Abstract
Polymorphisms within one-carbon metabolism genes have been largely studied in relation to cancer risk for the function of this pathway in nucleotide synthesis and DNA methylation. Aims of this study were to explore the possible link among several common functional gene polymorphisms within one-carbon metabolism and survival rate in primary liver cancers, i.e., hepatocellular carcinoma and cholangiocarcinoma, and to assess the additional effect of global DNA methylation on survival rate and mortality risk. Forty-seven primary liver cancer patients were genotyped for ten polymorphisms: DHFR 19bp ins/del, TS 2rpt-3rpt, MTHFD1 1958G>A, MTHFR 677C>T, MTR 2756A>G, MTRR 66A>G, RFC1 80G>A, SHMT1 1420C>T, BHMT 716 A>G, TC II 776C>G. Methylation was determined in peripheral blood mononuclear cells (PBMCs) DNA as methylcytosine (mCyt) content using LC/MS/MS. Among the polymorphisms analysed, the RFC1 80G>A (rs1051266) influenced the survival rate in primary liver cancers. The RFC1 80AA was associated to a significantly reduced survival rate (22.2%) as compared to both GG and GA genotypes (61.5% and 76% respectively, p = 0.005). When the cancer patients were stratified according to the mCyt median value as high (>5.34%) or low (≤5.34%), the concomitant presence of AA genotype and low mCyt level led to a significantly worse survival rate as compared to the G allele carriership (p<0.0001) with a higher Hazard Ratio (HR = 6.62, p = 0.001). The subjects carrying the AA genotype in association with high mCyt did not show a significant difference in survival rate as compared with the G allele carriers (p = 0.919). The RFC1 80G>A polymorphism influenced the survival rate, and the presence of RFC1 80AA genotype with low global methylation in PBMCs DNA was associated with poorer prognosis and higher mortality risk, therefore highlighting novel molecular signatures potentially helpful to define prognostic markers for primary liver cancers.
Introduction
One-carbon metabolism is essential for several intracellular reactions including those involved in nucleotide synthesis and biological methylation, in particular methylation of DNA, the major epigenetic mechanism in mammalian cells [1].
Genetic variants of one-carbon pathway genes have been investigated mainly in colon cancer as potential markers of cancer susceptibility [2–5] for the involvement of this metabolism in cellular development, proliferation and differentiation [6, 7]. The molecular mechanisms underlying the possible association among polymorphic variants of one-carbon metabolism genes, cancer survival rate and mortality risk are, however, not completely clarified.
DNA methylation is a heritable and reversible phenomenon that consists in the covalent binding of a methyl group to the 5’carbon of a cytosine in CpG dinucleotide sequences and it plays a role in gene expression regulation and maintenance of genomic stability [8–10]. An aberrant DNA methylation, both global and gene-specific, is involved throughout all the phases of cancer development and progression, and a status of global DNA hypomethylation is an almost universal finding in cancer tissues [11, 12]. A global hypomethylation in peripheral blood mononuclear cells (PBMCs) DNA was associated to an increased cancer risk [13] and a shorter survival rate in patients affected by different types of cancer [14–16]. In recent studies, we observed significantly reduced methylcytosine (mCyt) levels in PBMCs DNA of cancer patients [17, 18] and lower levels were directly associated to an unfavourable prognosis [18].
As for the possible relationship among one-carbon metabolism gene variants, global DNA methylation and cancer, we previously reported that the TT genotype of the MTHFR 677C>T polymorphism associated with low plasma folate levels presented a decreased DNA methylation [19] and higher incidence of cancer [17]. Global DNA hypomethylation in PBMCs in association with the carriership of MTHFR T allele represents, therefore, a predicting factor of cancer development and an unfavourable prognostic factor in cancer disease [17]. The potential association among other polymorphic variants of one-carbon metabolism genes, global DNA methylation in PBMCs and survival rate in cancer is still poorly investigated as it is the role of aberrant DNA methylation in clinical outcome and prognosis in cancer disease.
Cancer is a major public health issue [20] and among the different cancer types, primary liver cancers, i.e. hepatocellular carcinoma and cholangiocarcinoma, are prevailing malignancies with a high worldwide mortality rate [21].
Aims of the present study were: i) to explore the possible link among the most common variants of one carbon-related genes and survival rate in primary liver cancers, i.e. hepatocellular carcinoma and cholangiocarcinoma, and ii) to assess the additional effect of global DNA methylation on survival rate and mortality risk.
Materials and Methods
Study subjects and survival data collection
The study was approved by the Institutional Review Board Ethical Committee of the University of Verona School of Medicine Hospital (Verona, Italy). Written informed consent was obtained from each patient after a detailed explanation of the study.
Forty-seven patients affected by primary liver cancers, 31 hepatocellular carcinoma and 16 cholangiocarcinoma, were enrolled from April 2009 to March 2013 among those referring to the Division of Surgery of the Verona University Hospital for curative surgery intervention. Inclusion criteria were age ≥18 years with the following surgical resectability criteria: preserved liver function, class A Child-Pugh score, absence of extrahepatic metastases. Exclusion criteria were a coexisting human immunodeficiency (HIV), hepatitis B (HBV) or hepatitis C (HCV) viruses infection; presence of relevant concurrent medical conditions such as chronic inflammatory diseases or haematological disorders, including autoimmune liver diseases and hereditary hemochromatosis; presence of an acute inflammatory disease, decompensate liver cirrhosis (Child-Pugh B, C). A trained physician recorded a detailed clinical history data including lifestyle habits. All subjects under B vitamins supplementation and/or using drugs known to interfere with folate-related one-carbon metabolism in the month before enrolment were excluded.
