Abstract
We have previously characterized the unique organization of the U14 small nucleolar ribonucleic acid (snoRNA) gene in Chinese hamster HA-1 cells. The single copy of the hsc70/U14 gene is the only source for the production of both U14 snoRNA species and hsc70 messenger ribonucleic acid (mRNA) in these cells. Here we report that the accumulations of U14 snoRNA and hsc70 mRNA are different in response to various stress conditions, although both of them are transcribed in a single primary transcript. Heat shock induced an increased accumulation of both U14 snoRNA and hsc70 mRNA. On the other hand, exposure to sodium arsenite or azetidine induced an increased accumulation of hsc70 mRNA, but did not lead to a concomitant increase in the level of U14 snoRNA. Under normal growth conditions, the variations in the levels of U14 snoRNA and hsc70 mRNA, in the different phases of the cell cycle, are correlated. The increased expression of U14 snoRNA and hsc70 mRNA, and the hsc70 protein induced specifically by heat shock suggest that they participate in the repair process of heat-induced damage to macromolecular complexes involved in the synthesis and processing of ribosomal RNA.
INTRODUCTION
Eukaryotic cells contain diverse populations of metabolically stable small nucleolar ribonucleic acids (snoRNAs) (reviewed by Maxwell and Fournier 1995; Smith and Steitz 1997; Tollervey and Kiss 1997; Eddy 1999). Most of the snoRNA species identified so far are encoded by the introns of genes, which often encode proteins with roles in either the biogenesis of ribosomal RNA or the functioning of ribosomes (Liu and Maxwell 1990; Sollner-Webb 1993; Tollervey and Kiss 1997; Barneche et al 2000). U14 snoRNA is one of the most extensively studied small nucleolar RNAs. It is an essential RNA and is required for the synthesis and processing of 18S ribosomal RNA (Zagorski et al 1988; Li et al 1990; Liang and Fournier 1995). In human and Xenopus cells, the U14 snoRNA species are encoded by the introns of 2 different genes, the hsc70 gene and the ribosomal protein S13 gene (Xia et al 1995; Kenmochi et al 1996). However, in Chinese hamster cells, the organization of the U14 snoRNA genes is different. The ribosomal protein S13 gene, in Chinese hamster HA-1 cells, does not encode a functional U14 snoRNA (Chen and Laszlo 1999), and therefore the only source for the production of U14 snoRNA is from the transcription of the unique hsc70 gene in which 3 of the 8 introns encode U14 snoRNA species (Chen et al 1996a, Chen and Laszlo 1999). Even though there are some variations in the sequence of the U14 snoRNA species encoded by the different introns, all the 3 U14 snoRNA species encoded in the mammalian hsc70 gene contain 4 highly conserved regions, referred to as region A, region B, box C, and box D, that are essential for U14 snoRNA function (Balakin et al 1996; Tollervey and Kiss 1997). None of the 3 U14 snoRNA encoding introns of the mammalian hsc70 gene contain a U14 snoRNA–specific promoter (Leverette et al 1992; Chen et al 1996a). The 3 U14 snoRNA species and the hsc70 messenger RNA (mRNA) are transcribed as a part of an hsc70/U14 primary transcript (Leverette et al 1992). After transcription, the 3 U14 snoRNA species are excised from the individual introns, and the rest of the primary transcript is processed to produce a single mature hsc70 mRNA (Leverette et al 1992; Xia et al 1995).
The hsc70 protein is expressed abundantly under normal growth conditions, and it is further induced after exposure to various stresses (Laszlo and Li 1985; Laszlo 1988a, 1988b). The promoter of the mammalian hsc70 gene contains a set of basic transcriptional elements, including a TATA box, 2 CCAAT boxes, and 2 SP1 elements, and 2 sets of heat shock transcriptional elements (HSEs) to which the heat shock transcriptional factors (HSFs) bind, as a consequence of exposure to various stresses, resulting in an increased transcription of the hsc70 gene (Sarge et al 1993; Chen et al 1996a). The positioning of U14 snoRNA species in the 3 separate introns of the hsc70 gene strongly suggests that the host gene had been selected to allow some form of coregulation of the expression of both hsc70 mRNA and U14 snoRNA (Maxwell and Fournier 1995; Smith and Steitz 1997; Tollervey and Kiss 1997). Because the expression of both U14 snoRNA and hsc70 mRNA are regulated by the same promoter, it would be expected that the expression patterns of both U14 snoRNA and hsc70 mRNA are similar, provided that the turnover rates for both RNAs remain constant.
Here we report the differential accumulation of U14 snoRNA and hsc70 mRNA after exposure to various stress conditions, even though the levels of these RNAs are correlated under normal growth conditions in different phases of the cell cycle. Heat shock increased the levels of both U14 snoRNA and hsc70 mRNA. However, exposure of cells to sodium arsenite and azetidine only increased the level of hsc70 mRNA, without a concomitant increase in the level of U14 snoRNA.
