Abstract
We have isolated a complementary deoxyribonucleic acid clone that encodes the protein disulfide isomerase of Bombyx mori (bPDI). This protein has a putative open reading frame of 494 amino acids and a predicted size of 55.6 kDa. In addition, 2 thioredoxin active sites, each with a CGHC sequence, and an endoplasmic reticulum (ER) retention signal site with a KDEL motif were found at the C-terminal. Both sites are typically found in members of the PDI family of proteins. The expression of bPDI messenger ribonucleic acid (mRNA) was markedly increased during ER stress induced by stimulation with calcium ionophore A23187, tunicamycin, and dithiothreitol, all of which are known to cause an accumulation of unfolded proteins in the ER. We also examined the tissue distribution of bPDI mRNA and found pronounced expression in the fat body of insects. Hormonal regulation studies showed that juvenile hormone, insulin, and a combination of juvenile hormone and transferrin (although not transferrin alone) affected bPDI mRNA expression. A challenge with exogenous bacteria also affected expression, and the effect peaked 16 hours after infection. These results suggest that bPDI is a member of the ER-stress protein group, that it may play an important role in exogenous bacterial infection of the fat body, and that its expression is hormone regulated.
INTRODUCTION
All eukaryotic cells secrete several types of specialized plasma membrane proteins as well as secretory proteins from the lumen of the endoplasmic reticulum (ER). One of the most important functions of the ER is to provide an intracellular environment to facilitate the proper folding and assembly of newly synthesized exportable proteins. The ER also contains numerous chaperones that determine the final destination of proteins (Gething and Sambrook 1992). These ER chaperones have 2 important functions in protein folding. The first is the catalysis of folding reactions by the enzymes, protein disulfide isomerase (PDI, EC5.3.4.1) and cis-trans prolyl isomerase (Noiva 1999). The second is the prevention of self-aggregation of protein-folding intermediates and the stabilization of energetically unfavorable conformations by Bip and the glucose-regulated protein (GRP) family (Gomord et al 1999).
Protein disulfide bond formation is a rate-limiting step in protein folding and is catalyzed by enzymes belonging to the protein disulfide oxidoreductase superfamily, which includes PDI in eukaryotes and DsbA in bacteria (Noiva 1994). PDI catalyzes the oxidation of disulfides and the isomerization of incorrect disulfides on newly synthesized polypeptides during folding in the oxidizing environment of the ER. It is now accepted that PDI is a multifunctional protein involved in the folding, assembly, and posttranslational modification of many proteins. It also appears to be involved in actin filament polymerization, gene expression, cell-cell interactions, and the regulation of receptor function (Tabb et al 1998; Frand et al 2000). PDI is present as a subunit in 2 ER enzymes, prolyl 4-hydroxylase (P4H) and microsomal triglyceride transfer protein (MTP). Like other ER chaperones, PDI expression is enhanced when cells are exposed to external stimuli that increase the need for ER chaperones, such as heat shock, calcium ionophore A23187, and tunicamycin (Dorner et al 1990; Saloheimo et al 1999). PDI expression is controlled transcriptionally by insulin, which participates in cell signaling through interactions with receptors for interleukins, transferrin, insulin, tumor necrosis factor, thyroid-stimulating hormone, insulin growth factor, and colony-stimulating factor I (Hensel et al 1994; Marcus et al 1996; Yoshikawa et al 2000). Although the deoxyribonucleic acid (DNA) sequences of the PDI family show some diversity, PDIs are known to share high amino acid (aa) homology, and are abundantly expressed in various cell types and in many different organisms. According to the 5-domain model that has been proposed for PDI activity, there are 2 protein-thiol oxidoreductase active sites with a CGHC sequence in both the C- and the N-terminal regions, and an ER retention signal with a KDEL sequence in the C-terminal. The latter finding is evidence that PDI is located and retained in the ER lumen and functions as an ER chaperone (Chivers et al 1996; Noiva 1999; Ciaffi et al 2001; Warsame et al 2001).
