidemic MRSA genotypes led to shifts in AME and methylase gene distribution, with a decreased proportion of multidrug-resistant MRSA strains. Over the years, the proportion of patients who carried MRSA strains on admission has risen in hospitalized patients. These trends show that MRSA strains represent a major burden of nosocomial infections in Belgium, with an increasingly large patient reservoir that challenges infection control efforts.
Acknowledgments
This study was organized under the auspices of the Groupement pour le Dépistage, l'Etude et la Prévention des Infections Hospitalières (GDEPIH-GOSPIZ). This work was supported by grants from the Ministry of Public Health, Brussels, Belgium and from Pharmacia Belgium (now Pfizer, Inc.).
We thank our microbiologist colleagues for their participation in this surveillance program: Françoise Brancart for performing MLST analysis; Katokolo Tshibangu for PCR testing of MLS resistance determinants; and Ayaba Brenner, Magali Nesterenko and Christine Thiroux for performing phenotypic susceptibility tests. We thank Erik Hendrickx and Yves Dupont, Institute of Public Health, for logistical help with the survey and for reviewing the manuscript.
REFERENCES
- 1.Andrews, J. M. 2001. BSAC standardized disc susceptibility testing method. J. Antimicrob. Chemother. 48(Suppl. 1):43-57. [DOI] [PubMed] [Google Scholar]
- 2.Comité de l'Antibiogramme de la Société Française de Microbiologie. 4 February 2004, posting date. Communiqué 2003. [Online.] http://www.sfm.asso.fr/nouv/general/php?pa=2.
- 3.Denis, O., A. Deplano, R. De Ryck, C. Nonhoff, and M. J. Struelens. 2003. Emergence and spread of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus in Belgian hospitals. Microb. Drug Resist. 9:61-71. [DOI] [PubMed] [Google Scholar]
- 4.Denis, O., J. Magdalena, A. Deplano, C. Nonhoff, E. Hendrickx, and M. J. Struelens. 2002. Molecular epidemiology of resistance to macrolides-lincosamides-streptogramins in methicillin-resistant Staphylococcus aureus (MRSA) causing bloodstream infections in patients admitted to Belgian hospitals. J. Antimicrob. Chemother. 50:755-757. [DOI] [PubMed] [Google Scholar]
- 5.Deplano, A., W. Witte, W. J. van Leeuwen, Y. Brun, and M. J. Struelens. 2000. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighboring countries. Clin. Microbiol. Infect. 6:239-245. [DOI] [PubMed] [Google Scholar]
- 6.EARSS Management Team, Bilthoven, The Netherlands. EARSS annual report 2002. [Online.] http://www.earss.rivm.nl/.
- 7.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fluit, A. C., C. L. Wielders, J. Verhoef, and F. J. Schmitz. 2001. Epidemiology and susceptibility of 3,051 Staphylococcus aureus isolates from 25 university hospitals participating in the European SENTRY study. J. Clin. Microbiol. 39:3727-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoefnagels-Schuermans, A., L. Niclaes, F. Buntinx, C. Suetens, B. Jans, J. Verhaegen, and J. Van Eldere. 2002. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in nursing homes: a cross-sectional study. Infect. Control Hosp. Epidemiol. 23:546-549. [DOI] [PubMed] [Google Scholar]
- 11.Ida, T., R. Okamoto, C. Shimauchi, T. Okubo, A. Kuga, and M. Inoue. 2001. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J. Clin. Microbiol. 39:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lina, G., A. Quaglia, M. E. Reverdy, R. Leclercq, F. Vandenesch, and J. Etienne. 1999. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43:1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maes, N., J. Magdalena, S. Rottiers, Y. De Gheldre, and M. J. Struelens. 2002. Evaluation of a triplex polymerase chain reaction (PCR) assay to discriminate Staphylococcus aureus from coagulase-negative staphylococci (CoNS) and determine methicillin resistance from blood cultures. J. Clin. Microbiol. 40:1514-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing, twelfth informational supplement. Approved standard MS100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 16.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 19.Schmitz, F. J., A. C. Fluit, M. Gondolf, R. Beyrau, E. Lindenlauf, J. Verhoef, H. P. Heinz, and M. E. Jones. 1999. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 43:253-259. [PubMed] [Google Scholar]
- 20.Schmitz, F. J., R. Sadurski, A. Kray, M. Boos, R. Geisel, K. Kohrer, J. Verhoef, and A. C. Fluit. 2000. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 45:891-894. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz, F. J., J. Verhoef, A. C. Fluit, C. von Eiff, R. R. Reinert, M. Kresken, J. Brauers, D. Hafner, G. Peters, M. C. Roberts, J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, H. Seppala, B. Vester, and S. Douthwaite. 1999. Prevalence of resistance to MLS antibiotics in 20 European university hospitals participating in the European SENTRY surveillance programme. J. Antimicrob. Chemother. 43:783-792. [DOI] [PubMed] [Google Scholar]
- 22.Vanhoof, R., C. Godard, J. Content, H. J. Nyssen, E. Hannecart-Pokorni, and the Belgian Study Group of Hospital Infections (GDEPIH/GOSPIZ). 1994. Detection by polymerase chain reaction of genes encoding aminoglycoside-modifying enzymes in methicillin-resistant Staphylococcus aureus isolates of epidemic phage types. J. Med. Microbiol. 41:282-290. [DOI] [PubMed] [Google Scholar]
- 23.Wildemauwe, C., C. Godard, R. Vanhoof, E. V. Bossuyt, and E. Hannecart-Pokorni. 1996. Changes in major populations of methicillin-resistant Staphylococcus aureus in Belgium. J. Hosp. Infect. 34:197-203. [DOI] [PubMed] [Google Scholar]