MATERIALS AND METHODS
Cell culture and stress conditions
Chinese hamster HA-1 and mouse NIH 3T3 cells were cultured in monolayers at 37°C in the presence of 5 % CO2, as previously described (Goswami et al 1994; Chen et al 1996a). Briefly, HA-1 cells were grown in minimum essential medium (MEM), supplemented with 10 % fetal calf serum and antibiotics, whereas the 3T3 cells were grown in Ham's F10 medium supplemented with 10 % calf serum and antibiotic. Exponentially growing cells were used throughout these experiments. Cells were heated in a precisely controlled water bath (± 0.05°C). The culture medium was changed before heating, using medium prewarmed at 37°C, followed by incubation in a 37°C water bath for 10 minutes. The cells were then exposed to various elevated temperatures by immersion into a water bath for various lengths of time. Following heating, the cell monolayers were incubated in a 37°C water bath for 10 minutes, the medium was changed again, and cells were allowed to recover at 37°C for various lengths of time. Sodium arsenite treatment involved a 55-minute exposure to 100 μM sodium arsenite (Sigma, St Louis, MO, USA) in a growth medium at 37°C, followed by 5 rinses with fresh medium and recovery at 37°C for various lengths of time. For the treatment with the proline analog 2-azetidine-4-carboxylic acid (azetidine), exponentially growing cells were washed with warm (37°C) phosphate-buffered saline (PBS) 3 times, followed by the addition of prewarmed, fresh medium containing 2.5 mg/mL azetidine (Sigma), and then incubated for various lengths of time at 37°C.
Cell synchronization
Synchronous cell populations were obtained by the simultaneous selective mechanical detachment of mitotic cells from 10 T175 flasks, as described (Goswami et al 1996). 3 × 106 synchronized cells were plated per plate. The plates were then incubated at 37°C and harvested at different points in the cell cycle. Viability of the selected cells as measured by a colony-forming assay was more than 95 %, and the mitotic index was more than 97 % as determined by microscopic examination. Cell cycle progression was monitored by 1-dimensional flow cytometric analysis of ethanol-fixed cells collected at various points during the cell cycle. Samples were first treated with RNase A, stained with propidium iodide (10 μg/mL in PBS), and then analyzed on a FACS 440 flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA, USA). Data from 1 × 104 cells were acquired, and the fraction of cells in each phase of the cell cycle was calculated using Multicycle software (Phoenix Flow System, San Diego, CA, USA). The synchrony was >90 % up through the S phase and >70 % up through the G2 phase.
Northern blot analysis
Total cellular RNA from cells exposed to various experimental conditions was isolated using TRI reagent (Molecular Research Center Inc, Cincinnati, OH, USA), following the protocol provided by the manufacturer. Equal amounts (∼5 μg) of the total RNA were separated on a 1.2 % agarose gel containing formaldehyde or on a 15 % polyacrylamide gel (for the analysis of U14 snoRNA), blotted onto a GeneScreen filter, baked at 80°C for 2 hours after transfer, and hybridized with individual probes made by random primer labeling of purified deoxyribonucleic acid (DNA) fragments as described (Chen et al 1996a). An hsc70 specific probe was made from the 3′-noncoding region of the hsc70 gene (Chen et al 1996a), and a U14 probe was made from the U14 coding region of intron 5 of the same gene. The prehybridization and hybridization reactions were carried out in 10 % dextran sulfate, 1 % sodium dodecyl sulfate (SDS), and 1 M NaCl at 65°C overnight. The filters were washed twice with 2× standard saline citrate (SSC) at room temperature for 30 minutes, twice with 2× SSC and 1 % SDS at 65°C, and once with 0.1× SSC and 0.1 % SDS at 65°C. The filters were analyzed, using a Storm 840 Fluorescence Imager (Molecular Dynamics, Sunnyvale, CA, USA) or exposed to a Kodak SR-5 X-ray film. The probe made from the hamster intron 5–encoded U14 snoRNA presumably detects the intron 6– and the intron 8–encoded U14 snoRNAs in the hamster hsc70 gene because, under the same hybridization stringency conditions, we detected the mouse hsc70 intron 5–encoded sequences (Figs 1 and 3), and the sequence differences between the mouse and the hamster intron 5–encoded snoRNAs are greater than the differences between the hamster intron 5–, intron 6–, and intron 8–encoded snoRNAs (Liu and Maxwell 1990; Chen et al 1996a).