A periodic evaluation of the patients consisting in a complete medical examination or a telephone interview was performed during a follow-up period of 60 months. The follow-up period was calculated from the date of the surgical intervention up to the date of death or to the latest recorded medical examination.
Biochemical analyses
Samples of venous blood were drawn from each subject after an overnight fasting and analysed by routine laboratory test analysis for a complete blood count and determination of serum C-reactive protein (CRP), creatinine, aspartate transaminase (AST), alanine transaminase (ALT), gamma-glutamyltranspeptidase (gGT), alkaline phosphatase (ALP), total bilirubin, albumin, glycemic level, total cholesterol, triglycerides, prothrombin time-international ratio (PT-INR), ferritin, serological tests for hepatitis B and C viruses. Plasma folate and vitamin B12 were measured by an automated chemiluminescence method (ChironDiagnostics, East Walpole, MA, USA) and total plasma homocysteine concentrations was determined by high-performance liquid chromatography (HPLC) with fluorescent detection [22].
Genotyping
From each subject venous blood was drawn into Vacutainer® tubes containing EDTA as anticoagulant after an overnight fast and DNA was extracted from PBMCs by Wizard Genomic DNA Purification Kit (Promega Corporation, Fitchburg, WI, USA). One carbon metabolism gene variants were analysed by different methods, as follows: DHFR 19bp ins/del [23] and TS 2rpt-3rpt [24] by PCR; MTHFD1 1958G>A (rs2236225) [25], MTHFR 677C>T (rs1801133) [26], MTR 2756A>G (rs12749581) [27], MTRR 66A>G (rs1801394) [28], RFC1 80G>A (rs1051266) [29] and SHMT1 1420C>T (rs1979277) [30] by PCR followed by restriction fragment length polymorphism assays, BHMT 716 A>G (rs3733890) and TC II 776C>G (rs1801198) by allelic discrimination Real Time-PCR technology using the assay C_11646606_20 and the assay C_325467_10, respectively (ABI Prism 7500, Applied Biosystems, Carlsbad, CA, USA).
Global DNA methylation
Global DNA methylation was determined using a liquid chromatography/mass spectrometry (LC/MS/MS) method and mCyt levels expressed as percent (%)mCyt = [(mCyt)/(mCyt + Cyt)] x 100, as previously described [19, 31], with slight modifications [18]. Briefly, global DNA was extracted from PBMCs and hydrolyzed to nucleosides using 2 units of nuclease P1, 0.002 units of venom phosphodiesterase I and 0.5 units of alkaline phosphatase. Isotope-labelled internal standards for deoxycytidine and 5-methyl-deoxycytidine were added to samples before the run in a 3200 Q Trap MS-MS system coupled with an Agilent 1100 Series liquid chromatograph (Agilent, Santa Clara, CA, USA).
Statistical analysis
All the statistical computations were performed by using the IBM SPSS Statistics software version 22 for Windows (IBM Inc., Armonk, NY, USA). Continuous variables were expressed as mean values ± standard deviations (SD), parameters showing a skewed distribution were log-transformed and thus expressed as geometric means with 95% confidence intervals (CIs). Continuous variables were tested by analysis of variance (ANOVA) with Tukey's post-hoc comparison when appropriate. All genotype distributions were verified to be in agreement with Hardy-Weinberg equilibrium.
Survival rate was analysed by Kaplan-Meier curves with Log Rank test and pairwise comparison when indicated. Hazard ratio (HR) of mortality with 95% CI was estimated by Cox regression analysis adjusted for age and gender, and by including folate plasma concentration in the regression model. A p-value <0.05 was considered statistically significant.
Results
Clinical and biochemical characteristics of primary liver cancer patients
Table 1 reports the main clinical and biochemical characteristics of the 47 primary liver cancer patients (31 hepatocellularcarcinoma, 16 cholangiocarcinoma). The main biochemical analyses were within the normal range and, in particular, the indexes of hepatic function confirmed a compensated liver function status in all patients (Table 1). Viral serologic tests for HBV and HCV were negative in all patients according to the enrolment criteria. As for neoplastic markers, alpha-fetoprotein values were within the normal range (<7 g/l) in 39% of the HCC patients, whereas normal levels of CA 19.9 (<25 U/ml) and CEA (<5 ng/ml) were observed in 50% and 100% of CC patients, respectively.
Table 1. Clinical and biochemical characteristics of primary liver cancer patients.
| References values | Cancer patients (n = 47) | |
|---|---|---|
| Clinical characteristics | ||
| Age, years | 67.1 ± 9.0 | |
| Gender, % male | 76.6% | |
| Smoking | 68.1% | |
| Alcohol drinking | 72.3% | |
| Laboratory tests | ||
| CRP (mg/L)* | <5 | 7.70 (5.13–11.55) |
| Hb (g/dL) | 13.5–16.0 | 13.5 ± 1.62 |
| MCV (fL) | 86–98 | 91.9 ± 7.49 |
| WBCs (109/L) | 4.3–10.0 | 7.01 ± 2.96 |
| PLTs (109/L) | 150–400 | 235.6 ± 119.9 |
| AST (U/L)* | 8–50 | 41.7 (32.8–53.2) |
| ALT (U/L)* | 8–45 | 40.8 (30.2–55.0) |
| ALP (U/L)* | 30–130 | 92.4 (77.9–109.6) |
| gGT (U/L)* | <50 | 76.7 (58.6–100.3) |
| Total bilirubin (mg/dL)* | 0.11–1.05 | 0.71 (0.59–0.86) |
| Direct bilirubin (mg/dL)* | <0.35 | 0.25 (0.20–0.31) |
| PT (INR)* | 0.82–1.14 | 1.11 (1.07–1.15) |
| Albumin (g/L) | 35–50 | 39.9 ± 5.54 |
| CHE (U/L) | 4650–14400 | 6515 ± 1890 |
| Total cholesterol (mg/dL) | <200 | 159.5 ± 48.9 |
| Triglycerides (mg/dL)* | <150 | 112.7 (100.1–126.7) |
| Creatinine (mg/dL)* | 0.59–1.29 | 0.86 (0.74–1.00) |
| Glucose (mmol/L) | 3.5–5.5 | 6.39 ± 1.81 |
| Folate (nmol/L)* | 10.4–42.4 | 8.52 (9.95–10.45) |
| tHcy (μg/L)* | < 15 | 13.2 (10.4–16.7) |
| Vitamin B12 (pmol/L)* | 142–724 | 337.0 (288.4–393.8) |
| Vitamin B6 (nmol/L)* | 25–128 | 19.7 (15.4–25.2) |
| Ferritin (μg/L)* | 30–400 | 249.4 (178.2–349.0) |
Values are expressed as mean ± SD.
