Abstract
Powdery mildew is a major fungal disease on squash and pumpkin (Cucurbita spp.) in the US and throughout the world. Genetic resistance to the disease is not known to occur naturally within Cucurbita pepo and only infrequently in Cucurbita moschata, but has been achieved in both species through the introgression of a major resistance gene from the wild species Cucurbita okeechobeensis subsp. martinezii. At present, this gene, Pm-0, is used extensively in breeding, and is found in nearly all powdery mildew-resistant C. pepo and C. moschata commercial cultivars. In this study, we mapped C. okeechobeensis subsp. martinezii-derived single nucleotide polymorphism (SNP) alleles in a set of taxonomically and morphologically diverse and resistant C. pepo and C. moschata cultivars bred at Cornell University that, by common possession of Pm-0, form a shared-trait introgression panel. High marker density was achieved using genotyping-by-sequencing, which yielded over 50,000 de novo SNP markers in each of the three Cucurbita species genotyped. A single 516.4 kb wild-derived introgression was present in all of the resistant cultivars and absent in a diverse set of heirlooms that predated the Pm-0 introgression. The contribution of this interval to powdery mildew resistance was confirmed by association mapping in a C. pepo cultivar panel that included the Cornell lines, heirlooms, and 68 additional C. pepo cultivars and with an independent F2 population derived from C. okeechobeensis subsp. martinezii x C. moschata. The interval was refined to a final candidate interval of 76.4 kb and CAPS markers were developed inside this interval to facilitate marker-assisted selection.
Introduction
Powdery mildew, caused by the obligate biotrophic pathogens Podosphaera xanthii and Golovinomyces cichoracearum, is one of the most prevalent and destructive fungal diseases globally of Cucurbita species, and especially of C. pepo, the most economically important species of squash and pumpkin [1–5]. In the US, P. xanthii (syn. Podosphaera fusca, Sphaerotheca fuliginea) is the most common powdery mildew pathogen species on Cucurbita [6]. P. xanthii can infect numerous species in the Asteraceae, Cucurbitaceae, Lamiaceae, Scrophulariaceae, Solanaceae, and Verbenaceae families and is easily spread between hosts via windborne asexual conidia [7, 8]. Powdery mildew on squash and pumpkin is easily identified by white mycelial growth on stems, petioles, and leaf surfaces that appear four to seven days post-infection [9]. Symptoms include chlorotic lesions that can eventually lead to whole plant death due to inhibition of photosynthesis [8]. Fruit yield and quality may be reduced in infected plants due to disease-induced sunscald, incomplete ripening, or poor storability [9].
Genetic resistance is an important tool for controlling powdery mildew on squash and pumpkin. Although regular foliar applications of fungicide can be used to manage the disease, fungicide-resistant strains of P. xanthii have reduced or eliminated the efficacy of many formerly effective fungicides [8, 10, 11]. Additionally, the most effective fungicides can be costly, especially when used repeatedly over the course of a long growing season [12]. Growers can deploy resistant varieties as part of an integrated management approach that requires less frequent, effective, and expensive fungicide applications [13]. Organic growers rely even more heavily on robust genetic resistance. Out of 105 respondents from a survey of vegetable farmers in the northeastern US who managed at least part of their farm in accordance with organic standards, 89% responded that genetic resistance to powdery mildew on cucurbits was important, and 37% said that genetic resistance to powdery mildew should be considered a critical priority of breeding programs [14].
To date, genetic resistance to powdery mildew has never been identified in C. pepo, and is found in only a few wild accessions of C. moschata. In a screen of the entire USDA collection of C. pepo during the late 1960s, none of the 292 accessions were resistant [15]. More recent evaluations of cultivars and accessions belonging to the USDA C. pepo collection grown under field-infected and growth chamber-inoculated conditions have resulted in the identification of accessions with partial resistance, although none with a degree of resistance that is alone sufficient for control [16–18]. Additionally, robust resistance to powdery mildew in C. pepo has not been reported from accessions held internationally. For C. moschata, accessions with resistance have been reported, but resistance from these sources is not common in mainstream commercial cultivars [4, 15, 19–22].
Resistant wild Cucurbita species with which C. pepo and C. moschata are sparingly cross-compatible have been used to introgress resistance genes into cultivated material [23]. The wild Cucurbita species C. lundelliana contains a dominant resistance gene that was introgressed into C. pepo through a C. moschata bridge [24–27]. Cultivars with these introgressions have not been commercialized, however, due to linkage drag associated with the introgression and incompleteness of resistance in cultivated backgrounds [4, 20]. A breakthrough occurred when the resistance gene Pm-0, from the wild species C. okeechobeensis subsp. martinezii (Fig 1), was successfully introgressed into squash and pumpkin at Cornell University. This was achieved first in C. moschata with a cross to ‘Butternut’ beginning in 1974, and later in C. pepo through the interspecific hybrid cross: (((C. pepo ‘Yankee Hybrid’ x C. moschata ‘Butternut’) x 'Yankee Hybrid) x (C. moschata ‘Butternut 23’ x C. okeechobeensis subsp. martinezii F1)) [4, 20, 28–30]. Following the initial crosses, the gene was incorporated into the open-pollinated C. moschata butternut cultivars ‘Bugle’ and ‘PMT Large Butternut’ and into open-pollinated cultivars of multiple morphotypes of both cultivated C. pepo subspecies. These included: ‘Success PM’, ‘PMR Bush Delicata’ and ‘Sweet REBA’, representing the straightneck, delicata, and acorn morphotypes, respectively, in the subspecies C. pepo subsp. texana, and ‘Romulus’, ‘PMR Caserta’, ‘Improved Costata’, and ‘PMR Naked Seeded’, representing the zucchini, vegetable marrow, cocozelle, and pumpkin morphotypes, respectively, in the subspecies C. pepo subsp. pepo [31, 32]. These Cornell cultivars or their progenitors have been used widely by other public and private breeding programs. At present, the Pm-0 gene is responsible for resistance in nearly all powdery mildew resistant (PMR) commercial cultivars of C. moschata and C. pepo [20], barring the possible exception of certain cultivars from Hollar Seeds [33]. The inheritance of Pm-0 in most cultivated backgrounds is incompletely dominant. In many contexts, even without conferring complete resistance, the Pm-0 gene in the homozygous or even heterozygous condition in C. pepo has been adequate for practical disease control [4, 22, 28, 34].
Fig 1. Cucurbita okeechobeensis subsp. martinezii.
The wild inedible gourd, native to the Gulf Coast of Mexico [35], is depicted growing in Ithaca, NY. C. okeechobeensis subsp. martinezii is central in the Cucurbita clade and interfertile with other Cucurbita [36]. C. okeechobeensis subsp. martinezii is the original source of powdery mildew resistance now found in C. pepo.
Resistant inbred C. pepo cultivars which contain the Pm-0 introgression but are otherwise genetically diverse as a result of directional breeding efforts can be considered a community-generated shared-trait introgression panel. When combined with susceptible and especially heirloom cultivars (for this study defined as those pre-dating the Pm-0 introgression event), these cultivars represent a powerful resource for mapping Pm-0. With the shared-trait introgression panel mapping approach, molecular markers are identified that define interspecific differences, e.g. markers that are monomorphic between diverse heirloom C. pepo cultivars, but polymorphic between the heirloom group and C. okeechobeensis subsp. martinezii. Subsequently, the genotypes for these markers are determined for all cultivars. Genomic regions in modern cultivars that contain alleles identical to the wild species are presumed derived from the wild species. Any wild species-derived introgression common among resistant cultivars becomes a candidate interval for the gene of interest. In the case of single, historic, and widely used alleles such as Pm-0, the potential for historical recombination events in at least some cultivars to have reduced the size of the candidate interval around the gene of interest is high, barring chromosomal inversions or other rearrangements present in the region containing the introgression. Previously, this approach has been used to map other major resistance genes derived from wild species in tomato [37, 38]. Our study has advantages over previous efforts in that only one gene from one wild donor species is known to be widespread among current cultivars for the trait of interest. Additionally, the original source of resistance is still available, pedigree records tracing Pm-0 back to its original donor exist for a suite of university-bred diverse cultivars, and high-throughput genotyping enables saturation of the genome with high-density molecular markers.