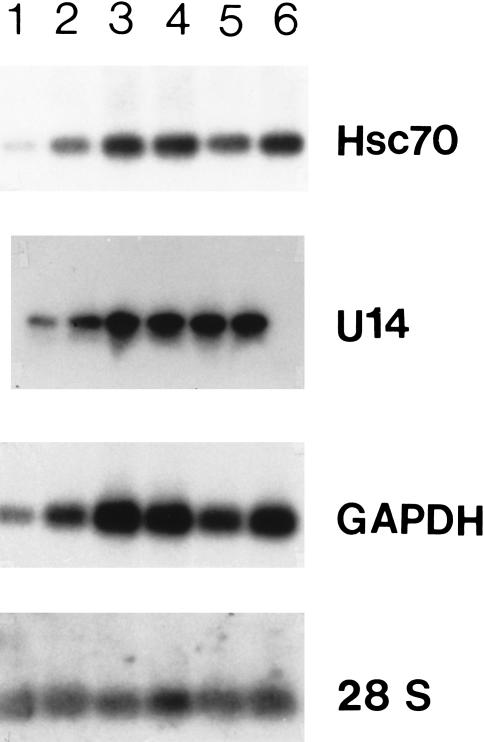
Fig 1.
Expression of U14 small nucleolar ribonucleic acid (snoRNA) and the hsc70 messenger RNA (mRNA) in different phases of the cell cycle under normal growth conditions. RNA samples were prepared from cells in the M (lane 1), early G1 (lane 2), G1 (lane 3), S (lane 4), late S (lane5), and G2 (lane 6) phases of the cell cycle, respectively. Five micrograms of the total RNA were separated on a 1.2 % agarose gel containing formaldehyde and blotted onto nitrocellulose, and hybridized with the hsc70 probe. For U14 analysis, equal amounts of total RNA were separated on a 10 % polyacrylamide gel, electro-blotted onto a GeneScreen filter, and hybridized with a U14 probe made from intron 5 of the hamster hsc70/U14 gene (Chen et al 1996a). Hybridization conditions are described under “Materials and Methods”. The figure represents an autoradiogram obtained after a 48-hour exposure
Nuclear-run-off analysis
Nuclear-run-off transcription assays were conducted following standard procedures (Ausubel et al 1987). The assay was performed with nuclei isolated from 3 independent, complete experiments. Briefly, cell monolayers were exposed to heat shock, sodium arsenite, or azetidine, respectively, as described earlier. In each experiment, about 1 × 108 cells were dislodged into 20 mL ice-cold PBS from the surface of four 175-cm2 tissue culture flasks by scraping with a rubber policeman, and collected by centrifugation for 5 minutes at 500 × g at 4°C. The cell pellets were lysed for 5 minutes in 4 mL lysis buffer (10 mM Tris·Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5 % non-idet P40 (NP40)) on ice. The lysate was then centrifuged for 5 minutes at 500 × g at 4°C, and the supernatant (cytoplasmic fraction) and pellet (nuclear fraction) were saved separately. The nuclear fraction was again treated with a lysis buffer and centrifuged as described earlier. The nuclei were then resuspended in 200 μL storage buffer (50 mM Tris·Cl, pH 8.3, 40 % glycerol, 5 mM MgCl2, and 0.1 mM ethylenediaminetetraacetic acid [EDTA]). This fraction was then frozen immediately in a dry ice ethanol bath and stored in liquid nitrogen.
The cytoplasmic fraction was treated with 80 μL of 20 mg/mL proteinase K in a solution containing 0.1 M Tris·Cl (pH 7.5), 0.22 M NaCl, 1 % SDS, and 12.5 mM EDTA for 30 minutes at 37°C. The resulting mixture was rapidly extracted twice with an equal volume of phenol-chloroform mixture (1:1). The RNA was precipitated by adding 2.5 volumes of ethanol to the aqueous phase and analyzed as described earlier for Northern blots.
For the nuclear-run-off transcription assay, 200 μL of frozen nuclei were thawed by incubating at 30°C for 15 minutes. Then 200 μL of the reaction buffer (10 mM Tris·Cl [pH 8.0], 5 mM MgCl2, 0.3 M KCl, 5 mM dithiothreitol [DTT], and 1 mM of each nucleoside triphosphate [NTP] except uridine triphosphate [UTP]) plus 0.1 mCi of [α-32P]UTP (760 Ci/mmol, 10 mCi/mL) (New England Nuclear, Boston, MA, USA (NEN)) was added to the nuclei, and the reaction mixture was incubated further at 30°C for 30 minutes. The reaction was stopped by the addition of 24 μg RNase-free DNase I (Worthington Biochemical, Lakewood, NJ, USA) and the incubation of the resulting mixture for 5 minutes at 37°C. The mixture was then extracted twice with an equal volume of phenol-chloroform, and the RNA was precipitated with an equal volume of isopropanol. Nonincorporated isotope was removed from the radioactive RNA sample by passing through a NucTrap pushing column (Stratagene, La Jolla, CA, USA).