This study examined bPDI, a PDI homolog from Bombyx mori that also has 2 thiol oxidoreductase sites and a KDEL motif in the C-terminal. The complementary DNAs (cDNAs) encoding the PDI family have been isolated from a number of organisms and tissues. However, so far only 1 PDI cDNA has been reported from the insect Drosophila melanogaster (Diptera), and reports on PDI function in insects are very limited (McKay et al 1995). To further our understanding of the role of PDI in insects, we report the isolation of a full-length B mori (Lepidoptera) cDNA encoding PDI, and the characterization of its expression in response to various ER stresses, hormones, and exogenous pathogens.
MATERIALS AND METHODS
Experimental insects and cells
B mori (Jam 306) silkworms were reared on an artificial diet at 24–27°C in 70–90 % humidity. Cells from the Bm5 line derived from B mori were cultured at 27°C in TC-100 medium (Sigma, St Louis, MO, USA) with 10 % fetal bovine serum (GIBCO Life Technologies, Gland Island, NY, USA) using the standard method (Wang et al 2000).
Differential screening for the bPDI gene
A cDNA library was obtained from Bm5 cells in which N-linked glycosylation was inhibited by tunicamycin treatment for 5 hours (5 μg/mL constructed in the Uni-ZAP XR vector Kit [Stratagene, La Jolla, CA, USA]), and 768 randomly selected cDNA fragments were fixed in duplicate on nitrocellulose membranes. Other cDNA fragments derived from normal and tunicamycin-treated poly(A+) ribonucleic acids (RNAs) were labeled with [α-32P]deoxyadenosine triphosphate (dATP) and used as molecular probes. The 2 membranes were hybridized in ExpressedHyb Hybridization Solution (Clontech, Palo Alto, CA, USA), and the selected cDNA fragments were cloned and amplified. The resulting positive cDNA fragments were partially sequenced. One of these fragments shared a high homology with the PDI gene family, and it was fully sequenced after obtaining a full cDNA sample by 3′–rapid amplification of cDNA ends (RACE) polymerase chain reaction (PCR), as described in a subsequent section (RACE PCR analysis).
Nucleotide sequence analysis
The selected cDNA clones were partially sequenced using T3 and T7 primers and an ABI 377 automatic sequencer (Perkin-Elmer, Watsonville, CA, USA). Double-stranded DNA was prepared using a Wizard Plus SV Miniprep DNA Purification System (Promega, Madison, WI, USA). Each DNA sample (300–500 ng) was mixed with primer (3.2 pmol) and Termination Reaction Mix (Perkin-Elmer), and was sequenced following 25 PCR cycles (96°C for 30 seconds, 50°C for 15 seconds, and 60°C for 4 minutes). The resulting PCR products were separated on a 4.5 % denaturing polyacrylamide gel and analyzed with DNA Sequencing Analysis Software (Perkin-Elmer). Both strands of the cDNA clones were sequenced. The aa sequence was deduced from the cDNA sequence, and homology with other species was analyzed through GenBank database searches.
RACE PCR analysis
To generate the 3′-translated sequence of the bPDI messenger RNA (mRNA), 3′-RACE PCR was carried out with Bm5 cells, poly(A+), and a Marathon cDNA Amplification Kit (Clontech) according to the manufacturer's instructions. The adapter sequence attached to the ends of the cDNA enabled it to be used in 5′- and 3′-RACE. Three gene-specific primers were designed using the sequence of the bPDI fragment. These primers were used in 3′-RACE in conjunction with the anchor primer (AP1) to amplify the 3′-ends of the gene from cDNA. The PCR conditions were as follows: initial denaturation at 94°C for 3 minutes followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, elongation at 72°C for 1 minute, and a final elongation step at 72°C for 10 minutes. The PCR products were subcloned into a pGEM-T Easy Vector (Promega), and the nucleotide sequence of the bPDI cDNA clone was determined.
RNA dot blot analysis
Dot blot analysis (Hoog 1991) was performed using the total cellular RNA extracted using a SV Total RNA Isolation System Kit (Promega) according to the manufacturer's instructions (Sondergaard 1993). Approximately 5 μg of total RNA per well was fixed overnight on a Hybond-N nylon membrane (Amersham Pharmacia Biotech, UK) in ExpressedHyb Hybridization Solution (Clontech) with [α-32P]dATP-labeled bPDI cDNA at 65°C. The membrane was subsequently washed at 65°C and subjected to autoradiography. For the control studies, 18S ribosomal RNA was used. Dot densities were analyzed with the Image Analysis System.