Genotyping-by-sequencing (GBS), which has been used to genotype other cucurbits [39], is an increasingly popular and cost-effective option for the de novo generation of thousands of high-density single nucleotide polymorphism (SNP) markers. In brief, GBS is the sequencing of multiplexed reduced-representation libraries that are generated by the enzymatic digestion of whole genomic DNA [40]. GBS is highly flexible to user requirements in order to achieve a read-depth sufficient for SNP-calling in populations of different types and genomes of varying sizes. Additionally, an array of restriction enzymes can be used to enrich for regions containing particular DNA patterns, including methylation-sensitive enzymes that enrich for non-repetitive, gene-rich genomic regions [41].
The objective of this research was to map the location of Pm-0, the primary resistance gene in C. pepo, through introgression mapping of a shared-trait introgression panel. Our results were validated by association mapping in a panel of C. pepo cultivars, and by testing the predictive ability of Pm-0-linked GBS SNP markers in an independent F2 population from a cross of C. okeechobeensis subsp. martinezii PI 532363 x C. moschata ‘Burpee’s Butterbush’. Finally, we developed CAPS markers predictive of powdery mildew resistance from Pm-0 in both C. pepo and C. moschata backgrounds that can be used for marker-assisted breeding efforts in further development of powdery mildew-resistant squash and pumpkin cultivars.
Materials and Methods
Plant Material
Introgression Mapping
Accessions and cultivars from three Cucurbita species were used to map the Pm-0-containing introgression. The original source of C. okeechobeensis subsp. martinezii, now PI 406680 [42], was regenerated from Cornell seed stocks and used to define “wild” alleles for SNP markers. A set of six C. pepo heirloom cultivars advertised in seed catalogs prior to the introgression of Pm-0 into C. pepo and belonging to multiple morphotypes and subspecies were used to define “C. pepo” alleles for SNP markers. The heirlooms and morphotypes were: ‘Black Beauty’ (zucchini), ‘Green Bush Vegetable Marrow’ (vegetable marrow), ‘Costata Romanesco’ (cocozelle), ‘Spirit’ (pumpkin), ‘Table King’ (acorn), and ‘Early Golden Summer Crookneck’ (crookneck). The shared-trait introgression panel consisted of a set of nine Cornell lines of C. pepo and C. moschata described in the introduction and listed in Table 1. Alleles in the resistant C. moschata cultivars were compared with the powdery mildew-susceptible C. moschata heirloom ‘Burpee’s Butterbush’.
Table 1. Germplasm used for introgression and association mapping of Pm-0.
| Name | Sp. | Subsp. | Type | Source | PMR | Name | Sp. | Subsp. | Type | Source | PMR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PI 406680 | o | mar | go | Cornell | R | PL5124-1 | p | pepo | pn | Rupp | UD |
| Bugle | m | bn | Cornell | R | Segev F1 | p | pepo | vm | High Mowing | R | |
| PMT Lg. Butternut | m | bn | Cornell | R | Caliph F1 | p | pepo | vm | Harris Moran | IR | |
| Success PM | p | tex | sn | Cornell | R | Citlali F1 | p | pepo | vm | Harris Moran | IR |
| PMR Bush Delicata | p | tex | de | Cornell | R | Hurakan F1 | p | pepo | vm | Harris Moran | IR |
| Sweet REBA | p | tex | ac | Cornell | R | Cha-Ching F1 | p | pepo | zu | High Mowing | R |
| Romulus | p | pepo | zu | Cornell | R | Emerald Delight F1 | p | pepo | zu | Territorial | R |
| PMR Caserta | p | pepo | vm | Cornell | R | Dunja F1 | p | pepo | zu | Johnny's | IR |
| Improved Costata | p | pepo | cz | Cornell | S | Elegance F1 | p | pepo | zu | Harris Moran | IR |
| PMR Nkd. Sd. Pkn. | p | pepo | pn | Cornell | R | Golden Glory F1 | p | pepo | zu | Johnny's | IR |
| Black Beauty | p | pepo | zu | Baker Creek | S | Midnight Lightning | p | pepo | zu | High Mowing | IR |
| Green Bush Veg. Mw. | p | pepo | vm | Baker Creek | S | Paycheck F1 | p | pepo | zu | Stokes | IR |
| Costata Romanesco | p | pepo | cz | High Mowing | S | Payroll F1 | p | pepo | zu | Stokes | IR |
| Spirit | p | pepo | pn | Jung | S | Preference F1 | p | pepo | zu | Harris Moran | IR |
| Table King | p | tex | ac | Olds | S | Prestige F1 | p | pepo | zu | Harris Moran | IR |
| Early Gn. Smr. Cknk. | p | tex | cn | Baker Creek | S | Quirinal F1 | p | pepo | zu | Stokes | IR |
| Burpee's Butterbush | m | bn | Rupp | S | Sebring F1 | p | pepo | zu | Fedco | IR | |
| Camaro F1 | p | pepo | pn | Hollar | R | Spineless Perfctn. F1 | p | pepo | zu | Johnny's | IR |
| Charisma F1 | p | pepo | pn | Johnny's | R | Wildcat F1 | p | pepo | zu | Harris Moran | IR |
| Hijinks F1 | p | pepo | pn | Osborne | R | Partenon F1 | p | pepo | zu | Johnny’s | IR |
| Mustang F1 | p | pepo | pn | Hollar | R | Ambassador F1 | p | pepo | zu | Osborne | S |
| WeeeeeOne F1 | p | pepo | pn | Rupp | R | Caserta | p | pepo | zu | Baker Creek | S |
| Bumpkin F1 | p | pepo | pn | Harris | IR | Zucchini Elite F1 | p | pepo | zu | Harris | S |
| Diablo F1 | p | pepo | pn | Fedco | IR | Honey Bear F1 | p | tex | ac | Johnny's | R |
| Gargoyle F1 | p | pepo | pn | Harris | IR | Sugar Dumpling F1 | p | tex | ac | High Mowing | R |
| Gladiator F1 | p | pepo | pn | Harris Moran | IR | TipTop PMR F1 | p | tex | ac | Johnny's | R |
| Gold Dust F1 | p | pepo | pn | Rupp | IR | Autumn Delight F1 | p | tex | ac | Osborne | IR |
| Iron Man F1 | p | pepo | pn | Harris | IR | Royal Ace PM F1 | p | tex | ac | Harris Moran | IR |
| Magic Lantern F1 | p | pepo | pn | Harris | IR | Table Star F1 | p | tex | ac | Rupp | IR |
| Magician F1 | p | pepo | pn | Harris Moran | IR | Table Treat F1 | p | tex | ac | Rupp | IR |
| Merlin F1 | p | pepo | pn | Osborne | IR | Taybelle PM F1 | p | tex | ac | Stokes | IR |
| Mischief F1 | p | pepo | pn | Harris Moran | IR | Celebration F1 | p | tex | ac | Rupp | IR |
| Owl's Eye F1 | p | pepo | pn | High Mowing | IR | Ebony | p | tex | ac | Reimer | S |
| Prankster F1 | p | pepo | pn | Rupp | IR | Sweet Lightning F1 | p | tex | ac | Rupp | IR |
| Warlock F1 | p | pepo | pn | Harris | IR | Delicata | p | tex | de | Baker Creek | S |
| Rival PMR F1 | p | pepo | pn | Johnny's | IR | Delta F1 | p | tex | cn | Territorial | IR |
| Chucky F1 | p | pepo | pn | Johnny's | S | Sunglo F1 | p | tex | cn | Osborne | IR |
| G.bumps Spr. Frk. F1 | p | pepo | pn | Territorial | S | Gold Star F1 | p | tex | cn | Osborne | IR |
| Howden | p | pepo | pn | High Mowing | S | Dk. Gn. Scall. F1 | p | tex | sc | High Mowing | R |
| Sorceror F1 | p | pepo | pn | Harris Moran | S | Yellow Scallopini F1 | p | tex | sc | High Mowing | IR |
| PL3602-2 | p | pepo | pn | Rupp | UD | Cheetah F1 | p | tex | sn | Harris Moran | IR |
| PL3517-3 | p | pepo | pn | Rupp | UD | Cougar F1 | p | tex | sn | Harris | S |
| PL3885-1 | p | pepo | pn | Rupp | UD |
Cucurbita spp. used for Pm-0 mapping: C. okeechobeensis subsp. martinezii PI 406680, the original source of Pm-0, the Cornell-bred shared-trait introgression panel (bolded and underlined), Cucurbita heirlooms (bolded and italicized), and assorted C. pepo cultivars. For species (“Sp.”), p = C. pepo, m = C. moschata, o = C. okeechobeensis subsp. martinezii. For C. pepo subspecies (“Subsp.”), pepo = C. pepo subsp. pepo, tex = C. pepo subsp. texana. For morphotype (“Type”), ac = acorn, bn = butternut, cn = crookneck, cz = cocozelle, de = delicata, go = gourd, pn = pumpkin, sc = scallop, sn = straightneck, vm = vegetable marrow, zu = zucchini. Subspecies and morphotypes are as defined by Paris et al. and Gong et al. [31, 32]. For “PMR”, resistance phenotypes are listed as described/inferred from the vendor’s website. R = resistant, IR = intermediately resistant (sometimes described as “tolerant”), S = susceptible, UD = undefined. These classifications were used for cultivar selection only and not for downstream analysis. Selected cultivars are abbreviated as follows: PMT Lg. Butternut = Large Powdery Mildew Tolerant Butternut, PMR Nkd. Sd. Pkn. = Powdery Mildew-Resistant Naked-Seeded Pumpkin, Green Bush Veg. Mw. = Green Bush Vegetable Marrow, Early Gn. Smr. Cknk. = Early Golden Summer Crookneck, G.bumps Spr. Frk. F1 = Goosebumps Super Freak F1, Spineless Perfctn. F1 = Spineless Perfection F1, Dk. Gn. Scall. = Dark Green Scallopini.