Equal amounts (about 200 μg) of various DNA templates, including the 3′-noncoding region of the hsc70 gene (Chen et al 1996a), U14 coding sequence from intron 5 of the hsc70/U14 gene (generated by polymerase chain reaction [PCR]), β-actin complementary DNA (cDNA), Bluescript II (BSII, Stratagene), the coding region of the human glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH, obtained from American Type Culture Collection, Manassas, VA, USA), were slot blotted onto a GeneScreen-plus membrane. The DNA templates were denatured by autoclaving the membrane for 3 minutes. The filter was then prehybridized and hybridized with the RNA probes obtained from nuclear-run-off transcription, under the same conditions as described earlier for the Northern blots. The filter was then exposed to X-ray film for 2 weeks. Desitometric analysis of the autoradiograms was performed on a personal densitometer (Molecular Dynamics), using the Image Quant Software supplied by the manufacturer.
RESULTS
Expression of U14 snoRNA and hsc70 mRNA under normal growth conditions
The levels of both U14 snoRNA and hsc70 mRNA in the different phases of the cell cycle were examined under normal growth conditions. Cells were synchronized by mitotic shakeoff, and RNA was prepared from cells in different phases of the cell cycle. Northern blot analysis of these RNA samples indicated that the levels of U14 snoRNA and hsc70 mRNA are correlated in each phase of the cell cycle (Fig 1). Although the absolute levels of both U14 snoRNA and hsc70 mRNA exhibited some variation in different phases of the cell cycle, they covaried. Cells in the M phase had the lowest level of both U14 snoRNA and hsc70 mRNA, followed by early G1 and late S phases. Cells in G1, early S, and G2 phases contained the highest levels of both U14 snoRNA and hsc70 mRNA. Even though the levels of U14 snoRNA and hsc70 mRNA vary in the different phases of the cell cycle in comparison with the level of the 28S ribosomal RNA species, the expression patterns of U14 snoRNA and hsc70 mRNA are quite similar to that of the GAPDH mRNA, which is often used as a control on Northern blots. The pattern of the laminin receptor mRNA (Chen et al 1996b) expression was also similar on the same blot (data not shown).
Expression of U14 snoRNA and hsc70 mRNA after exposure to various stress conditions
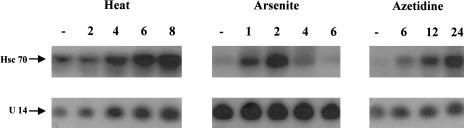
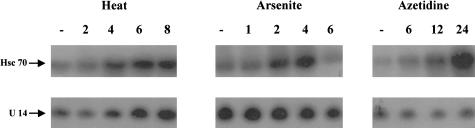
Exposing cells to various stress conditions increased the levels of hsc70 mRNA (Chen et al 1996a). In order to determine the effects of exposure to such stress conditions on the levels of U14 snoRNA and on the correlation between the levels of hsc70 and U14 snoRNA, RNA was isolated from cells exposed to various stresses. The analysis of these RNAs indicated that there was differential accumulation of U14 snoRNA and hsc70 mRNA in response to exposure to various stresses in both Chinese hamster HA-1 cells (Fig 2) and mouse NIH 3T3 cells (Fig 3). (Because Northern blots were exposed for different times to assess the effect of different stresses on the levels of U14 snoRNA, we compared the effect of the different stresses on the levels of U14 snoRNA relative to the level of the U14 snoRNA found in the nontreated cells in each group. The figure does not imply that there is a significant variation in the levels of U14 snoRNA in nontreated cells in the various groups.) In Chinese hamster HA-1 cells, exposure to sodium arsenite (100 μM), induced a prompt elevation of the level of hsc70 mRNA level, but did not induce any significant change in the relative level of U14 snoRNA over that found in the nontreated cells in this group (Fig 2, arsenite). An increase in the level of hsc70 mRNA was detected after 1 hour of the sodium arsenite treatment. The level of hsc70 mRNA reached a maximum at around 2 hours and returned to the level observed in control cells after 6 hours. On the other hand, the level of U14 snoRNA stayed constant under these conditions. Similar results were obtained after exposure of the cells to other concentrations of sodium arsenite, ranging from 50 μM to 400 μM (data not shown). Exposure to azetidine (an analog of proline) led to the accumulation of elevated levels of hsc70 mRNA, which was higher than that observed with either heat or sodium arsenite treatments (Fig 2, azetidine). Again, there was no significant change in the levels of U14 snoRNA under these conditions, when compared with that found in the nontreated cells in this group, similar to what was observed after exposure to sodium arsenite. On the other hand, exposure of HA-1 cells to a heat shock of 45°C for 15 minutes led to a parallel increase in the levels of both U14 snoRNA and hsc70 mRNA, which was detected initially after 4 hours of recovery from the heat shock treatment (Fig 2, heat). Both the RNAs reached the maximum level about 8 hours after the treatment. The induction patterns of U14 snoRNA and hsc70 mRNA were also examined in cells exposed to other heat doses, including 43°C for 30 minutes, 45°C for 5 minutes, and 45°C for 30 minutes, with similar results, namely a parallel increase in the levels of both RNAs and the persistence of elevated levels during the recovery period following heat shock (Chen and Laszlo, unpublished observations). Similar results were obtained with NIH 3T3 cells exposed to heat shock (Fig 3, heat), arsenite (Fig 3, arsenite), and azetidine (Fig 3, azetidine). Again, in the case of arsenite and azetidine, there was no increase in the levels of U14 snoRNA relative to the levels found in the nontreated cells in the respective groups. In the case of arsenite, the induction of hsc70 mRNA was slightly lower than that observed in HA-1 cells following sodium arsenite treatment, and was slightly delayed (Fig 3, arsenite). This difference was also observed after exposure to other doses of sodium arsenite, indicating that there is a slight difference in the response to this agent between HA-1 and 3T3 cells (Chen and Laszlo, unpublished observations).