*: log-transformed variables are shown as geometric mean with 95% confidence interval
Alcohol drinking defined as ≥ 36 g ethanol/day for males and ≥ 24 g ethanol/day for females
Abbreviations: CRP, C-reactive protein; Hb, Hemoglobin; MCV, Mean Corpuscular Volume; WBCs, White Blood Cells; PLTs, Platelets; AST, aspartate aminotransferase; ALT, aspartate alanine aminotransferase; ALP, Alkaline phosphatase; gGT, gamma glutamyl transferase; PT (INR), Prothrombin International Ratio; CHE, cholinesterase; tHcy, Homocysteine.
Survival rate according to one-carbon polymorphic variants
The polymorphic variants distribution of all evaluated one-carbon metabolism genes was in agreement with the Hardy-Weinberg equilibrium in the study subjects.
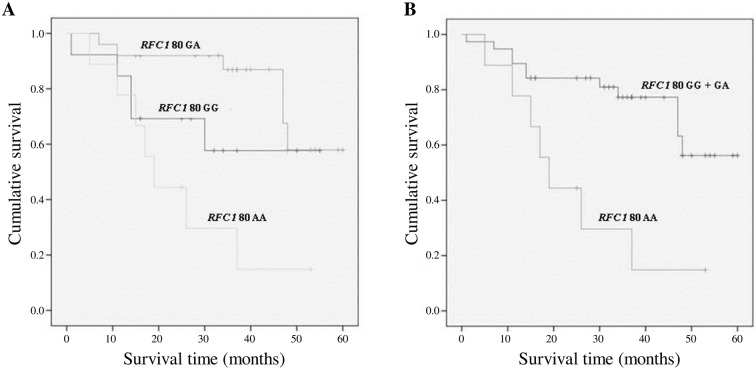
The Kaplan-Meier analysis to test the survival rate was performed in accordance to the ten gene polymorphic variants object of the study. Among the analysed polymorphisms, only the RFC1 80G>A polymorphism influenced the survival rate in primary liver cancers. The RFC1 80G>A genotypes frequencies are reported in Table 2. The homozygous variant RFC1 80AA was associated to a lower survival rate (22.2%) as compared to both RFC1 80 GG and GA genotypes (survival rate 61.5% and 76% respectively, p = 0.005) (Fig 1A). The statistical significance increased when the RFC1 80AA subjects were compared to the G allele carriers (RFC1 80 GG + GA) (Fig 1B) with a survival rate of 22.2% versus 71.1%, respectively (p = 0.002). When the main biochemical parameters, in particular plasma folate levels, were analysed according to the RFC1 80G>A variants, no significant differences were observed among the three genotypes except for platelet count, higher in the AA group, and triglycerides concentration that was lower in the GG genotype group (data not shown).
Table 2. RFC1 80G>A genotypes frequencies in primary liver cancer patients (n = 47).
| RFC1 80GG | RFC1 80GA | RFC1 80AA | |
|---|---|---|---|
| Number of patients | 13 | 25 | 9 |
| Percentage of patients | 27.7% | 53.2% | 19.1% |
Fig 1. Survival curves plotted by Kaplan-Meier analysis according to RFC1 80G>A genotypes.
(A) The survival rate was worse in RFC1 80AA (22.2%) patients as compared to the RFC1 80GA (76%) and RFC1 80GG (61.5%) genotypes (p = 0.005). (B) The survival rate was lower among the RFC1 80 AA patients as compared with the G allele carriers (RFC1 80GG+GA) (p = 0.002). The percentage of survivors was 22.2% and 71.1%, respectively.
RFC1 80G>A genotypes and survival rate according to DNA global methylation
The observed association of RFC1 80G>A polymorphism with survival rate in primary liver cancer (Fig 1) suggested to evaluate mCyt levels according to RFC1 80G>A genotypes. The comparison of mCyt levels showed no statistically significant differences among the genotypes (AA 5.41%, GA 5.36% and GG 5.25%, p = 0.607). The methylation status did not differ also when comparing 80AA subjects with G allele carriers (RFC1 80GG + GA) (5.41% versus 5.32%, respectively, p = 0.553) (data not shown).