Association Mapping
The Pm-0-containing genomic region identified through introgression mapping was confirmed by association mapping in a panel of 81 C. pepo cultivars that included 68 C. pepo commercial cultivars in addition to the seven Cornell-bred C. pepo cultivars in the shared-trait introgression panel and the six heirlooms used for introgression mapping. The species, subspecies, morphotype, seed source, and putative resistance based on catalog description of each cultivar are listed in Table 1.
Biparental Population
A biparental F2 population consisting of 177 individuals from a cross between C. okeechobeensis subsp. martinezii PI 532363 and the powdery mildew susceptible C. moschata ‘Burpee’s Butterbush’ was used to generate a genetic map to anchor SNP markers, and to test Pm-0-linked SNPs for predictiveness of resistance in a segregating population.
Genotyping
DNA Extraction
DNA was extracted and diluted in preparation for GBS. Two to three meristematic leaves from single plants of each cultivar, or in the case of the F2 population, from each plant, were collected in the field. Samples were then lyophilized for at least 48 hours. DNA was extracted using the frozen/lyophilized plant tissue protocol starting on page 35 of the 2012 Qiagen DNeasy Plant Handbook (https://www.qiagen.com/us/resources/resourcedetail?id=95dec8a9-ec37-4457-8884-5dedd8ba9448&lang=en) but eluted with 30 μL of Buffer AE twice for a final volume of 60 μL. Samples were then quantified using the Invitrogen Quant-iT PicoGreen kits. One microliter from each sample was pipetted into 198 μL of 1x TE buffer and 0.5 μL of 200x PicoGreen. Samples were quantified in a black, flat-bottomed 96-well plate with a SpectraMax plate reader using an excitation wavelength of 480 nm and emission wavelength of 520 nm. Fluorescence units were converted to concentrations based on a standard curve calculated using eight different concentrations of Lambda DNA from 0 to 200 ng/μL. DNA was diluted to a final concentration of 10 ng/μL.
GBS Library Preparation
Genotyping-by-sequencing was used to genotype all samples. 96-plex libraries were prepared according to the protocol described by Elshire et al. [40]. All distinct genotypes were sequenced individually except the parents of the F2 mapping population and C. okeechobeensis subsp. martinezii PI 406680, which were sequenced in replicate. The partially methylation-sensitive restriction enzyme ApeKI, which recognizes a degenerate five base pair sequence, was chosen for the digestion step due to its potential to enrich for gene-rich regions. Excess primer dimers in the library were removed using 1.8X volumes of the Agencourt AMPure beads (Beckman Coulter). Each GBS library was sequenced on one lane of a HiSeq 2000 Illumina Sequencing System.
Calling SNPs
SNPs were called using the TASSEL-GBS pipeline build 5.2.10 [43]. Bowtie2 was used to align Illumina reads to the C. pepo zucchini genome draft v3.2 pre-released by a joint effort of the Genomics and Bioinformatics and Cucurbits Breeding Groups of the COMAV–Polytechnic University of Valencia (www.cucurbigene.net). To accommodate formatting constraints within the TASSEL pipeline, the first 19 largest scaffolds in the draft genome were left unmodified, and all remaining scaffolds were concatenated into a superscaffold with 80 “N”s inserted between each of the original scaffolds. Default TASSEL pipeline parameters were used with the exception that the parameter “c” (minimum number of times a tag must be present to be output) was set at five for the MergeMultipleTagCount and TagCountToFastq plugins.
Genetic Map Construction
A genetic map was created using stringently filtered markers called in the C. okeechobeensis subsp. martinezii PI 532363 x C. moschata ‘Burpee’s Butterbush’ F2 population in order to anchor markers for downstream analyses. Using a custom python script, genotypes represented by less than seven reads were converted to missing data in order to reduce errors associated with under-calling or the false identification of heterozygous loci, common problems for low-coverage loci [44, 45]. Seven reads is the minimum number required to call a heterozygote using at least two reads of the “less tagged allele” based on the binomial likelihood ratio employed in TASSEL and assuming a sequencing error rate of 1%, a conservative estimate for Illumina sequencing [43, 46]. TASSEL was subsequently used to filter SNPs by a minimum minor allele frequency of 0.25, a locus call rate of 0.95 and a taxa call rate of 0.85 [47]. SNPs characterized by different alleles between the parents were selected using the ABH Genotype plugin in TASSEL [48]. The package R/qtl in the R statistics environment was used to generate the genetic map [49, 50]. Duplicate individuals and markers were removed, as well as markers showing segregation distortion, as determined by a p-value less than 1 x 10−8. Recombination frequencies between all pairs of markers were estimated using the function “est.rf”. Linkage groups (LGs) were formed using the “formLinkageGroups” function with a maximum recombination frequency of 0.15 and minimum lod of 25. The single marker that was not placed on the 20 primary LGs was discarded. Markers were ordered on LGs with the “OrderMarkers” function, and marker order was evaluated over a sliding window of 6 using the “ripple” option. Linkage disequilibrium between all pairs of markers for each chromosome were plotted, and in regions visually suggestive of incorrect ordering, markers were manually reordered if the new order increased the LOD score and decreased the length of the LGs. Sixteen markers were removed that in the majority of individuals were flanked by non-like genotypes, and genotypes with a high probability of being errors as defined by an error LOD score greater than 2 using the “calc.errorlod” function were changed to missing data using a custom Perl script.
Introgression Mapping
For each of the heirlooms and Cornell-bred shared-trait introgression panel cultivars, GBS marker genotypes were plotted along all 20 linkage groups using the genetic map to anchor markers with common SNP identification numbers. Alleles were shaded blue if the locus genotype was homozygous for the “wild” allele, identical to C. okeechobeensis subsp. martinezii, gray if the marker genotype was homozygous for the “C. pepo” allele, identical to all C. pepo heirlooms, or light blue if in the heterozygous state. Any markers that were not represented on the C. okeechobeensis subsp. martinezii PI 532363 x C. moschata ‘Burpee’s Butterbush’ F2 genetic map by common SNP ID numbers or that displayed interspecific monomorphism or intraspecific polymorphism were filtered out using the TASSEL ABH plugin and a custom Perl script. Missing genotypes that were doubly flanked by markers with identical genotype were imputed to the flanking genotype. Loci genotypes that were positioned at least 20 cM distant from an identical genotype, and which was positioned no more than 3 cM distant from flanking genotypes that were different to the locus under consideration but identical to each other, were considered errors and converted to the flanking genotypes.
After a genomic Pm-0-containing introgression region was identified, this region was mapped at higher resolution using all called SNP markers in the region ordered by their scaffold positions, regardless of whether the markers were represented in the genetic map. Markers were filtered by a locus call rate of 0.50 and missing genotypes were imputed using default settings in Beagle 4.0 [51]. SNPs defined by alternate alleles between C. okeechobeensis subsp. martinezii and C. pepo were selected as described for the whole genome introgression map. Marker genotypes were considered errors and converted to flanking genotypes if they were within 5 kb of flanking markers with different genotypes which were in turn part of a long string of identical marker genotypes that extended more than 10 kb in each direction. A Pm-0-containing candidate interval was identified by the common area of overlap between the introgressions in all resistant cultivars.
