Abstract
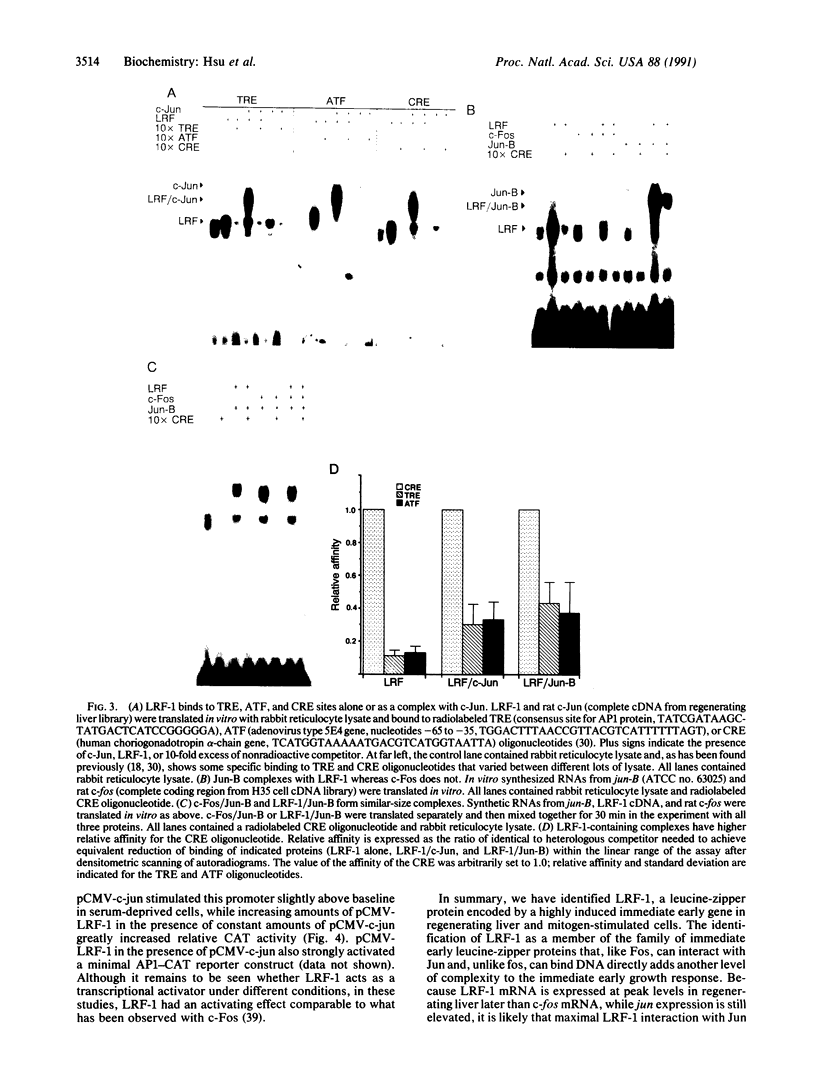
Liver regeneration provides one of the few systems for analysis of mitogenesis in the fully developed, intact animal. Several proteins have been identified as part of the primary growth response in regenerating liver and in mitogen-stimulated cells. Some of these proteins, such as the Jun and Fos families of transcription factors, are thought to have a role in activating transcription of genes expressed subsequently in the growth response. Through differential screening of a regenerating-liver cDNA library, we have identified a rapidly and highly induced gene encoding a 21-kDa leucine-zipper-containing protein that we have designated liver regeneration factor 1 (LRF-1). LRF-1 has no homology with other leucine-zipper proteins outside the basic and leucine-zipper domains. LRF-1 alone can bind DNA, but it preferentially forms heteromeric complexes with c-Jun and Jun-B and does not interact with c-Fos. In solution, it binds with highest affinity to cAMP response elements but also has affinity for related sites. In cotransfection studies, LRF-1 in combination with c-Jun strongly activates a c-Jun-responsive promoter. The induction of the LRF-1 gene in regenerating liver greatly increases the potential variety of heterodimeric combinations of leucine-zipper transcription factors. While LRF-1 mRNA is rapidly induced in the absence of protein synthesis, its peak induction is later than c-fos mRNA, suggesting that LRF-1 may regulate responsive genes at a later point in the cell cycle. As such, LRF-1 may have a unique and critical role in growth regulation of regenerating liver and mitogen-stimulated cells.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alcorn J. A., Feitelberg S. P., Brenner D. A. Transient induction of c-jun during hepatic regeneration. Hepatology. 1990 Jun;11(6):909–915. doi: 10.1002/hep.1840110602. [DOI] [PubMed] [Google Scholar]
- Almendral J. M., Sommer D., Macdonald-Bravo H., Burckhardt J., Perera J., Bravo R. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol. 1988 May;8(5):2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S., Davis D. L., Dahlbäck H., Jörnvall H., Russell D. W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989 May 15;264(14):8222–8229. [PubMed] [Google Scholar]
- Chavrier P., Zerial M., Lemaire P., Almendral J., Bravo R., Charnay P. A gene encoding a protein with zinc fingers is activated during G0/G1 transition in cultured cells. EMBO J. 1988 Jan;7(1):29–35. doi: 10.1002/j.1460-2075.1988.tb02780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Christy B. A., Lau L. F., Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with "zinc finger" sequences. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D. R., Curran T. fra-1: a serum-inducible, cellular immediate-early gene that encodes a fos-related antigen. Mol Cell Biol. 1988 May;8(5):2063–2069. doi: 10.1128/mcb.8.5.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comb M., Birnberg N. C., Seasholtz A., Herbert E., Goodman H. M. A cyclic AMP- and phorbol ester-inducible DNA element. 1986 Sep 25-Oct 1Nature. 323(6086):353–356. doi: 10.1038/323353a0. [DOI] [PubMed] [Google Scholar]