RESULTS AND DISCUSSION
Screening and analysis of bPDI cDNA
A differential screening method was used to identify novel genes involved in the unfolded protein response in B mori–derived Bm5 cells (Hoog 1991). The 768 clones identified in the screening were partially sequenced. One of these fragments shared high homology with the PDI gene family, and the cDNA fragment was retrieved and cloned. Further cDNA sequencing was carried out after obtaining a full cDNA sample by 3′-RACE PCR, as described in Materials and Methods. It was named B mori bPDI, in deference to the nomenclature used for the dPDI previously described in D melanogaster (McKay et al 1995).
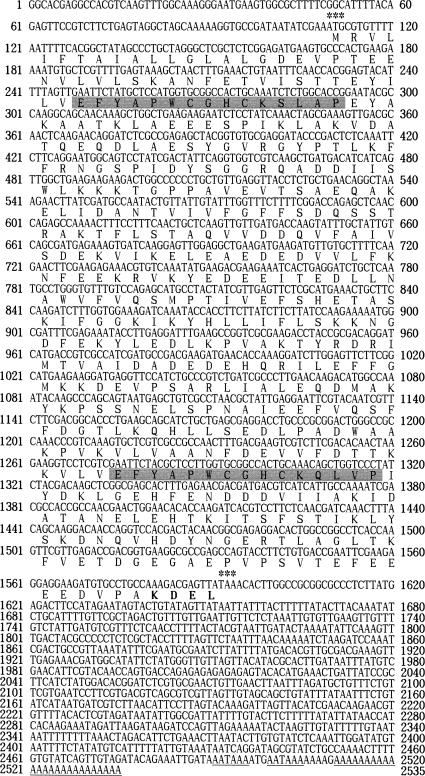
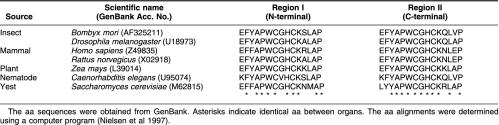
As shown in Figure 1, the bPDI gene contains a 5′-untranslational region of 109 nucleotides followed by an initiating ATG codon. The TAA termination codon is at nucleotide 1592, a consensus polyadenylation signal is present at 2491–2506, and the poly(A) tail is 5 nucleotides downstream of the polyadenylation signal sequence. A bPDI cDNA encoding an open reading frame of 494 aa was seen, which was estimated to have a molecular size of 55.6 kDa. The bPDI sequence was submitted to GenBank under the accession number AF325211. PDI family proteins typically have 2 thioredoxin active site sequences (CGHC), with the exception of ERp72 and Mpd1 from Saccharomyces cerevisiae, which have only 1 conserved domain (Tachikawa et al 1995; Benham et al 2000). The bPDI protein also contains 2 conserved thioredoxin catalytic domains, EFYAPWCGHCKSLAP (aa 47–61) and EFYAPWCGHCKQLVP (aa 389–403), at the N- and C-terminals, respectively. These are marked in gray in Figure 1 and are compared with the other known PDIs in Table 1. Other sites illustrated in Figure 1 include a cyclic adenosine monophosphate– or a cyclic guanosine monophosphate–dependent protein kinase phosphorylation site at aa 127–130 (KKKT), a tyrosine kinase phosphorylation site at aa 261–269 (KKNGDFEKY), and 2 N-myristoylation sites at aa 13–18 (GLALGD) and aa 115–120 (GGRQAD). ER-resident proteins typically have an ER retention signal at the C-terminal with the sequence KDEL (HDEL in yeast) (Burke et al 1996). A KDEL motif was also found at the C-terminal of bPDI (aa 491–494), which is evidence that bPDI is a reticuloplasmin with similarities to calreticulin, endoplasmin, and Bip. No potential glycosylation site was detected in the bPDI protein sequence. However, the aa sequence of bPDI from B mori shared 50–57 % identity and 68–74 % positivity with the PDIs of Drosophila (U18973), rat (X02918), and human (Z49835), in which functional regions, such as thioredoxin domains and ER retention signals, were very well conserved (Edman et al 1985; McKay et al 1995). On the basis of the cDNA sequence analysis described previously, we propose that the bPDI gene is a member of the PDI gene family.