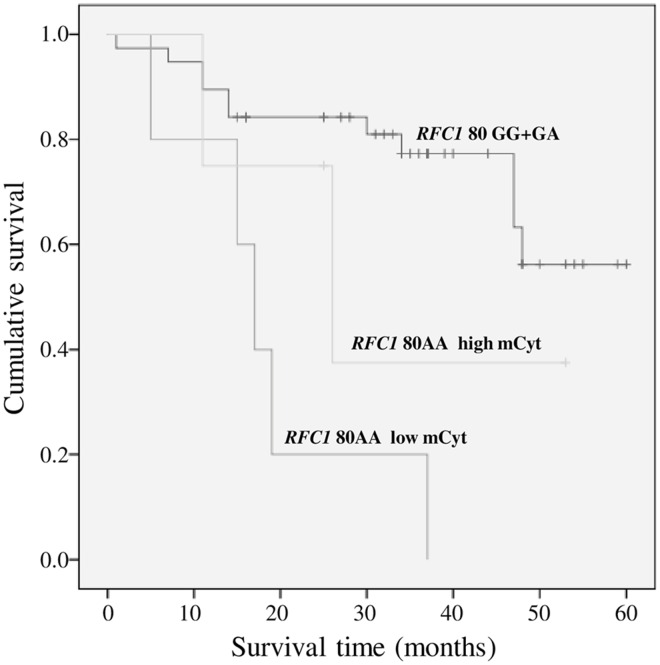
Subsequently the study group was stratified in two subsets according to high (>5.34%) or low (≤5.34%) global DNA methylation, defined on mCyt median value, and the survival rate evaluated by Kaplan—Meier analysis. RFC1 80AA patients with low mCyt had a significantly worse survival in comparison both with GA and GG subjects, either with low or high mCyt levels (p = 0.002) (data not shown). Since the RFC1 80G carriers exhibited similar survival rate curves, the two groups were merged and the Kaplan—Meier analysis repeated. The plotted curves highlighted that RFC1 80AA patients with low mCyt levels had a poorer survival rate, as compared with G allele carriers (p<0.0001) (Fig 2). Noteworthy, all the five patients characterized by RFC1 80AA with low mCyt were deceased at the time of observation, whereas both the RFC1 80G carriers and RFC1 80AA with high mCyt showed better survival rates (71.1% and 50%, respectively). No statistically significant differences were found when comparing the RFC1 80G carriers and the RFC1 80AA with high mCyt (p = 0.919) and when comparing RFC1 80AA with low mCyt and RFC180AA with high mCyt (p = 0.209) (Fig 2).
Fig 2. Survival curves plotted by Kaplan-Meier analysis according to RFC1 80G>A genotypes and mCyt levels.
RFC1 80AA patients with low mCyt levels (≤5.34%) were associated to a lower survival as compared to carriers of the RFC1 80G allele (71.1%) (p<0.0001). The comparison between RFC1 80G carriers and RFC1 80AA with high mCyt (>5.34%) was not statistically significant (p = 0.919), as it was the comparison between RFC1 80AA with low mCyt and with high mCyt (p = 0.209).
RFC1 80G>A genotypes and mortality risk according to global DNA methylation
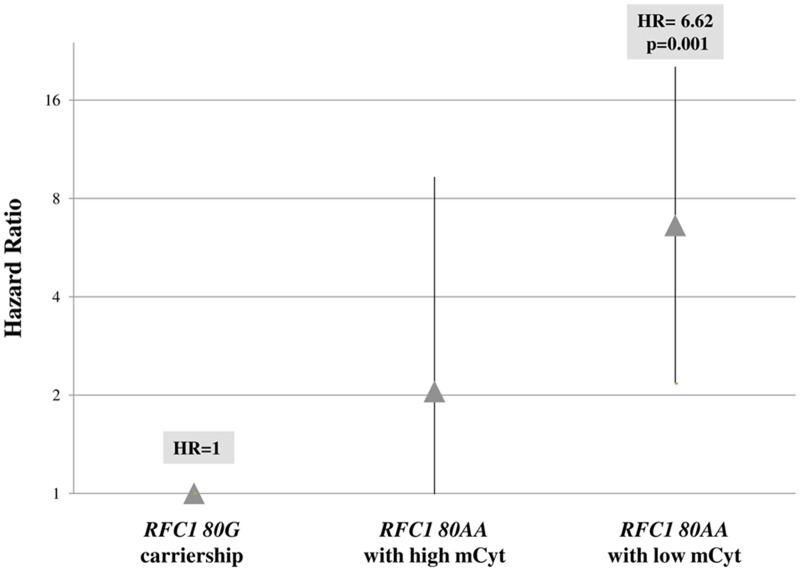
The analysis of mortality risk was performed by setting the RFC1 80G carriership as the reference group with Hazard Ratio (HR) = 1 and the mortality risk associated to RFC1 80AA genotype with high or low mCyt levels was calculated. Subjects carrying the RFC1 80AA genotype and high mCyt had an HR = 2.05, not statistically different from the reference group, whereas the concomitant presence of RFC1 80AA genotype and low mCyt was associated to a 6.62-fold higher HR, as compared to RFC1 80G carriers (p = 0.001) (Fig 3).
Fig 3. Mortality risk by Hazard Ratio for RFC1 80AA genotype with either high or low mCyt levels.
The RFC1 80AA genotype with low mCyt levels (≤5.34%), had a higher Hazard Ratio (HR) as compared to RFC1 80G carriership (RFC1 80GA plus RFC1 80GG) (HR = 6.62, 95% CI 2.17–20.25, p = 0.001). The HR for RFC1 80AA genotype with high mCyt levels (>5.34%) did not differ from the RFC1 80G carriership group (HR = 2.05, 95% CI 0.45–9.32, p = 0.351).