Fig 2.
Northern blot analysis of total cellular ribonucleic acid (RNA) isolated from Chinese hamster HA-1 cells exposed to a heat shock of 15 minutes at 45°C (heat), 100 μM sodium arsenite for 55 minutes (arsenite), or continuous exposure to a medium containing 2.5 mg/mL of azetidine for up to 24 hours (azetidine). The numbers above each lane indicate the number of hours of recovery at 37°C following exposure to heat shock or sodium arsenite, or the number of hours of treatment with azetidine-containing medium at 37°C, whereas (–) represents the control, nontreated cells. Exposure to the various stresses, and preparation and analysis of total cellular RNAs were performed as described under “Materials and Methods”
Fig 3.
Northern blot analysis of total cellular ribonucleic acid (RNA) isolated from mouse NIH 3T3 cells exposed to a heat shock of 15 minutes at 45°C (heat), 100 μM sodium arsenite for 55 minutes (arsenite), or continuous exposure to a medium containing 2.5 mg/mL azetidine for up to 24 hours (azetidine). The numbers above each lane indicate the number of hours of recovery at 37°C following exposure to heat shock or sodium arsenite, or the number of hours of treatment with azetidine at 37°C, whereas (–) represents the control, nontreated cells. Exposure to the various stresses, and preparation and analysis of total cellular RNAs were performed as described under “Materials and Methods”
Nuclear-run-off analysis
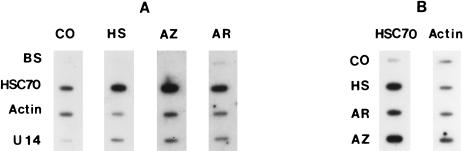
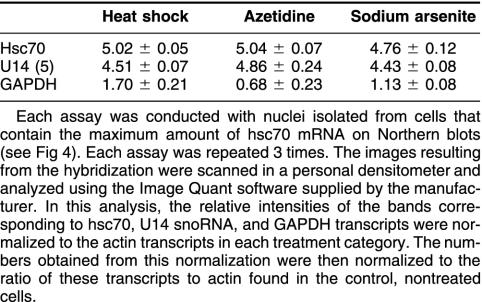
In order to examine the mechanisms involved in the increased levels of hsc70 mRNA and U14 snoRNA after exposure to various stresses, we performed nuclear-run-off experiments. To obtain comparable results, HA-1 cells exhibiting the maximum induction of hsc70 mRNA (as indicated by Northern blots illustrated in Fig 2) after each stress treatment were chosen for the nuclear-run-off assay. Namely, cells were collected at 2 hours, 8 hours, or 24 hours after treatment with sodium arsenite, heat shock, or azetidine, respectively, using the same experimental conditions as in the experiments illustrated in Figure 2. The results of these experiments are shown in Figure 4. The data indicate that there is an increase in the transcription of the hsc70/U14 snoRNA gene in HA-1 cells after exposure to heat shock, sodium arsenite, or azetidine. The relative increase of the rate of transcription observed with the different stresses was similar for both hsc70 mRNA and U14 snoRNA (Table 1).
Fig 4.