- Fausto N., Mead J. E. Regulation of liver growth: protooncogenes and transforming growth factors. Lab Invest. 1989 Jan;60(1):4–13. [PubMed] [Google Scholar]
- Gonzalez G. A., Yamamoto K. K., Fischer W. H., Karr D., Menzel P., Biggs W., 3rd, Vale W. W., Montminy M. R. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature. 1989 Feb 23;337(6209):749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Hai T. W., Liu F., Coukos W. J., Green M. R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989 Dec;3(12B):2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- Halazonetis T. D., Georgopoulos K., Greenberg M. E., Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988 Dec 2;55(5):917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- Henthorn P., Zervos P., Raducha M., Harris H., Kadesch T. Expression of a human placental alkaline phosphatase gene in transfected cells: use as a reporter for studies of gene expression. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6342–6346. doi: 10.1073/pnas.85.17.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv L. B., Liou H. C., Kara C. J., Lamph W. W., Verma I. M., Glimcher L. H. mXBP/CRE-BP2 and c-Jun form a complex which binds to the cyclic AMP, but not to the 12-O-tetradecanoylphorbol-13-acetate, response element. Mol Cell Biol. 1990 Apr;10(4):1609–1621. doi: 10.1128/mcb.10.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara C. J., Liou H. C., Ivashkiv L. B., Glimcher L. H. A cDNA for a human cyclic AMP response element-binding protein which is distinct from CREB and expressed preferentially in brain. Mol Cell Biol. 1990 Apr;10(4):1347–1357. doi: 10.1128/mcb.10.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz J. W., Iwahashi M. Insulin as a potent, specific growth factor in a rat hepatoma cell line. Science. 1981 Feb 27;211(4485):947–949. doi: 10.1126/science.7008195. [DOI] [PubMed] [Google Scholar]
- Kruijer W., Skelly H., Botteri F., van der Putten H., Barber J. R., Verma I. M., Leffert H. L. Proto-oncogene expression in regenerating liver is simulated in cultures of primary adult rat hepatocytes. J Biol Chem. 1986 Jun 15;261(17):7929–7933. [PubMed] [Google Scholar]
- Macgregor P. F., Abate C., Curran T. Direct cloning of leucine zipper proteins: Jun binds cooperatively to the CRE with CRE-BP1. Oncogene. 1990 Apr;5(4):451–458. [PubMed] [Google Scholar]
- Matsui M., Tokuhara M., Konuma Y., Nomura N., Ishizaki R. Isolation of human fos-related genes and their expression during monocyte-macrophage differentiation. Oncogene. 1990 Mar;5(3):249–255. [PubMed] [Google Scholar]
- Michalopoulos G. K. Liver regeneration: molecular mechanisms of growth control. FASEB J. 1990 Feb 1;4(2):176–187. [PubMed] [Google Scholar]
- Mohn K. L., Laz T. M., Hsu J. C., Melby A. E., Bravo R., Taub R. The immediate-early growth response in regenerating liver and insulin-stimulated H-35 cells: comparison with serum-stimulated 3T3 cells and identification of 41 novel immediate-early genes. Mol Cell Biol. 1991 Jan;11(1):381–390. doi: 10.1128/mcb.11.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn K. L., Laz T. M., Melby A. E., Taub R. Immediate-early gene expression differs between regenerating liver, insulin-stimulated H-35 cells, and mitogen-stimulated Balb/c 3T3 cells. Liver-specific induction patterns of gene 33, phosphoenolpyruvate carboxykinase, and the jun, fos, and egr families. J Biol Chem. 1990 Dec 15;265(35):21914–21921. [PubMed] [Google Scholar]