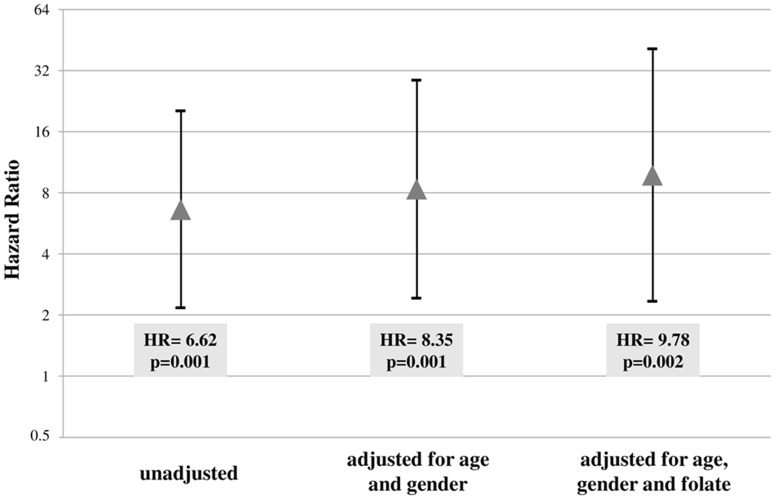
The HR significantly increased when the analysis was performed after adjustments for age and gender (HR = 8.35 CI 2.42–28.67, p = 0.001) and for age, gender and plasma folate concentrations (HR = 9.78 CI 2.34–40.94, p = 0.002) (Fig 4).
Fig 4. Mortality risk associated to carriership of the RFC1 80AA genotype with low mCyt using crude Hazard Ratio and after adjustments for age, gender and folate.
The unadjusted Hazard Ratio (HR) was 6.62 (95% CI 2.17–20.25, p = 0.001) and the value significantly increased after adjustments for age and gender (HR = 8.35, 95% CI 2.42–28.67, p = 0.001) and for age, gender and plasma folate concentrations (HR = 9.78, 95% CI 2.34–40.94, p = 0.002).
Discussion
The results of this study indicates that primary liver cancer patients carrying the RFC1 80AA genotype associated with low DNA methylation have a significantly poorer survival rate with a higher mortality risk, as compared with G allele carriers.
The Reduced Folate Carrier 1 (RFC1, official symbol SLC19A1) gene is located on chromosome 21 and encodes for an ubiquitously expressed transmembrane protein that serves as a bi-directional transporter of reduced folate species such as 5-methyl THF, the main circulating active form of folate [32, 33] and it is involved in both methylation pathway and nucleotides synthesis.
Chango et al., described a RFC1 polymorphic variant (rs 1051266) consisting in the substitution of a guanine to an adenine (G>A) at position 80 of gene sequence, causing an arginine to histidine substitution at amino acid position 27 [29]. The exact functional relevance of this variant is, however, still unclear [34, 35]. In several human and animal studies no significant differences were observed both in plasma and red blood cell folate concentration in association with RFC1 80G>A genotypes [29, 36–42]. Results from the present study confirmed the lack of differences in plasma folate concentrations according to the RFC1 80G>A genotypes. The RFC1 80G>A polymorphism might be associated to a lower availability of methyl groups at tissue level for methylation reaction rather than to an evidently reduced level of plasma folate. The relationship among the RFC1 80G>A polymorphism, cancer risk and clinical outcome have been the objects of several studies, though with contrasting reports.
RFC1 80G allele was associated with an increased risk of head and neck carcinoma [43, 44], whereas an increased risk of gastric and esophageal cancer was found in association with the RFC180AA genotype [45]. The RFC1 80AA genotype was also associated with increased risk for acute lymphoid leukaemia with worse outcome [46], higher chance of relapse and poorer survival [47, 48] as compared to GA and GG genotypes. On the contrary, in patients affected by rectal cancer the RFC1 80AA genotype was associated with a more favourable prognosis [49]. On the other hand, no association was found between RFC1 80G>A genotypes and breast cancer risk [50] or survival [51].
Global DNA hypomethylation is associated with genomic instability and compromised gene repression in genomic regions that are usually silenced in normal cells [11, 12, 52]. The consequence of this event may entail the expression of proto-oncogenes as well as the activation of viral and parasitic transposons [52, 53]. Moreover, it has been consistently demonstrated that DNA hypomethylation is able to increase the immunogenicity and the immune recognition of cancer cells, thus affecting transcriptional deregulation and tumour aggressiveness [52]. Most of the studies showing an aberrant DNA methylation evaluated this epigenetic feature of DNA at the level of the specific tissue affected by the malignant process. One key point for discussion is, in fact, the finding of low methylation levels measured in PBMCs DNA. Considering that epigenetic mechanisms including methylation are usually considered tissue-specific, some speculations in this regard are needed. It has been suggested that a global DNA hypomethylation status in PBMCs might represent a potential epigenetic marker for cancer [13] and low levels of mCyt are usually associated to increased cancer risk and to poor prognosis [13–16]. In a previous study we demonstrated that patients affected by different types of cancer had a significantly lower global methylation in PBMCs DNA as compared to cancer-free controls and that DNA hypomethylation was also associated with a higher risk for cancer development [17]. Moreover, in a recent study in primary liver cancer patients we demonstrated that low mCyt levels in PBMCs DNA were related to a worse prognosis, suggesting a putative prognostic relevance of DNA methylation [18]. A recent prospective case-control study reported that RFC1 80G>A is associated with LINE-1 undermethylation as measured in peripheral blood DNA of women who developed breast cancer as compared with breast cancer-free controls [54]. Nevertheless, whilst the RFC1 80G>A variant was associated to lower methylation, no relationship was found between the presence of the variant and cancer risk [54].