Pm-0 Validation by Association Mapping
Association mapping was used to validate the Pm-0-containing genomic interval identified by introgression mapping. Cultivars were grown and phenotyped in Ithaca, NY in the summer of 2013. Cultivars were transplanted in six-plant plots in a randomized complete block design with three replicates. Plants were transplanted near a squash field with high loads of natural inoculum; disease pressure was increased two weeks after transplanting by inoculating a mixture of cultivars planted around the perimeter of the field and throughout the field at five row intervals with a suspension of P. xanthii conidia from nearby squash plants and diluted to 10,000 spores mL -1 in a .002% Tween 20 solution. The pathogen of powdery mildew was determined by amplifying and sequencing rRNA ITS4 and ITS5 regions as described by White et al. and aligning them to NCBI sequences in the non-redundant (nr) database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [52,53]. Early-fruiting summer squash cultivars were stripped of harvestable fruit on a weekly basis to remove resistance effects associated with maturity and fruit load. After six weeks, petioles of fully-expanded leaves were rated on a per-plot basis, averaged over three plots, using a scale described in Fig 2. Petiole ratings were chosen based on our previous observations in both cultivar panels and biparental populations that petiole symptoms at this stage of development were the most straightforward and reliable predictors of Pm-0 dosage and presence/absence of powdery mildew resistance in the rest of the plant. In addition to “high”, “medium”, and “low” disease ratings, which might be expected for a single incompletely dominant gene, intermediary classifications were also included, which accounted for observed variations in the field and the likely presence of small-effect modifier genes.
Fig 2. Petiole rating using a 1–5 scale.
1—No pathogen colonies visible on petioles. 2—A small number of colonies limited to the base of some petioles. 3—Colonies on nearly all petioles near the base, and extending halfway up the petiole. 4—Colonies on all petioles, extending the full length of the petiole to the leaf blade, but lacking colony density of fully susceptible cultivars, especially near the leaf blade. 5—All petioles covered with pathogen colonies from petiole base to the leaf blade at high density; most individual colonies have coalesced into larger colonies.
For the analysis, we used a mixed linear model approach using the SUPER GWAS method as implemented in GAPIT, controlling for population structure with kinship and three principal components generated by the software [54, 55]. Markers from C. pepo cultivars were filtered for a minor allele frequency of 0.05 and a locus call rate of 0.50 and were drawn from scaffold locations within 30kb of markers identified on the F2 genetic map through common SNP ID numbers; they were subsequently assigned the genetic map position of their anchor marker using a custom python script. A Manhattan plot was generated in R using the qqman package [56].
Refining the Interval
The Pm-0-containing genomic interval was reduced to a smaller interval by analyzing co-segregation between resistance phenotypes and selected marker genotypes for the shared-trait introgression panel and selected proprietary commercial cultivars. The interval was continuously narrowed based on absence of universal co-segregation of genotypes and phenotypes until an interval of 76.4 kb was reached with the flanking markers S9_1474683 and S9_1551065. CAPS primers were designed from 1000 bp sequences from the C. pepo draft 3.2 genome that surrounded GBS markers using Primer3Plus and filtered for single alignment to the genome using a custom python script [57]. The forward and reverse primers for S9_1474683 were: 5´-TGTCGCAGCATGACATCTAGTT-3´ and 5´-TGTCAGATATGGCGTCTGGATG-3´, respectively. The forward and reverse primers for S9_1551065 were 5´-ACGATCCATCCTCATTGACC-3´ and 5´-TGAGGACAGAGCAGCGAGTA-3´, respectively. CAPS markers were amplified with the following PCR reagents: 10 μL of 2 ng/μL DNA, 2 μL of 10x PCR buffer, 1 μL of 2.5 mM dNTPs, 0.25 μL of 10 μM forward primer, 0.25 μL of 10 μM reverse primer, 0.25 μL Taq polymerase, and 6.25 μL of sterile distilled water using the following thermocycler program: initial denaturation at 94°C for 3 minutes, 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 90 seconds, and a final extension at 72°C for 15 minutes. PCR products were sequenced on an Applied Biosystems Automated 3730xl DNA Analyzer and analyzed with Sequencher version 4.9 to form consensus sequences [58]. The Sol Genomics Network (SGN) CAPS designer was used to select RsaI and PvuII as restriction enzymes to digest markers S9_1474683 and S9_1551065, respectively [59]. Samples were digested at 37°C for 2 hours using the following reagents: 10 μL of PCR product, 2 μL of 10x NEB CutSmart restriction buffer, 0.1 μL of 50 Unit/μL restriction enzyme, and 7.9 μL of sterile distilled water. The result was visualized on a 1.5% agarose gel.
Pm-0 Validation in a Segregating Population
GBS markers within the Pm-0-containing candidate genomic interval were validated within the C. okeechobeensis subsp. martinezii PI 532363 x C. moschata ‘Burpee’s Butterbush’ F2 population grown in Wauseon, OH by Rupp Seeds, Inc. Natural inoculum was prevalent in the field two months after transplanting, and ratings were taken approximately four months after transplanting near the end of the season. Petioles of F2 plants were scored with a binary rating, where 0 indicated no powdery mildew signs or symptoms, and 1 indicated presence of pathogen colonies and/or lesion symptoms. The Pm-0-containing interval identified by introgression mapping was divided into 10 bins spaced 50 kb apart. For the first GBS marker in each bin that showed no segregation distortion and a 95% call rate, a one-way ANOVA as implemented in the agricolae package in R was used to determine statistical difference between the genotype classes [60].
Identification of Candidate Genes
The validated 76.4 kb Pm-0-containing genomic interval was aligned to the nr database by nucleotide BLAST using the NCBI web-interface and the megablast and discontiguous megablast options (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [53].
CAPS Marker Development and Validation for Marker-Assisted Selection
A CAPS marker in a putative NBS-LRR gene within the newly refined interval and displaying complete co-segregation of genotypes with phenotypes in the shared-trait introgression panel was developed for use in marker-assisted breeding using the same protocol used to develop the interval-defining CAPS markers. The forward and reverse PCR primers for the marker, labeled NBS_S9_1495924, were 5´-TCAACGGATATCTCCACCAAG-3´ and 5´-TACAGAGCAGCCTGGATGAGT-3´, respectively. The PCR products were digested with restriction enzyme HaeIII using the aforementioned described digest conditions. A secondary marker was developed as an additional resource. This marker was developed near the predicted Cucumis melo uncharacterized LOC103484742. The forward and reverse primers for this marker, S9_1539675 were 5´-ACTTAGAGAATGGTTCGACCTCTG-3´ and 5´-CTGGAGAGCTGTAAGTGAAGATCA-3´, respectively. The PCR products were digested with restriction enzyme MspI under the same restriction digest conditions as the previous enzymes.
Results and Discussion
Genotyping
GBS was used to call over 50,000 conservatively filtered markers in each species and in the F2 population, resulting in one of the largest SNP data sets to date for Cucurbita. Raw Illumina reads were trimmed to 64 bases and filtered for the presence of an expected cut site remnant, barcode sequence, and no missing bases with the TASSEL-GBS pipeline. For C. pepo cultivars, C. moschata cultivars, and the C. okeechobeensis subsp. martinezii PI 532363 x C. moschata ‘Burpee’s Butterbush’ F2 population, the number of filtered barcoded reads, reads aligning to the physical scaffolds, number of unique markers, average read depth, and missing data are reported in Table 2 for all GBS markers as well as for a subset with an average minimum read depth of five. GBS in 96-plex using the enzyme ApeKI is effective for generating high numbers of deep-coverage markers for the Cucurbita species included in this study.
Table 2. GBS sequencing read and marker statistics for genotyped Cucurbita.
| C. pepo | C. moschata | C. okee. | F2 | ||
|---|---|---|---|---|---|
| Individuals | 81 | 3(4)** | 2(6)** | 177 | |
| Filtered Barcoded Sequencing Reads | 115,452,288 | 5,918,285 | 6,769,505 | 226,188,080 | |
| Reads Aligned to Physical Scaffolds | 106,503,712 | 5,433,639 | 5,583,919 | 197,758,572 | |
| All GBS Markers | 254,760 | 190,579 | 194,730 | 252,090 | |
| Avg. Read Depth | 5.62 | 7.87 | 5.59 | 4.91 | |
| Proportion Missing Data | 0.42 | 0.27 | 0.33 | 0.45 | |
| GBS Markers with avg. min. read depth ≥ 5 | 61,090 | 63,058 | 53,796 | 57,151 | |
| Avg. Read Depth | 19.63 | 20.66 | 16.43 | 17.39 | |
| Proportion Missing Data | 0.04 | 0.02 | 0.03 | 0.07 |
C. pepo includes the cultivar panel. C. moschata includes ‘PMT Large Butternut’, ‘Bugle’, and ‘Burpee’s Butterbush’. “C. okee.” includes two C. okeechobeensis subsp. martinezii accessions: PI 406680, the original source of Pm-0, and PI 532363, one of the parents of the F2 population. The F2 population is derived from C. okeechobeensis subsp. martinezii PI 532363 and C. moschata ‘Burpee’s Butterbush’
** The number outside of the parentheses is the number of distinct genotypes. The number inside of the parenthesis includes the total number of individuals sequenced in the case where some genotypes were sequenced in multiple technical replicates. Values in the table represent all technical replicates.
