Summary
No one knows yet how to organize, in a simple yet predictive form, the knowledge concerning the anatomical, biophysical, and molecular properties of neurons that are accumulating in thousands of publications every year. The situation is not dissimilar to the state of Chemistry prior to Mendeleev’s tabulation of the elements. We propose that the patterns of presence or absence of axons and dendrites within known anatomical parcels may serve as the key principle to define neuron types. Just as the positions of the elements in the Periodic Table indicate their potential to combine into molecules, axonal and dendritic distributions provide the blueprint for network connectivity. Furthermore, among the features commonly employed to describe neurons, morphology is considerably robust to experimental conditions. At the same time, this core classification scheme is suitable for aggregating biochemical, physiological, and synaptic information.
Keywords: axons, circuits, classification, dendrites, neurons
Introduction
Every day neuroscientists around the world characterize the morphology, electrophysiology, and biochemistry of neurons in various brain regions of many species. However, no one yet knows how to express this knowledge in a standardized form that is both simple and predictive. This lack of organization hampers both conceptual progress and day-to-day research. The current inability to structure methodically new and established information about neurons limits ongoing attempts to assemble a comprehensive “list of parts” for the nervous system, making it challenging to uncover patterns and identify gaps in our understanding [1–3]. A robust neuronal classification framework would provide the foundation for a much needed computational theory of neural function [4–6].
At a practical level, the present state of confusion in the community is fueling endless terminological inconsistencies for neuron types and their properties, and multiple names are employed for the same concept or a single name used to refer to different concepts [7]. The ensuing “Tower of Babel” is especially apparent in the ambiguous nomenclature of GABAergic cortical interneurons, which reflects both real disagreements (e.g. do Ivy and Neurogliaform cells constitute distinct neuron types [8]?) and common misunderstandings (e.g. using the word “bistratified” to indicate a neuron type [9] or an arbor feature [10]). Rectifying these ambiguities is essential to facilitate scientific communication, enable effective literature mining, and minimize the risk of incorrect interpretations.
The root of this organizational impasse is buried deep in the diversity and complexity of neurons. Ideally, we would measure all relevant aspects in a representatively large number of neurons. In practice, existing techniques in most cases only provide a partial window on neuronal identity that depends on the measurement instrument. Thus, three broad categories of neuronal properties are typically distinguished [11]: anatomical (most often probed by microscopy), electrophysiological (usually recorded with electrodes), and molecular (normally characterized through biochemical assays).
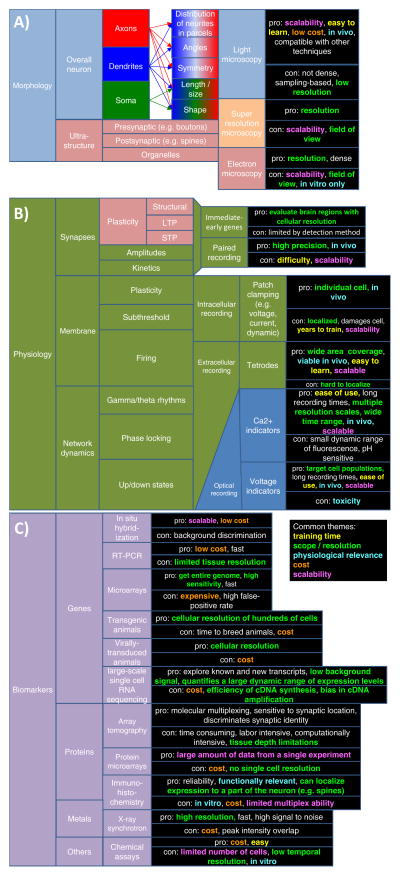
Several alternative schemes for neuronal classification are possible even within a single domain (Fig. 1). For example, the morphometric quantification of axonal or dendritic length across the entire neuronal arbor by light microscopy on the one hand, and the ultrastructural characterization of synaptic contacts by electron microscopy on the other, depict largely complementary views of neurons, yet both fall under the anatomical category. Similarly, electrophysiology investigates intrinsic excitability (firing patterns and active membrane properties) as well as synaptic dynamics and plasticity, which are rather distinct phenomena. The same applies to molecular studies spanning from transcription factor regulation to the subcellular distribution of biomarkers. Each of these approaches to characterizing neurons has advantages and disadvantages, including cost, technical difficulty, invasiveness, scalability, and compatibility with multiple experimental designs (Figs. 1A–C).