The results of the present study demonstrate that the RFC1 80AA genotype is significantly associated with a poorer life expectancy in primary liver cancer patients. Moreover, when the mCyt levels were stratified as either high (>5.34%) or low (≤5.34%), patients with the RFC1 80AA genotype and low mCyt showed a significantly worse survival rate and a higher mortality risk as compared to the RFC1 80G carriers, even after adjustments for sex, age and folate concentrations. Noteworthy, the five patients characterized by the RFC1 80AA genotype and low mCyt levels were all deceased at the time of the observation.
The mechanism by which global DNA hypomethylation in the presence of the RFC1 80AA genotype may entail a poorer prognosis in patients affected by primary liver cancer remains to be explored. RFC1 is a key transmembrane bi-directional transporter of 5-methyl THF, the main circulating active form of folate, and it is involved in both methylation pathway and nucleotides synthesis. The present results showed that the AA genotype is associated to lower survival in primary liver cancer and that the AA genotype and concomitant low mCyt levels have a significantly poorer survival rate with a higher mortality risk. One hypothesis for the explanation of this finding is that RFC1 might be involved in carcinogenesis by affecting both methylation reactions and DNA repair capacity.
Even if the results of this study suggest that primary liver cancer patients showing RFC1 80AA genotype and low global DNA methylation at the enrolment, are at risk for poorer survival, further validation is required to assess whether this genetic-epigenetic fingerprint may be used as a clinically valuable prognostic tool. The reliable identification of prognostic molecular biomarkers may assume an important role in the clinical decision process for tailored therapeutic approaches in cancer treatment and in the improvement of cancer prognosis. Further investigations are certainly warranted to clarify the biological effect of RFC1 80G>A and the prognostic significance of PBMCs global DNA methylation assessment in primary liver cancers.
Acknowledgments
The work was performed in part in the LURM (Laboratorio Universitario di Ricerca Medica) Research Center, University of Verona, Verona, Italy.
Abbreviations
- HR
Hazard Ratio
- mCyt
methylcytosine
- PBMCs
peripheral blood mononuclear cells
- RFC1
Reduced Folate Carrier 1
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews Genetics. 2002;3(6):415–28. 10.1038/nrg816 [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ, Figueiredo JC, Lee W, Conti DV, Kennedy K, Duggan DJ, et al. A candidate gene study of folate-associated one carbon metabolism genes and colorectal cancer risk. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19(7):1812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazra A, Wu K, Kraft P, Fuchs CS, Giovannucci EL, Hunter DJ. Twenty-four non-synonymous polymorphisms in the one-carbon metabolic pathway and risk of colorectal adenoma in the Nurses' Health Study. Carcinogenesis. 2007;28(7):1510–9. 10.1093/carcin/bgm062 [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Kyte C, Valcin M, Chan W, Wetmur JG, Selhub J, et al. Polymorphisms in the one-carbon metabolic pathway, plasma folate levels and colorectal cancer in a prospective study. International journal of cancer Journal international du cancer. 2004;110(4):617–20. 10.1002/ijc.20148 [DOI] [PubMed] [Google Scholar]
- 5.Han SS, Sue LY, Berndt SI, Selhub J, Burdette LA, Rosenberg PS, et al. Associations between genes in the one-carbon metabolism pathway and advanced colorectal adenoma risk in individuals with low folate intake. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21(3):417–27. [DOI] [PubMed] [Google Scholar]
- 6.Choi SW, Mason JB. Folate status: effects on pathways of colorectal carcinogenesis. The Journal of nutrition. 2002;132(8 Suppl):2413S–8S. [DOI] [PubMed] [Google Scholar]
- 7.Kim YI. Role of folate in colon cancer development and progression. The Journal of nutrition. 2003;133(11 Suppl 1):3731S–9S. [DOI] [PubMed] [Google Scholar]
- 8.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33 Suppl:245–54. Epub 2003/03/01. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–40. Epub 2007/05/25. 10.1038/nature05919 [DOI] [PubMed] [Google Scholar]
- 10.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nature reviews Genetics. 2012;13(10):679–92. 10.1038/nrg3270 [DOI] [PubMed] [Google Scholar]
- 11.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–74. Epub 2006/08/17. [DOI] [PubMed] [Google Scholar]
- 12.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92. Epub 2007/02/27. 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo HD, Kim J. Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PloS one. 2012;7(4):e34615 Epub 2012/04/18. 10.1371/journal.pone.0034615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tangkijvanich P, Hourpai N, Rattanatanyong P, Wisedopas N, Mahachai V, Mutirangura A. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clinica chimica acta; international journal of clinical chemistry. 2007;379(1–2):127–33. 10.1016/j.cca.2006.12.029 [DOI] [PubMed] [Google Scholar]