Fig 1.
The nucleotide and deduced aa sequences of Bombyx mori protein disulfide isomerase (bPDI) complementary deoxyribonucleic acid (cDNA). The cDNA sequence of this bPDI gene was submitted to GenBank under accession number AF325211. The predicted amino acid sequence (single-letter abbreviations) is shown below the nucleotide sequence within the open reading frame. Two activation domains, EFYAPWCGHCKSLAP (aa 47–61) and EFYAPWCGHCKQLVP (aa 389–403), are indicated by shaded areas, and the endoplasmic reticulum retention signal (KDEL) is indicated in bold. The translation start and stop codons are indicated by asterisks. The underlined nucleotide sequences indicate the putative polyadenylation signal and poly(A) tail, respectively
Table 1.
Comparison of conserved thioredoxin-like active sites in the amino acid (aa) sequences of protein disulfide isomerases
Tissue distribution of bPDI expression
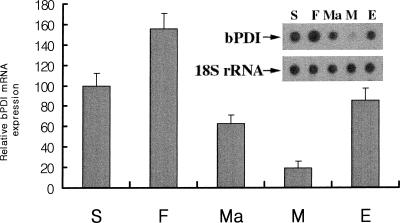
We used Northern dot blot analysis to identify where the bPDI gene was expressed in B mori (Hoog 1991). Whereas very weak bPDI mRNA expression was detected in the midgut, higher levels were found in the silk gland, fat body, Malpighian tubule, and epidermis (Fig 2). The relative expression of the bPDI gene was as follows: fat body > silk gland > epidermis > Malpighian tubule > midgut. Among the tissues tested, the highest level of bPDI expression was found in the fat body, with approximately 8-fold higher expression than in the midgut. This is the first report indicating the pronounced expression of a PDI family protein (bPDI) in the fat body of an insect. Recently, Abel et al (1992) and Sondergaard (1993) demonstrated that, physiologically, insect fat bodies were similar to the mammalian liver in that they played a role in the detoxification of metabolic and environmental stimuli. Our results imply that bPDI may have an important physiological function in the fat body, similar to that seen in the mammalian liver.
Fig 2.
Dot blot analysis of Bombyx mori protein disulfide isomerase (bPDI) messenger ribonucleic acid (RNA) expression in tissues. Total RNA was isolated from the following organs: S, silk gland; F, fat body; M, midgut; Ma, Malpighian tubules; E, epidermis. Total RNA (5 μg) from each sample was transferred onto a Hybond-N nylon membrane. Hybridization was carried out using [α-32P]deoxyadenosine triphosphate–labeled bPDI complementary deoxyribonucleic acid as a probe. The bPDI band intensities were measured using ImageQuant software. The bar graph shows the averages of the quantified data from 3 independent experiments, and the inset shows the results from 1 representative experiment. 18S ribosomal RNA was used as a quantitation marker
bPDI induction during ER stress
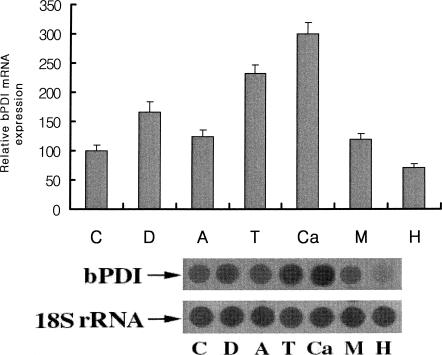
It is well known that the accumulation of unfolded proteins in the ER is induced by drugs such as dithiothreitol (DTT) (disruption of proper disulfide bond formation), antimycin A (ATP depletion), tunicamycin (N-glycosylation inhibition), calcium ionophore A23187 and monensin (disturbance of ER calcium stores), brefeldin A (prevention of secretory protein export from the ER), and H2O2 (oxidative stress) (Mkrtchian et al 1998). Earlier studies demonstrated that steady-state PDI mRNA levels were slightly increased in cells subjected to ER stress, whereas the levels of mRNA for other ER proteins (Grp78, GRP94, ERp72) were markedly increased (Dorner et al 1990). In this study we tested the effect of ER stress on bPDI expression. Bm5 cells derived from B mori were treated separately with DTT, ATP, tunicamycin, calcium ionophore A23187, monensin, and H2O2. As shown in Figure 3, bPDI expression was markedly increased following treatment with calcium ionophore A23187 (∼3-fold), tunicamycin (∼2.3-fold), and DTT (∼1.7-fold). Treatment with H2O2, however, produced only a small increase (∼0.7-fold), suggesting that H2O2 preferentially induces intracellular thioredoxin, which acts as a scavenger for excess free radicals. The production of bPDI is therefore relatively reduced during H2O2 stimulation.