Figure 1.
The three main experimental approaches to characterize neuron types. A: Morphology. B: Electrophysiology. C: Molecular biomarkers. Each approach is suitable to investigate a subset of general properties and specific features based on different techniques. The advantages and disadvantages of every technique are color-coded along themes that recur across dimensions: invasiveness, cost, scalability, training time, coverage scope, and resolution. D: The GABAergic interneurons of hippocampal area CA1 can be grouped on the basis of selected properties across three dimensions in a highly simplified binary classification scheme. In this example, neuron types are divided along the vertical axis based on parvalbumin (PV) expression (one of many molecular biomarkers), along the depth axis based on firing threshold potential (Vthresh; one of many electrophysiological variables), and along the horizontal axis as to whether they can receive perforant path (PP) input (one of many morphological factors).
Although consensus is lacking on what the defining features are, separate neuronal classes typically share a subset of qualities. Thus, it is generally assumed that a detailed characterization combining multiple modalities is both necessary and sufficient for the positive identification of neuron types. For example, neurons from hippocampal area CA1 can be divided into parvalbumin-positive or negative, high- or low-threshold, or distinguished on the basis of whether they receive direct entorhinal input (Fig. 1D). The discriminating power of these categories increases further by considering additional molecular, morphological, and physiological properties. However, the long-standing argument between lumpers and splitters is far from settled, as neuroscientists still widely disagree on how many neuron types should be distinguished. If the same measurements were consistently extracted for a representatively large sample of neurons, classification granularity could in principle be optimized to maximize information content based on Bayesian theory [12].
The problem posed by the phenotypic diversity of neurons and the number of relevant properties is further complicated by additional challenges [13]. Neuronal classification also needs to contend with differences across species, developmental stages, and brain regions. Moreover, neuronal features may also depend on the specific experimental conditions: for example, molecular expression can vary with circadian rhythms, and electrophysiological measurements are sensitive to temperature and solution composition. Particular attention must be paid to the difference between in vivo and in vitro characterizations: while in vivo characterizations can reveal relevant behavioral or cognitive functions and underlying computational mechanisms, the in vitro studies account for the vast majority of available data to date, requiring a certain degree of extrapolation. For example, projection axons cannot be morphologically characterized from the slice preparation. Likewise, the natural firing behavior of neurons might be particularly distorted in vitro on account of missing background activity.
As a result of the absence of a consistent organizational framework, data on neuronal characteristics are accumulating in thousands of new articles every year scattered throughout hundreds of journals, yet even recent attempts to classify neurons based on morphology, electrophysiology, and biochemistry have been limited to restricted datasets or fairly broad classes [14–17]. By and large, a century after the advent of the neuron doctrine [18–22], a complete accounting of neuron types and their defining properties is still lacking [23].
Cajal like Lavoisier
The current situation in Neuroscience is not dissimilar to the state of Chemistry 150 years ago. Following Lavoisier’s experimental demonstration of the law of conservation of mass in the 1770’s, the knowledge for combining reactants in optimal ratios led to an exuberant accumulation of chemical data for nearly a century. Several new elements (including sodium, potassium, calcium, silicon, bromine, and iodine) and numerous reactions (such as the synthesis of acetic acid from non-organic materials) were discovered during this period [24]. Large collections of empirical recipes were developed, but a unifying principle was still missing. Mendeleev’s ordering of chemical elements in 1869 had tremendous explanatory power by fully accounting for the observed fixed ratios in the many-to-many molecular combinations of the elements. Specifically, the so-called octet rule corresponding to the elements’ periodicity integrated and expanded many known but seemingly unrelated facts, such as the unitary valence of alkalis and halogens, the dual valence (3 and 5) of nitrogen and phosphorous, and the inert behavior of noble gases. Moreover, the horizontal alignment of elements in Mendeleev’s original publication (commonly displayed in vertical orientation in modern times) predicted their similar reactivity, and missing entries in the table strongly suggested the existence of yet to-be-discovered elements. Several of these predictions were confirmed by the isolation of gallium, scandium, and especially germanium during Mendeleev’s lifetime, stimulating the thorough (and eventually successful) search for the remaining missing atoms for many decades thereafter.