Genetic Map Construction
The C. okeechobeensis subsp. martinezii PI 532363 x C. moschata ‘Burpee’s Butterbush’ F2 population was used to generate a high-density genetic map for anchoring C. pepo SNP markers. The order of C. pepo markers based on a population derived from non-C. pepo parents was considered accurate based on previous reports describing synteny, no major chromosomal rearrangements, and high rates of marker transferability between C. pepo and C. moschata [61, 62], and the lack of any marker pairs in the map separated by large genetic distances which would indicate large chromosomal rearrangements between C. moschata and C. okeechobeensis subsp. martinezii. With stringent filtering conditions, our map yielded 2,669 markers over a total map distance of 2,199.2 cM, summarized in S1 Table, approximating the C. pepo map distance reported by Gong et al. for the only other Cucurbita map consisting of 20 LGs (1936 cM) [62], and the C. pepo map distance reported by Esteras et al. for the only other Cucurbita map generated with SNP markers (1740.8 cM) [63]. Identification numbers, LGs, and genetic map position for all markers are available in S2 Table. LGs are ordered by map distance. For marker ID numbers, the number following “S” corresponds to the scaffold of alignment from the C. pepo draft genome v3.2, with the exception of scaffold 20, which represents the “superscaffold” as described in the methods section. The number after the underscore corresponds to the base position of the relevant scaffold. For the 19 largest scaffolds of the C. pepo draft genome, only two scaffolds: 11 and 19, were not collinear on a single LG in our map. This could reflect chimeric scaffolds of the draft genome or rearrangement between C. moschata and C. pepo. In either case, the LGs containing these split scaffolds did not contain C. okeechobeensis subsp. martinezii introgressions, and were not important for downstream introgression or association mapping in this study.
Introgression Mapping
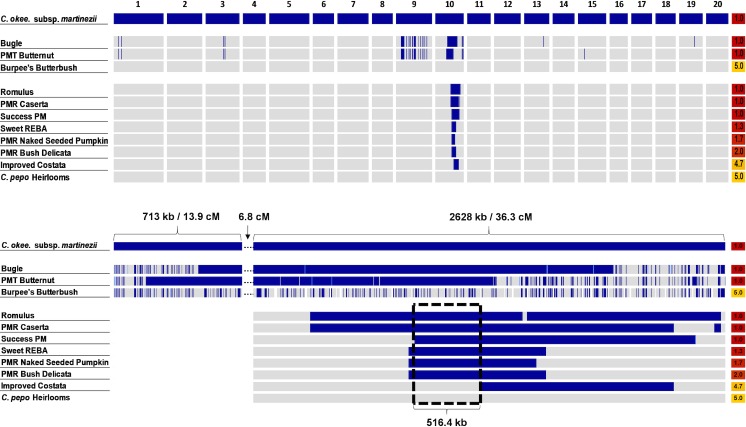
The Pm-0-containing introgression from C. okeechobeensis subsp. martinezii was mapped in a set of 17 Cornell-bred and heirloom C. moschata and C. pepo cultivars (Fig 3). Genotypes of 1,011 loci were plotted across 20 LGs; only loci present in the F2 genetic map and characterized by fixed, variant alleles between C. okeechobeensis subsp. martinezii and a set of six heirloom C. pepo cultivars were used. Heirloom cultivars, which were collectively used to define “C. pepo” allele genotypes, appeared true-to-type phenotypically and genotypically. One wild-derived introgression on LG 10 was common among all resistant cultivars and absent in all susceptible cultivars, identifying it as the Pm-0-containing region (Fig 3A). Of note is that the two Cornell-bred, powdery mildew-resistant C. moschata cultivars contain additional C. okeechobeensis subsp. martinezii introgressions absent in C. moschata ‘Burpee’s Butterbush’. Although these could contribute to resistance, it is likely that these introgressions are relicts from the breeding process, given that these cultivars are closely related to each other and are fewer generations removed from C. okeechobeensis subsp. martinezii than any of the C. pepo cultivars used in this study.
Fig 3. Introgression maps of Cornell-bred and heirloom Cucurbita inbreds.
Genomic regions homozygous for the C. pepo alleles, as defined by the heirlooms, are shaded gray; genomic regions homozygous for the C. okeechobeensis subsp. martinezii alleles are shaded dark blue, and heterozygotes are shaded light blue. Cultivars are ordered based on petiole rating, from most resistant to least resistant, and secondly by the size of the largest and most prevalent C. okeechobeensis subsp. martinezii introgression on LG 10. (A) Whole Genome Map. LG 10 contains the Pm-0-containing introgression. (B) LG 10 Map. A dotted box appears around the 516.4 kb region of the introgression that all resistant cultivars share in common, indicating the putative interval for Pm-0. The region spans two scaffolds from the v.3.2 draft genome.
A higher resolution map of the introgression region illuminated a Pm-0-containing region (Fig 3B). The marker order of the physical scaffolds corresponding to this region on the genetic map agreed with the genetic map positions, and so all markers with a locus call rate greater than 0.5 were plotted and physical scaffold positions used, regardless of whether the marker was present in the genetic map. One side of the interval was defined by ‘Success PM’ using marker S9_1150923 and the other side of the interval was defined by marker S9_1667287 by ‘Improved Costata’, which displayed C. okeechobeensis subsp. martinezii-derived powdery mildew resistance in early generations of breeding but lost the resistance in later generations, as demonstrated by high petiole ratings. The cultivar retained some of the wild introgression, but not the portion containing Pm-0. The total size of the interval is 516.4 kb.
The small size of the candidate interval and the loss of resistance from ‘PMR Costata’ indicates that recombination events have occurred around the Pm-0 gene as it has been incorporated into new cultivars. The capacity for recombination in this region to reduce the size of the wild introgression may be important to breeding efforts if the larger introgression contributes negatively to any non-disease-related horticultural and agronomic traits, as has been reported previously. For instance, C. pepo lines homozygous for the resistance gene have been reported as inherently lower-yielding when compared with susceptible commercial lines of the same fruit type [10]. Additionally, late-maturity has been associated with resistance in some cultivars [30]. However, these issues have been resolved in some cases by incorporating the resistance into new and especially highly productive backgrounds [20, 30], suggesting that either: large wild introgressions which contain alleles that retard yield or maturity can be decoupled from Pm-0 through recombination, or that epistatic interactions between Pm-0 or closely linked genes and certain genetic backgrounds may affect the pleiotropic expression of Pm-0 for other non-disease resistance traits.
Pm-0 Validation by Association Mapping
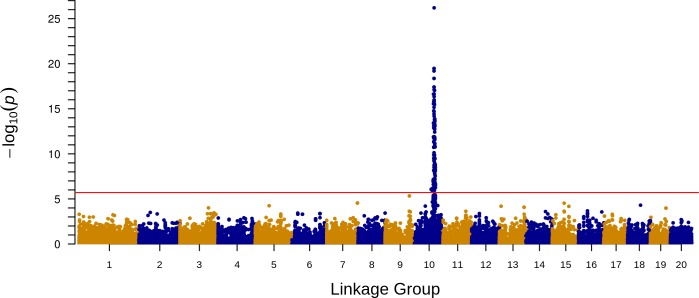
Association mapping validated the significance of the Pm-0 candidate interval using a set of 25,446 markers. The squash cultivar panel was phenotyped amidst heavy and uniform disease pressure throughout the field in Ithaca, NY in 2013. The pathogen of powdery mildew was confirmed to be P. xanthii by 99% homology of sequenced rRNA ITS4 and ITS5 regions to NCBI sequences of P. xanthii. No phenotypic variation was observed among or between plots of any given cultivar that would indicate genetic segregation for powdery mildew resistance or any other trait. Average petiole ratings with standard error for Cornell-bred cultivars, heirloom cultivars, and commercial cultivars are listed in S3 Table. The SUPER method as implemented in GAPIT was used for mapping, and principal components and kinship were used to account for population structure, which clearly existed between the C. pepo subspecies. A clear peak on the Manhattan plot occurs in the Pm-0 candidate interval (Fig 4), and the most significant p-value, 6.27e-27, is at marker S9_1551065 on LG 10.