- 15.Fabris S, Bollati V, Agnelli L, Morabito F, Motta V, Cutrona G, et al. Biological and clinical relevance of quantitative global methylation of repetitive DNA sequences in chronic lymphocytic leukemia. Epigenetics: official journal of the DNA Methylation Society. 2011;6(2):188–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu ZZ, Sparrow D, Hou L, Tarantini L, Bollati V, Litonjua AA, et al. Repetitive element hypomethylation in blood leukocyte DNA and cancer incidence, prevalence, and mortality in elderly individuals: the Normative Aging Study. Cancer causes & control: CCC. 2011;22(3):437–47. Epub 2010/12/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friso S, Udali S, Guarini P, Pellegrini C, Pattini P, Moruzzi S, et al. Global DNA hypomethylation in peripheral blood mononuclear cells as a biomarker of cancer risk. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2013;22(3):348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Udali S, Guarini P, Moruzzi S, Ruzzenente A, Tammen SA, Guglielmi A, et al. Global DNA methylation and hydroxymethylation differ in hepatocellular carcinoma and cholangiocarcinoma and relate to survival rate. Hepatology. 2015;62(2):496–504. 10.1002/hep.27823 [DOI] [PubMed] [Google Scholar]
- 19.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(8):5606–11. Epub 2002/04/04. 10.1073/pnas.062066299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer Journal international du cancer. 2010;127(12):2893–917. 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 21.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. Journal of the National Cancer Institute. 2004;96(19):1420–5. 10.1093/jnci/djh275 [DOI] [PubMed] [Google Scholar]
- 22.Araki A, Sako Y. Determination of free and total homocysteine in human plasma by high-performance liquid chromatography with fluorescence detection. Journal of Chromatography. 1987;422:43–52. [DOI] [PubMed] [Google Scholar]
- 23.Johnson WG, Stenroos ES, Spychala JR, Chatkupt S, Ming SX, Buyske S. New 19 bp deletion polymorphism in intron-1 of dihydrofolate reductase (DHFR): a risk factor for spina bifida acting in mothers during pregnancy? American journal of medical genetics Part A. 2004;124A(4):339–45. 10.1002/ajmg.a.20505 [DOI] [PubMed] [Google Scholar]
- 24.Ulrich CM, Bigler J, Bostick R, Fosdick L, Potter JD. Thymidylate synthase promoter polymorphism, interaction with folate intake, and risk of colorectal adenomas. Cancer research. 2002;62(12):3361–4. [PubMed] [Google Scholar]
- 25.Hol FA, van der Put NM, Geurds MP, Heil SG, Trijbels FJ, Hamel BC, et al. Molecular genetic analysis of the gene encoding the trifunctional enzyme MTHFD (methylenetetrahydrofolate-dehydrogenase, methenyltetrahydrofolate-cyclohydrolase, formyltetrahydrofolate synthetase) in patients with neural tube defects. Clinical genetics. 1998;53(2):119–25. [DOI] [PubMed] [Google Scholar]
- 26.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nature genetics. 1995;10(1):111–3. 10.1038/ng0595-111 [DOI] [PubMed] [Google Scholar]
- 27.Klerk M, Lievers KJ, Kluijtmans LA, Blom HJ, den Heijer M, Schouten EG, et al. The 2756A>G variant in the gene encoding methionine synthase: its relation with plasma homocysteine levels and risk of coronary heart disease in a Dutch case-control study. Thrombosis research. 2003;110(2–3):87–91. [DOI] [PubMed] [Google Scholar]
- 28.Jacques PF, Bostom AG, Selhub J, Rich S, Ellison RC, Eckfeldt JH, et al. Effects of polymorphisms of methionine synthase and methionine synthase reductase on total plasma homocysteine in the NHLBI Family Heart Study. Atherosclerosis. 2003;166(1):49–55. [DOI] [PubMed] [Google Scholar]
- 29.Chango A, Emery-Fillon N, de Courcy GP, Lambert D, Pfister M, Rosenblatt DS, et al. A polymorphism (80G->A) in the reduced folate carrier gene and its associations with folate status and homocysteinemia. Molecular genetics and metabolism. 2000;70(4):310–5. 10.1006/mgme.2000.3034 [DOI] [PubMed] [Google Scholar]
- 30.Heil SG, Van der Put NM, Waas ET, den Heijer M, Trijbels FJ, Blom HJ. Is mutated serine hydroxymethyltransferase (shmt) involved in the etiology of neural tube defects? Molecular genetics and metabolism. 2001;73(2):164–72. 10.1006/mgme.2001.3175 [DOI] [PubMed] [Google Scholar]
- 31.Friso S, Choi SW, Dolnikowski GG, Selhub J. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem. 2002;74(17):4526–31. Epub 2002/09/19. [DOI] [PubMed] [Google Scholar]
- 32.Dixon KH, Lanpher BC, Chiu J, Kelley K, Cowan KH. A novel cDNA restores reduced folate carrier activity and methotrexate sensitivity to transport deficient cells. The Journal of biological chemistry. 1994;269(1):17–20. [PubMed] [Google Scholar]
- 33.Brigle KE, Spinella MJ, Sierra EE, Goldman ID. Characterization of a mutation in the reduced folate carrier in a transport defective L1210 murine leukemia cell line. The Journal of biological chemistry. 1995;270(39):22974–9. [DOI] [PubMed] [Google Scholar]
- 34.DeVos L, Chanson A, Liu Z, Ciappio ED, Parnell LD, Mason JB, et al. Associations between single nucleotide polymorphisms in folate uptake and metabolizing genes with blood folate, homocysteine, and DNA uracil concentrations. The American journal of clinical nutrition. 2008;88(4):1149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matherly LH, Hou Z, Deng Y. Human reduced folate carrier: translation of basic biology to cancer etiology and therapy. Cancer metastasis reviews. 2007;26(1):111–28. 10.1007/s10555-007-9046-2 [DOI] [PubMed] [Google Scholar]