Fig 3.
Effects of various endoplasmic reticulum stresses on Bombyx mori protein disulfide isomerase (bPDI) messenger ribonucleic acid (RNA) expression. Bm5 cells were treated for 5 hours with 3 mM dithiothreitol (D), 8 μM antimycin A (A), 5 μg/mL tunicamycin (T), 10 μM calcium ionophore A23187 (Ca), 100 μM monensin (M) and 100 μM H2O2 (H). C indicates the control without tunicamycin treatment. Total RNA (5 μg) from each sample was transferred onto a Hybond-N nylon membrane. Hybridization was carried out using [α-32P]deoxyadenosine triphosphate–labeled bPDI complementary deoxyribonucleic acid as a probe. The bPDI band intensities were measured using ImageQuant software. The values in the bar graph are the averages of 3 repeated experiments. The bottom panel shows the results from 1 representative experiment. 18S ribosomal RNA was used as a quantitation marker
Other possible explanations for these findings include the increased biosynthesis of retention molecules, an increased affinity of the retention mechanism for PDI, and the increased association of bPDI with other cellular proteins in the ER. Further experimentation is required to clarify these results.
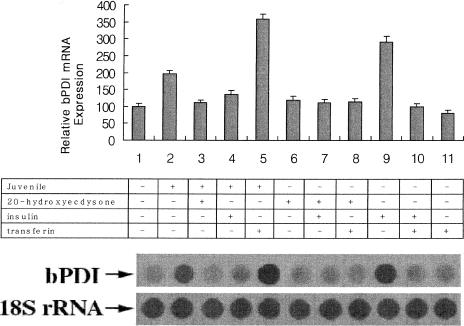
Hormonal regulation of bPDI expression
We examined the effects of growth factors essential for the growth of Bm5 cells (20-hydroxyecdysone, insulin, transferrin) on bPDI expression in the presence or absence of juvenile hormone. Following preincubation for 1 day in growth medium, cells were treated for 24 hours with 2 μg/mL 20-hydroxyecdysone, 1 μg/mL insulin, and 5 μg/mL transferrin, in the presence or absence of 2 μg/mL juvenile hormone. As shown in Figure 4, both juvenile hormone (∼2-fold) and insulin (∼3-fold) affected the constant bPDI mRNA levels in Bm5 cells. Recently, Graham et al (1996) demonstrated that in the mealworm beetle, the expression of a stress protein (dsp28) produced in the fat body increased in response to the juvenile hormone analog of methoprene. In contrast, Nieto et al (1990) reported that insulin down-regulated PDI expression in rats. Although transferrin alone or in combination with insulin had no effect, the highest level of bPDI expression was detected when juvenile hormone was added together with transferrin (∼4-fold) (Nieto et al 1990). Rybczynski and Gilbert (2000) demonstrated that hsc70 expression increased following 20-hydroxyecdysone treatment, although in our study 20-hydroxyecdysone had no effect on bPDI expression. These results show that juvenile hormone and insulin up-regulate bPDI expression in Bm5 cells, whereas juvenile hormone and transferrin play the most important role in bPDI expression.
Fig 4.