Fig 4. Mapping the Pm-0 gene in the cultivar panel.
Manhattan plot of negative log p-values for each marker across all 20 LGs. The threshold for significance was set at bonferonni-adjusted α = 0.05 of 1.96e-6. 145 markers on LG 10 are significant and the most highly significant markers fall within the Pm-0 candidate interval identified through introgression mapping.
The single GWAS signal, along with the lack of multiple wild-derived introgressions in the Cornell-bred resistance lines, suggests that powdery mildew-resistance in C. pepo varieties developed by U.S.-based seed companies is conferred largely by a single gene from C. okeechobeensis subsp. Martinezii PI 406680. However, resistance alleles from other sources may be important in some cultivars; petiole ratings between cultivars, even those representing the same market class and relative maturity, varied more than would be expected for a trait controlled entirely by a single incompletely dominant gene. Small-effect resistance-enhancing alleles may have been acquired in more resistant cultivars from susceptible ancestors that did not carry the alleles in the appropriate zygosity or genetic background for the expression of notable resistance [64].
Refining the Interval
All 20 markers that were most significantly associated with powdery mildew resistance as determined by association mapping localized within the 516.4 kb Pm-0-containing interval identified through introgression mapping. To further refine the interval, CAPS markers were developed within the interval and genotyped in the shared-trait introgression panel and selected proprietary commercial cultivars until no recombinational breakpoints could be identified in any cultivar. The final interval of 76.4 kb was flanked by markers S9_1474683 and S9_1551065. This 76.4 kb interval was used for the identification of candidate genes.
Pm-0 Validation in Segregating F2 Population
SNP markers in the LG 10 Pm-0-containing interval from C. pepo were also associated with wild-derived resistance in an F2 population generated from a cross between C. okeechobeensis subsp. martinezii PI 532363, and C. moschata ‘Burpee’s Butterbush’. ANOVA tests of significance on four GBS markers from 50 kb bins within and immediately flanking the refined interval confirmed the effect of an incompletely dominant gene for powdery-mildew resistance in a 2nd C. okeechobeensis subsp. martinezii accession–PI 532363 (Table 3).
Table 3. ANOVA of Pm-0 in interspecific F2 population.
| Marker Name | Genotype | Genotype Average | Tukey's HSD |
|---|---|---|---|
| S9_1473058 | |||
| A | 0.6522 | a | |
| H | 0.5769 | a | |
| B | 0.4902 | a | |
| S9_1498203 | |||
| A | 0.7021 | a | |
| H | 0.561 | ab | |
| B | 0.4565 | b | |
| S9_1547588 | |||
| A | 0.7143 | a | |
| H | 0.5676 | ab | |
| B | 0.46 | b | |
| S9_1604471 | |||
| A | 0.7021 | a | |
| H | 0.5443 | a | |
| B | 0.5106 | a |
For every marker tested in and near the Pm-0 candidate region, the class of individuals characterized by homozygous C. moschata-derived alleles (“A” genotypes) displayed higher scores for binary powdery mildew ratings on petioles when compared with the heterozygous class (“H” genotype) and the class with homozygous C. okeechobeensis subsp. martinezii-derived alleles (“B” genotypes). The markers inside of the refined interval were statistically significant at a p-value <0.05 as determined by a Tukey’s Honestly Significant Difference (HSD) test, while those outside of the refined interval were not statistically significant.
In phenotyping the interspecific F2 population, it was clear that in addition to Pm-0 on LG 10, additional genes were contributing to resistance in the most disease-free individuals. Out of a total of 173 F2 individuals phenotyped, 75 were given a rating of 0, indicating that no P. xanthii colonies or powdery mildew symptoms were observed, even though disease pressure was high and ratings were taken at the end of a long season. The absence of single-gene Mendelian segregation patterns confirms observational data that the resistance in C. okeechobeensis subsp. martinezii, which is characterized as complete, is multigenic and complex. With replicated families and a quantitative rating system, it may be possible to identify some of these additional resistance alleles in the future, and the incorporation of new resistance alleles from C. okeechobeensis subsp. martinezii into C. moschata and C. pepo may be valuable to future squash breeding efforts. Although Pm-0 continues to provide good control of powdery mildew in many trials of C. pepo in the US [34, 65, 66], additional control of powdery mildew may be needed in the future based on some recent reports indicating that the level of control provided by Pm-0 appears reduced or eliminated relative to previous years [67, 68], potentially a result of the emergence of new races of P. xanthii [69, 70].
Identification of Candidate Genes
BLAST alignment of the 76.4 kb Pm-0-containing interval to the NCBI nr database yielded 14 putative genes, listed in S4 Table. Several putative genes in the interval are homologous to genes in other genera that are known to be involved in disease resistance. Of particular interest is the probable homolog of At5g66900, a NBS-LRR protein in Arabidopsis thaliana that contains a domain with similarity to the RPW8 locus that confers resistance to powdery mildew. In addition to Arabidopsis, NBS-LRR proteins have been found in powdery mildew resistance-associated regions in watermelon, a relative of Cucurbita spp. in the Cucurbitaceae family [71–74]. In addition to the putative NBS-LRR locus, numerous other candidates exist in the interval. At position 4, homology to a predicted peroxidase gene was identified from C. melo. Peroxidase gene clusters have been found to co-localize with basal powdery mildew resistance QTL in barley [75]. The predicted salicylic acid binding protein 2 (SABP2) was identified at position 44,701 from C. melo. Salicylic acid-induced defense responses, important for resistance to many biotrophic pathogens, have been described for A. thaliana against G. cichoracearum, one of the powdery mildew pathogens that also infects cucurbits [76, 77]. Finally, homology to a predicted Dof zinc finger was identified at position 52,057 from C. melo. Dof zinc finger proteins are known to have diverse functions, including response to infection [78]. A Dof zinc finger protein in A. thaliana has been shown to be associated with the regulation of defense genes as a response to signals from the salicylic acid pathway [79].
Development of CAPS Markers for Marker-Assisted Selection
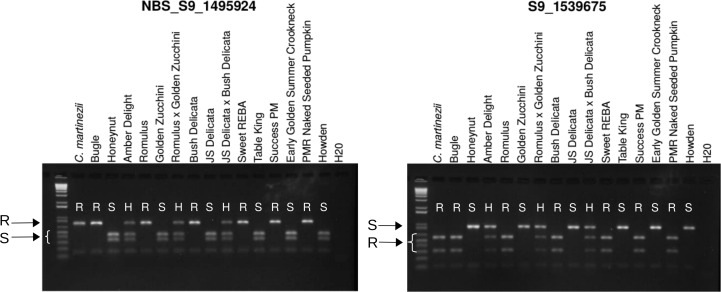
Two CAPS markers were developed for utility in marker-assisted selection. The first, NBS_S9_1495924, was located in the NBS-LRR gene. This marker distinguishes the resistance allele as a set of 134 and 759 bp fragments and the susceptible allele as a set of 134, 316, and 443 bp fragments. The marker fully co-segregates with the disease resistance phenotype as evaluated in the panel of Cornell-bred and heirloom C. okeechobeensis subsp. martinezii PI 406680, C. moschata, and C. pepo cultivars (Fig 5). A secondary marker with complete co-segregation, S9_1539675, is also reported (Fig 5). Both markers can be utilized for marker-assisted selection in breeding programs to screen and select for the presence of Pm-0 in C. pepo and C. moschata.
Fig 5. CAPS markers with complete co-segregation with Pm-0 in a panel of susceptible and resistant cultivars.
R = Homozygous for the C. okeechobeensis subsp. martinezii-derived resistance allele. S = Homozygous for the C. pepo/C. moschata susceptibility allele. H = Heterozygous. ‘Amber Delight’ is a hybrid of ‘Bugle’ and ‘Honeynut’. Left. NBS_S9_1495924 in a putative NBS-LRR gene with an A. thaliana powdery mildew resistance domain. Right. S9_1539675 in an unknown putative gene.