- 36.Winkelmayer WC, Eberle C, Sunder-Plassmann G, Fodinger M. Effects of the glutamate carboxypeptidase II (GCP2 1561C>T) and reduced folate carrier (RFC1 80G>A) allelic variants on folate and total homocysteine levels in kidney transplant patients. Kidney international. 2003;63(6):2280–5. 10.1046/j.1523-1755.2003.00025.x [DOI] [PubMed] [Google Scholar]
- 37.Yates Z, Lucock M. G80A reduced folate carrier SNP modulates cellular uptake of folate and affords protection against thrombosis via a non homocysteine related mechanism. Life sciences. 2005;77(22):2735–42. 10.1016/j.lfs.2005.02.029 [DOI] [PubMed] [Google Scholar]
- 38.Ulrich CM, Curtin K, Potter JD, Bigler J, Caan B, Slattery ML. Polymorphisms in the reduced folate carrier, thymidylate synthase, or methionine synthase and risk of colon cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2005;14(11 Pt 1):2509–16. [DOI] [PubMed] [Google Scholar]
- 39.Devlin AM, Clarke R, Birks J, Evans JG, Halsted CH. Interactions among polymorphisms in folate-metabolizing genes and serum total homocysteine concentrations in a healthy elderly population. The American journal of clinical nutrition. 2006;83(3):708–13. [DOI] [PubMed] [Google Scholar]
- 40.Stanislawska-Sachadyn A, Mitchell LE, Woodside JV, Buckley PT, Kealey C, Young IS, et al. The reduced folate carrier (SLC19A1) c.80G>A polymorphism is associated with red cell folate concentrations among women. Annals of human genetics. 2009;73(Pt 5):484–91. 10.1111/j.1469-1809.2009.00529.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotze T, Rocken C, Rohl FW, Wex T, Hoffmann J, Westphal S, et al. Gene polymorphisms of folate metabolizing enzymes and the risk of gastric cancer. Cancer letters. 2007;251(2):228–36. 10.1016/j.canlet.2006.11.021 [DOI] [PubMed] [Google Scholar]
- 42.Ma DW, Finnell RH, Davidson LA, Callaway ES, Spiegelstein O, Piedrahita JA, et al. Folate transport gene inactivation in mice increases sensitivity to colon carcinogenesis. Cancer research. 2005;65(3):887–97. [PMC free article] [PubMed] [Google Scholar]
- 43.Galbiatti AL, Ruiz MT, Rodrigues JO, Raposo LS, Maniglia JV, Pavarino EC, et al. Polymorphisms and haplotypes in methylenetetrahydrofolate reductase gene and head and neck squamous cell carcinoma risk. Molecular biology reports. 2012;39(1):635–43. 10.1007/s11033-011-0781-7 [DOI] [PubMed] [Google Scholar]
- 44.Galbiatti AL, da Silva LM, Ruiz-Cintra MT, Raposo LS, Maniglia JV, Pavarino EC, et al. Association between 11 genetic polymorphisms in folate-metabolising genes and head and neck cancer risk. European journal of cancer. 2012;48(10):1525–31. 10.1016/j.ejca.2011.09.025 [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Chen W, Wang J, Tan Y, Zhou Y, Ding W, et al. Reduced folate carrier gene G80A polymorphism is associated with an increased risk of gastroesophageal cancers in a Chinese population. European journal of cancer. 2006;42(18):3206–11. 10.1016/j.ejca.2006.04.022 [DOI] [PubMed] [Google Scholar]
- 46.de Jonge R, Tissing WJ, Hooijberg JH, Jansen G, Kaspers GJ, Lindemans J, et al. Polymorphisms in folate-related genes and risk of pediatric acute lymphoblastic leukemia. Blood. 2009;113(10):2284–9. 10.1182/blood-2008-07-165928 [DOI] [PubMed] [Google Scholar]
- 47.Laverdiere C, Chiasson S, Costea I, Moghrabi A, Krajinovic M. Polymorphism G80A in the reduced folate carrier gene and its relationship to methotrexate plasma levels and outcome of childhood acute lymphoblastic leukemia. Blood. 2002;100(10):3832–4. 10.1182/blood.V100.10.3832 [DOI] [PubMed] [Google Scholar]
- 48.Leyva-Vazquez MA, Organista-Nava J, Gomez-Gomez Y, Contreras-Quiroz A, Flores-Alfaro E, Illades-Aguiar B. Polymorphism G80A in the reduced folate carrier gene and its relationship to survival and risk of relapse in acute lymphoblastic leukemia. Journal of investigative medicine: the official publication of the American Federation for Clinical Research. 2012;60(7):1064–7. [DOI] [PubMed] [Google Scholar]
- 49.Jang MJ, Kim JW, Jeon YJ, Chong SY, Hong SP, Hwang SG, et al. Polymorphisms of folate metabolism-related genes and survival of patients with colorectal cancer in the Korean population. Gene. 2014;533(2):558–64. 10.1016/j.gene.2013.09.056 [DOI] [PubMed] [Google Scholar]
- 50.Kotsopoulos J, Zhang WW, Zhang S, McCready D, Trudeau M, Zhang P, et al. Polymorphisms in folate metabolizing enzymes and transport proteins and the risk of breast cancer. Breast cancer research and treatment. 2008;112(3):585–93. 10.1007/s10549-008-9895-6 [DOI] [PubMed] [Google Scholar]
- 51.Xu X, Gammon MD, Wetmur JG, Bradshaw PT, Teitelbaum SL, Neugut AI, et al. B-vitamin intake, one-carbon metabolism, and survival in a population-based study of women with breast cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17(8):2109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esteller M. Epigenetics in cancer. The New England journal of medicine. 2008;358(11):1148–59. Epub 2008/03/14. 10.1056/NEJMra072067 [DOI] [PubMed] [Google Scholar]
- 53.Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143(6):1442–60 e1 10.1053/j.gastro.2012.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deroo LA, Bolick SC, Xu Z, Umbach DM, Shore D, Weinberg CR, et al. Global DNA methylation and one-carbon metabolism gene polymorphisms and the risk of breast cancer in the Sister Study. Carcinogenesis. 2014;35(2):333–8. 10.1093/carcin/bgt342 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.