Effects of various growth factors on Bombyx mori protein disulfide isomerase (bPDI) messenger ribonucleic acid (RNA) expression. Confluent Bm5 cells were treated for 24 hours either with (+) or without (−) 2 μg/mL juvenile hormone, combined with 2 μg/mL 20-hydroxyecdysone, 1 μg/mL insulin, or 5 μg/mL transferrin. Total RNA (5 μg) from each sample was transferred onto a Hybond-N nylon membrane. Hybridization was carried out using [α-32P]deoxyadenosine triphosphate–labeled bPDI complementary deoxyribonucleic acid as a probe. The bPDI band intensities were measured using ImageQuant software. The results in the bar graph are the average of 3 separate experiments. The bottom panel shows the results from 1 representative experiment. 18S ribosomal RNA was used as a marker of quantitation
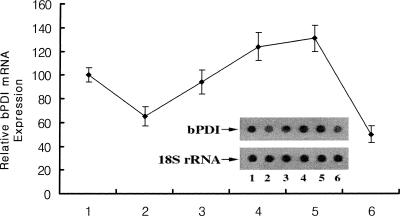
bPDI expression in response to bacterial infection
The fat body of an insect is an organ with multiple metabolic functions, including the metabolism of carbohydrates, lipids, and nitrogenous compounds, the storage of glycogen, fat, and protein, the synthesis and regulation of blood sugar, and the synthesis of major hemolymph proteins (Dean et al 1985; Keeley 1985). A number of observations demonstrated that the insect fat body shares functional homology with the mammalian liver (Abel et al 1992; Sondergaard 1993; Halarnkar and Schooley 1995). For example, the Drosophila transcription factor box B-binding factor 2 (BBF-2), which binds to fat body–specific enhancers of alcohol dehydrogenase, also binds specifically to regulatory elements required for the liver-specific expression of the human and rat tyrosine aminotransferase genes. This suggests that BBF-2 binding may be an important component of a tissue-specific regulatory mechanism conserved between Drosophila and man. Natori et al (1999) demonstrated that various antimicrobial proteins were inducible by the fat body of Sarcophaga peregrina (flesh fly) during an acute-phase response to bacterial infection. Our previous data also demonstrated that B mori transferrin was strongly induced only in the fat body by the injection of E coli, suggesting that the fat body plays an important role in self-defense. It is clear that insects have the ability to mount a potent defensive response against microbial parasites and that this response is different from mammalian immunity (Yun et al 1999). Figure 5 shows bPDI expression in the fat body of a silkworm that was injured on the epidermis by E coli infection according to Natori's method (Natori et al 1999). Although bPDI expression decreased by approximately 65 % 2 hours after infection, its expression gradually increased and reached a maximum (∼1.3-fold) 16 hours after infection. At 24 hours, expression had again fallen by 50 %. This suggests that although bPDI is currently best known as a chaperone, it may also function as a potent defense against microbial infection in invertebrate immunity.
Fig 5.
Temporal induction of Bombyx mori protein disulfide isomerase (bPDI) gene expression following infection. Total ribonucleic acid (RNA) extracted from the fat body of B mori infected with E coli for 0 hour (control, lane 1), 2 hours (lane 2), 4 hours (lane 3), 8 hours (lane 4), 16 hours (lane 5), and 24 hours (lane 6). Total RNA (5 μg each) per sample was transferred onto a Hybond-N nylon membrane and hybridized with a [α-32P]deoxyadenosine triphosphate–labeled bPDI complementary deoxyribonucleic acid as a probe. The bPDI band intensities were measured using ImageQuant software. The dot graph represents an average of 3 separate experiments. The inset shows the results from 1 representative experiment. 18S ribosomal RNA was used as a quantitation marker
In conclusion, protein disulfide bond formation is a rate-limiting step in protein folding and is catalyzed by PDI in eukaryotes and by DsbA in bacteria. Although PDI genes have been isolated from a number of organisms, their biological functions during protein folding in the ER and in insect physiology are not clear. Recently, it was demonstrated that the ER-resident glycoprotein Ero1p provided oxidizing equivalents to newly synthesized proteins via PDI, which contains an important CXXCXXC motif for the PDI-Ero1p complex (Benham et al 2000). This suggests that PDI is the primary donor of disulfide bonds to newly synthesized ER proteins, while Ero1p maintains PDI in an oxidized ER state. The results described here suggest that bPDI is a member of the ER-stress protein family and that it may play an important role in the defense against both ER and environmental stresses. We are now interested in the functional role of bPDI as an ER chaperone in the insect, which may help to further our understanding of insect physiology.
Acknowledgments
This work was supported by a grant for the G-7 project from the Ministry of Science and Technology, South Korea.