Conclusions
Using cultivars that comprised a shared-trait introgression panel and GBS to generate high-density genotype data, we have successfully mapped the major gene for powdery mildew resistance in squash, Pm-0, to a small genomic interval. The methods and tools presented here should be useful for elucidating other major genes, especially those derived from wild species, in squash and other crops. The CAPS markers presented here in addition to other sequence information should be useful to plant breeders seeking to employ marker-assisted selection towards the development of improved powdery mildew-resistant cultivars. Finally, we have identified a list of candidate genes that can be screened in future studies to definitively identify the Pm-0 gene.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Tim Dodge, Michael Glos, Maryann Fink, Nick Vail, and John Jantz for field and greenhouse assistance. We thank Christopher Hernandez for providing bioinformatics support. We thank Phil Rupp and Sheri Fry of Rupp Seeds, Inc. for their partnership. We thank H.M. Clause, Rupp Seeds, High Mowing, and Johnny’s Selected Seeds for contributing seeds of resistant and susceptible cultivars. We thank the Bioinformatics and Cucurbits Breeding groups of the COMAV- Polytechnic University of Valencia for use of their pre-publication C. pepo genome. We acknowledge the legacy of Henry M. Munger and his students, especially Max Contin, in creating the genetic resources that were the foundation of this study and Nancy and Martha Munger for support of continuing research on this plant breeding legacy.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Support for William Holdsworth was provided by USDA National Institute of Food and Agriculture, Plant Breeding and Education Grant No. 2010-85117-20551 and the USDA National Institute of Food and Agriculture, Agriculture and Food Research Initiative Competitive Grant No. 2013-67013-21232 and was later employed by Rupp Seeds Inc. (a company that develops and distributes powdery mildew-resistant squash and pumpkin cultivars) during the time in which the manuscript was edited and in review. Duane Bell was employed by Rupp Seeds Inc and received salary support during which time he generated and phenotyped the C. okeechobeensis subsp. martinezii PI 532363 x C. moschata 'Burpee's Butterbush' F2 population. Rupp Seeds Inc. contributed resources for the analysis of the F2 population and support in the form of salaries for authors WLH and DB, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the 'author contributions' section. Support for Kyle LaPlant was provided by the Calvin Noyes Keeney Endowment for Plant Breeding. Funding for the association analysis was provided via new faculty startup funds from Cornell University. Funding for marker development was provided by USDA National Institute of Food and Agriculture Specialty Crop Research Initiative Grant No. 2015-51181-24285. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Paris HS. Summer squash In: Prohens J, Nuez F, editors. Handbook of plant breeding vegetables I: Springer; 2008. pp. 351–379. [Google Scholar]
- 2.Ferriol M, Picó B. Pumpkin and winter squash In: Prohens J, Nuez F, editors. Handbook of plant breeding vegetables I: Springer; 2008. pp. 317–349. [Google Scholar]
- 3.Formisano G, Paris HS, Frusciante L, Ercolano MR. Commercial Cucurbita pepo squash hybrids carrying disease resistance introgressed from Cucurbita moschata have high genetic similarity. Plant Genet Res: Characterization and Utilization. 2010;8(3): 198–203. [Google Scholar]
- 4.Contin M. Interspecific transfer of powdery mildew resistance in the genus Cucurbita. Ph.D. Dissertation, Cornell University. 1978.
- 5.Navazio J. Cucurbitaceae The organic seed grower. White River Junction, VT: Chelsea Green Publishing; 2012. pp. 207–252. [Google Scholar]
- 6.McCreight JD. Notes on the change of the causal species of cucurbit powdery mildew in the U.S. Rep Cucurbit Genet Coop. 2004;27: 8–23. [Google Scholar]
- 7.McGrath MT. Heterothallism in Sphaerotheca fuliginea. Mycologia. 1994;86(4): 517–523. [Google Scholar]
- 8.Pérez-García A, Romero D, Fernández-Ortuño D, López-Ruiz F, De Vicente A, Torés JA. The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol Plant Pathol. 2009;10(2): 153–160. 10.1111/j.1364-3703.2008.00527.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zitter TA, Hopkins DL, Thomas CE, editors. Compendium of cucurbit diseases. St. Paul, MN: The American Phytopathological Society; 1996. [Google Scholar]
- 10.McGrath MT, Staniszewka H. Management of powdery mildew in summer squash with host resistance, disease threshold-based fungicide programs, or an integrated program. Plant Dis. 1996;80(9): 1044–1052. [Google Scholar]
- 11.O'Brien R, Vawdrey L, Glass R. Fungicide resistance in cucurbit powdery mildew Sphaerotheca fuliginea and its effect on field control. Aust J Exp Agric. 1988;28(3): 417–423. [Google Scholar]
- 12.McGrath MT. Guidelines for managing cucurbit powdery mildew with fungicides in 2005. Vegetable MD Online 2005. Available: http://vegetablemdonline.ppath.cornell.edu/NewsArticles/Cuc_PM_Update.htm
- 13.Coolong T, Seebold K. Impact of fungicide program and powdery mildew resistance in three varieties of pumpkin. Horttechnology. 2011;21(5): 533–538. [Google Scholar]
- 14.Hultengren R, Glos M, Mazourek M. Breeding research and education needs assessment for organic vegetable growers in the Northeast. 2016. Database: eCommons Digital Repository at Cornell University, Ithaca, NY. Available: http://hdl.handle.net/1813/.
- 15.Sowell FJ, Corley WL. Resistance of Cucurbita plant introductions to powdery mildew. HortScience. 1973;8: 492–493. [Google Scholar]
- 16.Křistková E, Lebeda A. Powdery mildew field infection on leaves and stems of Cucurbita pepo accessions. Acta Hortic. 2000;510. [Google Scholar]
- 17.Lebeda A, Křistková E. Genotypic variation in field resistance of Cucurbita pepo cultivars to powdery mildew (Erysiphe cichoracearum). Genet Resour Crop Evol. 1996;43: 79–84. [Google Scholar]
- 18.Cohen R, Leibovich G, Shtienberg D, Paris HS. Variability in the reaction of squash (Cucurbita pepo) to inoculation with Sphaerotheca fuliginea and methodology of breeding for resistance. Plant Pathol. 1993;42: 510–516. [Google Scholar]
- 19.Adeniji AA, Coyne DP. Genetics and nature of resistance to powdery mildew in crosses of butternut with calabaza squash and 'Seminole Pumpkin'. J Am Soc Hortic Sci. 1983;108(3): 360–368. [Google Scholar]
- 20.Jahn M, Munger HM, McCreight JD. Breeding cucurbit crops for powdery mildew resistance In: Bélanger RR, Bushnell WR, Dik AJ, Carver TLW, editors. The Powdery Mildews: A Comprehensive Treatise. St. Paul, MN: The American Phytopathological Society; 2002. pp. 239–248. [Google Scholar]
- 21.Zhou J, Hu H, Li X, Zhou R, Zhang H, editors. Identification of a resource of powdery mildew resistance in Cucurbita moschata. Proceedings of the 4th International Symposium on Cucurbits; 2010: Acta Hortic.
- 22.Paris HS, Cohen R. Powdery mildew-resistant summer squash hybrids having higher yields than their susceptible, commercial counterparts. Euphytica. 2002;124: 121–128. [Google Scholar]
- 23.Robinson RW, Decker-Walters DS. Cucurbits. New York, NY: CAB International; 1997. [Google Scholar]
- 24.Rhodes AM. Inheritance of powdery mildew resistance in the genus Cucurbita. Plant Dis Rep. 1964;48(1): 54–55. [Google Scholar]
- 25.Rhodes AM. Species hybridization and interspecific gene transfer in the genus Cucurbita. J Am Soc Hortic Sci. 1959;74: 546–551. [Google Scholar]
- 26.Sitterly WR. Breeding for disease resistance in cucurbits. Annu Rev Phytopathol. 1972;10: 471–490. [Google Scholar]
- 27.Whitaker TW. The origin of cultivated Cucurbita. Am Nat. 1956;90(852):171–6. [Google Scholar]
- 28.Cohen R, Hanan A, Paris HS. Single-gene resistance to powdery mildew in zucchini squash (Cucurbita pepo). Euphytica. 2003;130: 433–441. [Google Scholar]
- 29.Paris HS, Brown RN. The genes of pumpkin and squash. HortScience. 2005;40(6): 1620–1630. [Google Scholar]
- 30.Kyle M, editor. Breeding cucurbits for multiple disease resistance. International symposium on Cucurbitaceae '94: evaluation and enhancement of cucurbit germplasm. 1995; South Padre Island, TX.
- 31.Gong L, Paris HS, Nee MH, Stift G, Pachner M, Vollmann J, et al. Genetic relationships and evolution in Cucurbita pepo (pumpkin, squash, gourd) as revealed by simple sequence repeat polymorphisms. Theor Appl Genet. 2012;124(5): 875–891. 10.1007/s00122-011-1752-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paris HS, Yonash N, Portnoy V, Mozes-Daube N, Tzuri G, Katzir N. Assessment of genetic relationships in Cucurbita pepo (Cucurbitaceae) using DNA markers. Theor Appl Genet. 2003;106(6): 971–978. 10.1007/s00122-002-1157-0 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, inventor; Hollar Seeds, assignee. Cucurbita pepo pumpkins having a mutant allele for powdery mildew resistance patent US20130283463 A1. 2013.