Panel A: Nuclear-run-off transcription assays performed with nuclei isolated from control (CO), heated (HS), sodium arsenite–treated (AR), or azetidine-treated (AZ) HA-1 cells. The nuclei for each assay were isolated from cells that contained the maximal levels of hsc70 messenger ribonucleic acid (mRNA) as judged by Northern blots (see Fig 2): for heat shock, 8 hours after exposure to 45°C for 15 minutes; for sodium arsenite, 2 hours after exposure to 100 μM sodium arsenite; and for azetidine, 24 hours after exposure to the medium containing 2.5 mg/mL azetidine. The Bluescript (BS) KS II (Stratagene) DNA was used as a negative control, whereas the β-actin complementary deoxyribonucleic acid (cDNA) (actin) was used as a positive control. Both the U14- and the hsc70-specific DNA fragments were generated by polymerase chain reaction (PCR) from intron 5 and the 3′-noncoding region of the hsc70/U14 gene, respectively, as described under “Materials and Methods”. Equal amounts of each DNA sample were slot blotted onto GeneScreen filters, denatured by high temperature, and hybridized separately to individual 32P-labeled RNA samples derived from the nuclear-run-off transcription reactions. The autoradiogram represents a 14-day exposure. Panel B: Dot-blot analysis of the corresponding cytoplasmic RNA fractions with the hsc70 and β-actin probes, respectively, demonstrating the effectiveness of the stress treatments
Table 1.
Transcriptional induction of the hsc70 messenger ribonucleic acid (mRNA), the U14 small nucleolar RNA (snoRNA), and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA under various stress conditions
DISCUSSION
In mammalian cells, U14 snoRNA and hsc70 mRNA are first transcribed as a single RNA precursor, and then processed into 3 individual U14 snoRNA species (encoded in introns 5, 6, and 8 of the hsc70/U14 gene) and 1 mature hsc70 mRNA (Leverette et al 1992). The single-copy hsc70/U14 gene is the only source for the production of hsc70 mRNA and U14 snoRNA in Chinese hamster HA-1 cells (Chen et al 1996a, Chen and Laszlo 1999), and possibly in other rodent cells such as mouse (Trinh-Rohlik and Maxwell 1988; Liu and Maxwell 1990). Therefore, in HA-1 cells, the expression of both the 3 U14 snoRNA species and the hsc70 mRNA is regulated by the same sets of transcriptional elements located within the promoter of the hsc70/U14 gene. These elements include 2 HSEs (Chen et al 1996a), to which the HSFs bind, mediating an increased transcription after exposure to various stress conditions. Thus, one would expect that as the transcription of the U14 snoRNA species and hsc70 mRNA are linked, there would be a correlation between the levels of U14 snoRNA and hsc70 mRNA, both under normal conditions and after exposure to stress. Such correlation between U14 snoRNA and hsc70 mRNA levels was observed in different phases of the cell cycle under normal growth conditions (Fig 1). In comparison with the level of the ribosomal RNAs, the levels of both the U14 snoRNA species and the hsc70 mRNA exhibited similar variations in different phases of the cell cycle, namely, highest level in G1, S, and G2 phases, followed by late S and early G1 phases, and lowest in M phase. A similar cell-cycle dependent variation was also observed with several other polymerase II transcripts, including the GAPDH mRNA (Fig 1) and the laminin receptor mRNA (Chen et al 1996b) in the same blot (data not shown).
In contrast to the apparent correlation of their levels under normal growth conditions, the increase in the levels of U14 snoRNA and hsc70 mRNA, after exposure to different stresses, was more complex. Northern blots indicated that heat shock increased the levels of both U14 snoRNA and hsc70 mRNA in both hamster and mouse cells (Figs 2 and 3). Exposure to sodium arsenite or azetidine did not lead to a significant increase in the levels of U14 snoRNA, although the levels of hsc70 mRNA increased. In the case of exposure to azetidine, the levels of hsc70 mRNA were significantly higher than those observed in heated cells, without any significant increase in the levels of U14 snoRNA (Figs 2 and 3). Nuclear-run-off transcription assays indicated that the transcription of both of U14 snoRNA and hsc70 mRNA were induced to the same extent by these 3 stress agents (Fig 3 and Table 1) as expected. Therefore, the differential accumulation of U14 snoRNA and hsc70 mRNA under different stress conditions must be due to posttranscriptional processes.
As both U14 snoRNA and hsc70 mRNA are transcribed as a single pre-mRNA, and then processed to produce individual products, changes in the relative levels of both U14 snoRNA and hsc70 mRNA should be similar after alterations in their expression, if these 2 RNA species have similar stability. The difference in the levels between U14 snoRNA and hsc70 mRNA, after exposure to different stress conditions, could result from the altered stability of U14 snoRNA. The increase in the U14 snoRNA level after heat shock may also indicate that U14 snoRNA is involved in the recovery from and repair of heat-induced perturbations in cellular functions. Alternatively, the increased accumulation of U14 snoRNA may be because of a decreased efficiency in its degradation after heat shock. It is of interest to note, however, that in general, the snoRNAs are quite stable (reviewed by Smith and Steitz 1997).