- Morello D., Fitzgerald M. J., Babinet C., Fausto N. c-myc, c-fos, and c-jun regulation in the regenerating livers of normal and H-2K/c-myc transgenic mice. Mol Cell Biol. 1990 Jun;10(6):3185–3193. doi: 10.1128/mcb.10.6.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y., Ryder K., Nathans D. DNA binding activities of three murine Jun proteins: stimulation by Fos. Cell. 1988 Dec 2;55(5):907–915. doi: 10.1016/0092-8674(88)90146-8. [DOI] [PubMed] [Google Scholar]
- Ryder K., Lanahan A., Perez-Albuerne E., Nathans D. jun-D: a third member of the jun gene family. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1500–1503. doi: 10.1073/pnas.86.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder K., Lau L. F., Nathans D. A gene activated by growth factors is related to the oncogene v-jun. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1487–1491. doi: 10.1073/pnas.85.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryseck R. P., Hirai S. I., Yaniv M., Bravo R. Transcriptional activation of c-jun during the G0/G1 transition in mouse fibroblasts. Nature. 1988 Aug 11;334(6182):535–537. doi: 10.1038/334535a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P., Ransone L. J., Verma I. M. Cross-talk in signal transduction: TPA-inducible factor jun/AP-1 activates cAMP-responsive enhancer elements. Oncogene. 1990 Mar;5(3):427–431. [PubMed] [Google Scholar]
- Sonnenberg J. L., Rauscher F. J., 3rd, Morgan J. I., Curran T. Regulation of proenkephalin by Fos and Jun. Science. 1989 Dec 22;246(4937):1622–1625. doi: 10.1126/science.2512642. [DOI] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986 Oct 16;323(6089):640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- Sukhatme V. P., Cao X. M., Chang L. C., Tsai-Morris C. H., Stamenkovich D., Ferreira P. C., Cohen D. R., Edwards S. A., Shows T. B., Curran T. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988 Apr 8;53(1):37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- Taub R., Gould R. J., Garsky V. M., Ciccarone T. M., Hoxie J., Friedman P. A., Shattil S. J. A monoclonal antibody against the platelet fibrinogen receptor contains a sequence that mimics a receptor recognition domain in fibrinogen. J Biol Chem. 1989 Jan 5;264(1):259–265. [PubMed] [Google Scholar]
- Taub R., Roy A., Dieter R., Koontz J. Insulin as a growth factor in rat hepatoma cells. Stimulation of proto-oncogene expression. J Biol Chem. 1987 Aug 5;262(22):10893–10897. [PubMed] [Google Scholar]
- Zerial M., Toschi L., Ryseck R. P., Schuermann M., Müller R., Bravo R. The product of a novel growth factor activated gene, fos B, interacts with JUN proteins enhancing their DNA binding activity. EMBO J. 1989 Mar;8(3):805–813. doi: 10.1002/j.1460-2075.1989.tb03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel P. F., Irving S. G., Kelly K., Siebenlist U. Complexity of the primary genetic response to mitogenic activation of human T cells. Mol Cell Biol. 1989 Mar;9(3):1041–1048. doi: 10.1128/mcb.9.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]