REFERENCES
- Abel T, Bhatt R, Maniatis T. A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev. 1992;6:466–480. doi: 10.1101/gad.6.3.466. [DOI] [PubMed] [Google Scholar]
- Benham AM, Cabibbo A, Fassio A, Bulleid N, Sitia R, Braakman I. The CXXCXXC motif determines the folding, structure and stability of human Ero1-Lalpha. EMBO J. 2000;19:4493–4502. doi: 10.1093/emboj/19.17.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J, Lipari F, Igdoura S, Herscovics A. The Saccharomyces cerevisiae processing alpha 1,2-mannosidase is localized in the endoplasmic reticulum, independently of known retrieval motifs. Eur J Cell Biol. 1996;70:298–305. [PubMed] [Google Scholar]
- Chivers PT, Laboissiere MC, Raines RT. The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- Ciaffi M, Paolacci AR, Dominici L, Tanzarella OA, Porceddu E. Molecular characterization of gene sequences coding for protein disulfide isomerase (PDI) in durum wheat (Triticum turgidum ssp. durum) Gene. 2001;265:147–156. doi: 10.1016/s0378-1119(01)00348-1. [DOI] [PubMed] [Google Scholar]
- Dean RL, Locke M, and Collins JV 1985 Structure of fat body. In: Comprehensive Insect Physiology, Biochemistry, and Pharmacology, vol 3, ed Kerkut GA, Gilbert LI. Pergamon Press, Oxford, 155–210. [Google Scholar]
- Dorner AJ, Wasley LC, Raney P, Haugejorden S, Green M, Kaufman RJ. The stress response in Chinese hamster ovary cells. Regulation of ERp72 and protein disulfide isomerase expression and secretion. J Biol Chem. 1990;265:22029–22034. [PubMed] [Google Scholar]
- Edman JC, Ellis L, Blacher RW, Roth RA, Rutter WJ. Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature. 1985;317:267–270. doi: 10.1038/317267a0. [DOI] [PubMed] [Google Scholar]
- Frand AR, Cuozzo JW, Kaiser CA. Pathways for protein disulphide bond formation. Trends Cell Biol. 2000;10:203–210. doi: 10.1016/s0962-8924(00)01745-1. [DOI] [PubMed] [Google Scholar]
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Gomord V, Wee E, Faye L. Protein retention and localization in the endoplasmic reticulum and the golgi apparatus. Biochimie. 1999;81:607–618. doi: 10.1016/s0300-9084(99)80118-7. [DOI] [PubMed] [Google Scholar]
- Graham LA, Bendena WG, Walker VK. Juvenile hormone regulation and developmental expression of a Tenebrio desiccation stress protein gene. Dev Genet. 1996;18:296–305. doi: 10.1002/(SICI)1520-6408(1996)18:4<296::AID-DVG3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Halarnkar PP, Schooley DA. A comparative catabolism study of isoleucine by insect and mammalian tissues. Comp Biochem Physiol (B) 1995;110:357–365. doi: 10.1016/0305-0491(94)00152-k. [DOI] [PubMed] [Google Scholar]
- Hensel G, Assmann V, Kern HF. Hormonal regulation of protein disulfide isomerase and chaperone synthesis in the rat exocrine pancreas. Eur J Cell Biol. 1994;63:208–218. [PubMed] [Google Scholar]
- Hoog C. Isolation of a large number of novel mammalian genes by a differential cDNA library screening strategy. Nucleic Acids Res. 1991;19:6123–6127. doi: 10.1093/nar/19.22.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley LL 1985 Physiology and biochemistry of the fat body. In: Comprehensive Insect Physiology, Biochemistry, and Pharmacology, vol 3, ed Kerkut GA, Gilbert LI. Pergamon Press, Oxford, 211–248. [Google Scholar]
- Marcus N, Shaffer D, Farrar P, Green M. Tissue distribution of three members of the murine protein disulfide isomerase (PDI) family. Biochim Biophys Acta. 1996;1309:253–260. doi: 10.1016/s0167-4781(96)00133-9. [DOI] [PubMed] [Google Scholar]