- 34.McGrath MT, Fox GM, Menasha S. Powdery mildew resistant zucchini and yellow summer squash variety evaluation. New York 2008. Purdue University, 2008.
- 35.Nee M. The domestication of Cucurbita (Cucurbitaceae). Economic Botany. 1990;44(3): 56–68. [Google Scholar]
- 36.Gong L, Paris HS, Stift G, Pachner M, Vollmann J, Lelley T. Genetic relationships and evolution in Cucurbita as viewed with simple sequence repeat polymorphisms: the centrality of C. okeechobeensis. Genet Resour Crop Ev. 2013;60(4): 1531–1546. [Google Scholar]
- 37.Menda N, Strickler SR, Edwards JD, Bombarely A, Dunham DM, Martin GB, et al. Analysis of wild-species introgressions in tomato inbreds uncovers ancestral origins. BMC Plant Biol. 2014;14:287 10.1186/s12870-014-0287-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Beek JG, Verkerk R, Zabel P, Lindhout P. Mapping strategy for resistance genes in tomato based on RFLPs between cultivars: Cf9 (resistance to Cladosporium fulvum) on chromosome 1. Theor Appl Geneti. 1992;84(1–2): 106–112. [DOI] [PubMed] [Google Scholar]
- 39.Nimmakayala P, Levi A, Abburi L, Abburi V, Tomason Y, Saminathan T, et al. Single nucleotide polymorphisms generated by genotyping by sequencing to characterize genome-wide diversity, linkage disequilibrium, and selective sweeps in cultivated watermelon. BMC Genomics. 2014;15(1): 767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLOS ONE. 2011;6(5): e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonah H, Bastien M, Iquira E, Tardivel A, Légaré G, Boyle B, et al. An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLOS ONE. 2013;8(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornell University Experiment Station, Plant Variety Protection- "Bugle" 1999.
- 43.Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, et al. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLOS ONE. 2014;9(2): e90346 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoberman R, Dias J, Ge B, Harmsen E, Mayhew M, Verlaan DJ, et al. A probabilistic approach for SNP discovery in high-throughput human resequencing data. Genome Res. 2009;19(9): 1542–1552. 10.1101/gr.092072.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swarts K, Li H, Romero Navarro JA, An D, Romay MC, Hearne S, et al. Novel methods to optimize genotypic imputation for low-coverage, next-generation sequence data in crop plants. Plant Genome. 2014;7(3). [Google Scholar]
- 46.Quail M, Smith M, Coupland P, Otto T, Harris S, Connor T, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13(1): 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19): 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- 48.Reuscher S, Glaubitz J, Johnson L. First annual TASSEL hackathon. 2015.
- 49.Broman KW, Wu H, Sen Ś, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7): 889–890. [DOI] [PubMed] [Google Scholar]
- 50.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2015.
- 51.Browning Sharon R, Browning Brian L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Geneti. 2007;81(5):1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. New York: Academic Press; 1990. p. 315–22. [Google Scholar]
- 53.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33: D34–D38. 10.1093/nar/gki063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28(18): 2397–2399. 10.1093/bioinformatics/bts444 [DOI] [PubMed] [Google Scholar]
- 55.Wang Q, Tian F, Pan Y, Buckler ES, Zhang Z. A SUPER powerful method for genome wide association study. PLOS ONE. 2014;9(9): e107684 10.1371/journal.pone.0107684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv. 2014.
- 57.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35(suppl 2): W71–W4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sequencher® version 4.9 sequence analysis software, Gene Codes Corporation, Ann Arbor, MI USA http://www.genecodes.com
- 59.Fernandez-Pozo N, Menda N, Edwards JD, Saha S, Tecle IY, Strickler SR, et al. The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Mendiburu F. Una herramienta de analisis estadistico para la investigacion agricola: Universidad Nacional de Ingenieria; 2009.
- 61.Gong L, Pachner M, Kalai K, Lelley T. SSR-based genetic linkage map of Cucurbita moschata and its synteny with Cucurbita pepo. Genome. 2008;51:878–887. 10.1139/G08-072 [DOI] [PubMed] [Google Scholar]
- 62.Gong L, Stift G, Kofler R, Pachner M, Lelley T. Microsatellites for the genus Cucurbita and an SSR-based genetic linkage map of Cucurbita pepo L. Theor Appl Genet. 2008;117: 37–48. 10.1007/s00122-008-0750-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esteras C, Goómez P, Monforte AJ, Blanca J, Vicente-Dólera N, Roig C, et al. High-throughput SNP genotyping in Cucurbita pepo for map construction and quantitative trait loci mapping. BMC Genomics. 2012;13(1): 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moncada P, Martínez CP, Borrero J, Chatel M, Gauch H Jr, Guimaraes E, et al. Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet. 2001;102(1): 41–52. [Google Scholar]
- 65.McGrath MT, Davey JF. Managing powdery mildew with resistant squash and pumpkin cultivars. Phytopathology. 2007;97(7): S73–S4. [Google Scholar]
- 66.Lawson V. Evaluation of winter squash cultivars with resistance to powdery mildew. Iowa State University, Muscatine Island Research and Demonstration Farm, 2005. Available: http://lib.dr.iastate.edu/cgi/viewcontent.cgi?article=2076&context=farms_reports.
- 67.McGrath MT, Fox GM. Evidence of reduced suppression of powdery mildew (Podosphaera xanthii) provided by resistant squash (Cucurbita pepo) cultivars in NY. Phytopathology. 2009;99(6): S194. [Google Scholar]
- 68.McGrath MT, Hunsberger LK, Menasha S. Powdery mildew resistant pumpkin variety evaluation, New York, 2010. Purdue University, 2010. Available: https://www2.ag.purdue.edu/hla/fruitveg/MidWest%20Trial%20Reports/5-1_McGrath_Pumpkin_Powdery%20mildew%2010_LR.pdf. [Google Scholar]
- 69.Coffey MD, McCreight JD, Miller T. New races of the cucurbit powdery mildew Podosphaera xanthii present in California. Phytopathology. 2006;96(6): S25. [Google Scholar]
- 70.Cohen R, Burger Y, Shraiber S. Physiological races of Sphaerotheca fuliginea: Factors affecting their identification and the significance of this knowledge In: Maynard DN, editor. Cucurbitaceae 2002; 2002; Naples, Florida: ASHS Press; 2002. pp. 181–187. [Google Scholar]
- 71.Yang S, Li J, Zhang X, Zhang Q, Huang J, Chen J-Q, et al. Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc Natl Acad Sci USA. 2013;110(46): 18572–18577. 10.1073/pnas.1318211110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Srichumpa P, Susanne B, Keller B, Yahiaoui N. Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 2005;139: 885–895. 10.1104/pp.105.062406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coleman C, Copetti D, Cipriani G, Hoffmann S, Kozma P, Kovács L, et al. The powdery mildew resistance gene REN1 co-segregates with an NBS-LRR gene cluster in two Central Asian grapevines. BMC Genet. 2009;10(1): 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim K-H, Hwang J-H, Han D-Y, Park M, Kim S, Choi D, et al. Major quantitative trait loci and putative candidate genes for powdery mildew resistance and fruit-related traits revealed by an intraspecific genetic map for watermelon (Citrullus lanatus var. lanatus). PLOS ONE. 2015;10(12): e0145665 10.1371/journal.pone.0145665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.González AM, Marcel TC, Kohutova Z, Stam P, van der Linden CG, Niks RE. Peroxidase profiling reveals genetic linkage between peroxidase gene clusters and basal host and non-host resistance to rusts and mildew in barley. PLOS ONE. 2010;5(8): e10495 10.1371/journal.pone.0010495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, et al. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291(5501): 118–120. 10.1126/science.291.5501.118 [DOI] [PubMed] [Google Scholar]
- 77.Vlot AC, Liu P-P, Cameron RK, Park S-W, Yang Y, Kumar D, et al. Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 2008;56(3): 445–456. 10.1111/j.1365-313X.2008.03618.x [DOI] [PubMed] [Google Scholar]
- 78.Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7(12):555–60. [DOI] [PubMed] [Google Scholar]
- 79.Zhang B, Chen W, Foley RC, Büttner M, Singh KB. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell. 1995;7(12): 2241–2252. 10.1105/tpc.7.12.2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.