Although other possibilities cannot be ruled out at present, it is likely that there is a function associated with the increased accumulation of U14 snoRNA after exposure to heat shock. In yeast, U14 snoRNA is one of the essential components involved in the processing and modification of 18S ribosomal RNA, and its deletion is lethal (Li et al 1990). Vertebrate U14 snoRNA may have similar functions because a hybrid of the yeast and mouse U14 snoRNA (containing the entire mouse U14 snoRNA and a yeast-specific sequence of around 100 bp) can functionally complement the otherwise lethal deletion of the yeast U14 gene (Li and Fournier 1992). Moreover, all U14 snoRNA species are highly conserved from yeast to humans (Dworniczak and Mirault 1987; Zagorski et al 1988; Xia et al 1995; Kenmochi et al 1996). Disruption of the U14 snoRNA expression in yeast results in an alteration in the patterns of ribosomal RNA processing and in an underaccumulation of the 18S rRNA (Li et al 1990). Heat shock inhibits ribosomal RNA processing and results in an underaccumulation of the 18S rRNA in hamster cells (Bouche et al 1981a, 1981b). It is likely that the U14 snoRNA and hsc70 protein cooperate with each other, either directly or indirectly, to aid in the process of the recovery of the macromolecular machinery involved in ribosomal RNA synthesis and processing from heat-induced perturbations. The fact that both snoRNA and hsc70 are encoded by a single gene, and their expression is regulated by a stress-responsive promoter (Sarge et al 1993; Chen et al 1996a) supports this notion. Consistent with this possibility, the hsc70 protein is translocated into the nucleus from the cytoplasm promptly upon heat shock and localized in the nucleolus (Laszlo 1992; Hattori et al 1993; Morcillo et al 1997), where the U14 snoRNA is also located. Such a translocation of the hsc70 protein has not been observed after the treatments with either sodium arsenite or azetidine (Ohtsuka and Laszlo 1992), which also failed to induce an elevated accumulation of U14 snoRNA. There is a domain in the hsc70 protein, which is homologous to a group of RNA-binding proteins, suggesting that it is possible for U14 snoRNA and hsc70 protein to interact with each other in the recovery of the nucleolar function from heat-induced perturbation (Chen et al 1996b). We are currently investigating possible interactions between the U14 snoRNA species and the hsc70 protein under normal conditions and after heat shock.
Acknowledgments
We thank Ms Terri Davidson for excellent technical support. This research was supported by NIH grant CA-49018 (M.-S.C., A.L.). P.C.G. was supported by CA-69593.
REFERENCES
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, and Struhl K. eds. 1987 Current Protocols in Molecular Biology. Greene Publishing Associates/Wiley Interscience, New York. [Google Scholar]
- Balakin AG, Smith L, Fournier M. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- Barneche F, Steinmetz F, Echeverria M. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J Biol Chem. 2000;275:27212–27220. doi: 10.1074/jbc.M002996200. [DOI] [PubMed] [Google Scholar]
- Bouche G, Caizergues-Ferrer M, Amalric F, Zalta JP, Banville D, Simard R. Synthesis and behavior of small RNA species of CHO cells submitted to a heat shock. Nucleic Acids Res. 1981a;9:1615–1625. doi: 10.1093/nar/9.7.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche G, Raynal F, Amalric F, Zalta JP. Unusual processing of nucleolar RNA synthesized during a heat shock in CHO cells. Mol Biol Rep. 1981b;7:253–258. doi: 10.1007/BF00805762. [DOI] [PubMed] [Google Scholar]
- Chen MS, Laszlo A. Unique features of Chinese hamster S13 gene relative to its human and Xenopus analogs. DNA Cell Biol. 1999;18:463–470. doi: 10.1089/104454999315187. [DOI] [PubMed] [Google Scholar]
- Chen MS, Featherstone T, Laszlo A. Amplification and altered expression of the hsc70/U14 snoRNA gene in a heat resistant Chinese hamster cell line. Cell Stress Chaperones. 1996a;1:47–61. doi: 10.1379/1466-1268(1996)001<0047:aaaeot>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Laszlo A, Fornace AJ. Characterization of an hsp70 related clone encoding a 33 kDa protein with homology to a protein which associates with polysomes. Biochem Biophys Acta. 1996b;1297:124–126. doi: 10.1016/s0167-4838(96)00140-9. [DOI] [PubMed] [Google Scholar]