- McKay RR, Zhu L, Shortridge RD. A Drosophila gene that encodes a member of the protein disulfide isomerase/phospholipase C-alpha family. Insect Biochem Mol Biol. 1995;25:647–654. doi: 10.1016/0965-1748(95)00001-c. [DOI] [PubMed] [Google Scholar]
- Mkrtchian S, Fang C, Hellman U, Ingelman-Sundberg M. A stress-inducible rat liver endoplasmic reticulum protein, ERp29. Eur J Biochem. 1998;251:304–313. doi: 10.1046/j.1432-1327.1998.2510304.x. [DOI] [PubMed] [Google Scholar]
- Natori S, Shiraishi H, Hori S, Kobayashi A. The roles of Sarcophaga defense molecules in immunity and metamorphosis. Dev Comp Immunol. 1999;23:317–328. doi: 10.1016/s0145-305x(99)00014-2. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nieto A, Mira E, Castano JG. Transcriptional regulation of rat liver protein disulphide-isomerase gene by insulin and in diabetes. Biochem J. 1990;267:317–323. doi: 10.1042/bj2670317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiva R. Enzymatic catalysis of disulfide formation. Protein Expr Purif. 1994;5:1–13. doi: 10.1006/prep.1994.1001. [DOI] [PubMed] [Google Scholar]
- Noiva R. Protein disulfide isomerase: the multifunctional redox chaperone of the endoplasmic reticulum. Semin Cell Dev Biol. 1999;10:481–493. doi: 10.1006/scdb.1999.0319. [DOI] [PubMed] [Google Scholar]
- Rybczynski R, Gilbert LI. cDNA cloning and expression of a hormone-regulated heat shock protein (hsc 70) from the prothoracic gland of Manduca sexta. Insect Biochem Mol Biol. 2000;30:579–589. doi: 10.1016/s0965-1748(00)00031-x. [DOI] [PubMed] [Google Scholar]
- Saloheimo M, Lund M, Penttila ME. The protein disulphide isomerase gene of the fungus Trichoderma reesei is induced by endoplasmic reticulum stress and regulated by the carbon source. Mol Gen Genet. 1999;262:35–45. doi: 10.1007/s004380051057. [DOI] [PubMed] [Google Scholar]
- Sondergaard L. Homology between the mammalian liver and the Drosophila fat body. Trends Genet. 1993;9:193. doi: 10.1016/0168-9525(93)90113-v. [DOI] [PubMed] [Google Scholar]
- Tabb JS, Molyneaux BJ, Cohen DL, Kuznetsov SA, Langford GM. Transport of ER vesicles on actin filaments in neurons by myosin V. J Cell Sci. 1998;111:3221–3234. doi: 10.1242/jcs.111.21.3221. [DOI] [PubMed] [Google Scholar]
- Tachikawa H, Takeuchi Y, Funahashi W, Miura T, Gao XD, Fujimoto D, Mizunaga T, Onodera K. Isolation and characterization of a yeast gene, MPD1, the overexpression of which suppresses inviability caused by protein disulfide isomerase depletion. FEBS Lett. 1995;369:212–216. doi: 10.1016/0014-5793(95)00750-4. [DOI] [PubMed] [Google Scholar]
- Wang W, Swevers L, Iatrou K. Mariner (Mos1) transposase and genomic integration of foreign gene sequences in Bombyx mori cells. Insect Mol Biol. 2000;9:145–155. doi: 10.1046/j.1365-2583.2000.00172.x. [DOI] [PubMed] [Google Scholar]
- Warsame A, Vad R, Kristensen T, Oyen TB. Characterization of a gene encoding a Pichia pastoris protein disulfide isomerase. Biochem Biophys Res Commun. 2001;281:1176–1182. doi: 10.1006/bbrc.2001.4479. [DOI] [PubMed] [Google Scholar]
- Yoshikawa S, Kamada M, and Maegawa M. et al. 2000 Hormonal control of mRNA expression of immunoglobulin binding factor in uterine cervix. Biochem Biophys Res Commun. 279:898–903. [DOI] [PubMed] [Google Scholar]
- Yun EY, Kang SW, Hwang JS, Goo TW, Kim SH, Jin BR, Kwon OY, Kim KY. Molecular cloning and characterization of a cDNA encoding a transferrin homolog from Bombyx mori. Biol Chem. 1999;380:1455–1459. doi: 10.1515/BC.1999.188. [DOI] [PubMed] [Google Scholar]