- Dworniczak B, Mirault M-E. Structure and expression of a human gene coding for a 71 kd heat shock ‘cognate' protein. Nucleic Acids Res. 1987;15:5181–5197. doi: 10.1093/nar/15.13.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Noncoding RNA genes. Curr Opin Genet Dev. 1999;9:695–699. doi: 10.1016/s0959-437x(99)00022-2. [DOI] [PubMed] [Google Scholar]
- Goswami PC, He W, Higashikubo R, Roti Roti JL. Accelerated G1-transit following transient inhibition of DNA replication is dependent on two processes. Exp Cell Res. 1994;214:198–208. doi: 10.1006/excr.1994.1249. [DOI] [PubMed] [Google Scholar]
- Goswami PC, Roti Roti JL, Hunt CR. The cell cycle coupled expression of topoisomerase IIα during S-phase is regulated by mRNA stability and is disrupted by heat shock or ionizing radiation. Mol Cell Biol. 1996;16:1500–1508. doi: 10.1128/mcb.16.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori H, Kaneda T, Lokeshwar B, Laszlo A, Ohtsuka K. A stress-inducible 40 kDa (hsp40): purification by modified two-dimensional gel electrophoresis and co-localization with hsc70(p73) in heat-shocked HeLa cells. J Cell Sci. 1993;104:629–638. doi: 10.1242/jcs.104.3.629. [DOI] [PubMed] [Google Scholar]
- Kenmochi N, Higa S, Yoshihama M, Tanaka T. U14 snoRNAs are encoded in introns of human ribosomal protein S13 gene. Biochem Biophy Res Commun. 1996;228:371–374. doi: 10.1006/bbrc.1996.1668. [DOI] [PubMed] [Google Scholar]
- Laszlo A. Heat shock proteins, protein synthesis and thermotolerance. Exp Cell Res. 1988a;178:401–414. doi: 10.1016/0014-4827(88)90409-0. [DOI] [PubMed] [Google Scholar]
- Laszlo A. Regulation of the synthesis of heat-shock proteins in heat-resistant variants of Chinese hamster fibroblasts. Radiat Res. 1988b;116:427–441. [PubMed] [Google Scholar]
- Laszlo A. The effects of hyperthermia on mammalian cell structure and function. Cell Prolif. 1992;25:69–87. doi: 10.1111/j.1365-2184.1992.tb01482.x. [DOI] [PubMed] [Google Scholar]
- Laszlo A, Li GC. Heat-resistant variants of Chinese hamster fibroblasts altered in the expression of heat shock protein. Proc Natl Acad Sci U S A. 1985;82:8029–8033. doi: 10.1073/pnas.82.23.8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverette RD, Andrews MT, Maxwell ES. Mouse U14 snRNA is a processed intron of the cognate hsc70 heat shock pre-messenger RNA. Cell. 1992;71:1215–1221. doi: 10.1016/s0092-8674(05)80069-8. [DOI] [PubMed] [Google Scholar]
- Li H, Fournier MJ. U14 function in Saccharomyces cerevisiae can be provided by large deletion variants of yeast U14 and hybrid mouse-yeast U14 RNAs. EMBO J. 1992;11:683–689. doi: 10.1002/j.1460-2075.1992.tb05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HV, Zagorski J, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WQ, Fournier MJ. U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev. 1995;9:2433–2443. doi: 10.1101/gad.9.19.2433. [DOI] [PubMed] [Google Scholar]
- Liu J, Maxwell ES. Mouse U14 snRNA is encoded in an intron of the mouse cognate hsc70 heat shock gene. Nucleic Acids Res. 1990;18:6565–6571. doi: 10.1093/nar/18.22.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Morcillo G, Gorab E, Tanguay RM, Diez JL. Specific intra-nucleolar distribution of hsp70 during heat shock in polytene cells. Exp Cell Res. 1997;236:361–370. doi: 10.1006/excr.1997.3726. [DOI] [PubMed] [Google Scholar]
- Ohtsuka K, Laszlo A. The relationship between hsp70 localization and heat resistance. Exp Cell Res. 1992;202:519–539. doi: 10.1016/0014-4827(92)90106-i. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B. Novel intron-encoded small nuclear RNAs. Cell. 1993;75:403–405. doi: 10.1016/0092-8674(93)90374-y. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Kiss T. Functions and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Trinh-Rohlik Q, Maxwell ES. Homologous genes for mouse 4.5S hybRNA are found in all eukaryotes and their low molecular weight RNA transcripts intermolecularly hybridize with eukaryotic 18S ribosomal RNAs. Nucleic Acids Res. 1988;16:6041–6056. doi: 10.1093/nar/16.13.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Liu J, Sage C, Trexier EB, Andrews MT, Maxwell ES. Intronic U14 snoRNA of Xenopus laevis are located in two different parent genes and can be processed from their introns during early oogenesis. Nucleic Acids Res. 1995;23:4844–4849. doi: 10.1093/nar/23.23.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorski J, Tollervey D, Fournier MJ. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispersible and essential small nuclear RNAs. Mol Cell Biol. 1988;8:3282–3290. doi: 10.1128/mcb.8.8.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]