Interestingly, the success of Mendeleev’s approach was empirically driven rather than theoretically inspired. Nowadays quantum mechanics links the number of protons in the atomic nucleus to the shape of orbitals, explaining chemical valence and molecular bonding. Mendeleev’s original Table, however, ordered the elements by their mass, one among the more limited set of properties that could be systematically measured at those times. Since the number of neutrons equals the number of protons in most elements, the atomic mass is roughly twice the atomic number, thus approximately preserving the periodic relationship. Remarkably, Mendeleev noticed the residual mismatch, effectively predicting the atomic number 40 years prior to the discovery of the proton.
Can a set of similarly simple yet predictive features be identified that would enable the orderly arrangement of neurons?
Waiting for neuroscience’s Mendeleev
Co-localization of the axon of one neuron and the dendrite of another in the same anatomical parcel, such as an area, layer, or nucleus, provides the opportunity, and at the same time constitutes a prerequisite, to form a synapse [25, 26]. Therefore, just like the positions of the elements in the Periodic Table indicate their potential to combine into molecules, axonal and dendritic distributions provide the blueprint for network connectivity [27]. Following this premise, we propose using the patterns of presence or absence of axons and dendrites within distinguishable neuropil boundaries, along with the identity of the main neurotransmitter, as the core criteria to define neuron types.
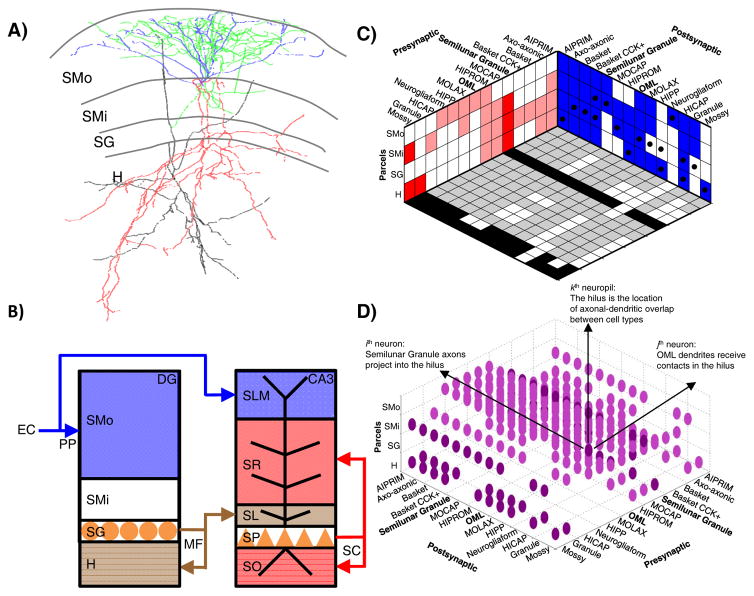
As an example of the proposed classification scheme, a rat dentate gyrus (DG) semilunar granule cell [28] (Fig. 2A) is a glutamatergic neuron with dendrites in the outer and inner stratum moleculare (SMo and SMi) and axons in SMi, stratum granulare (SG), and the hilus (H) (as well as in CA3 stratum lucidum). A cat neocortical layer 4 (L4) basket cell [29] is a GABAergic neuron with both axons and dendrites in L2/3 and L4. The proposed definition can also be applied to classify as yet-unnamed neurons from large-scale studies. For instance cell Gad1-F-700000 from the fly brain [30] is categorized (together with 4 out of over 16,000 other cells from the same study) as a GABAergic neuron with axons in the ventrolateral protocerebrum (VLP) and dendrites in the lobula (LOB).
Figure 2.
Axonal-dendritic patterns and anatomical regions: two sides of the same coin. A: Morphological reconstructions (from NeuroMorpho.Org [50]) of a rat dentate gyrus semilunar granule cell (dendrites in blue, axons in red; NMO_10035 from Ref. [28]) and of an OML cell (dendrites in black, axons in green; NMO_00178 from Ref. [31]). B: The layers of the dentate gyrus (DG) and hippocampal area CA3 are defined by the somatic locations and axonal terminals of principal neurons. The perforant path (PP) axonal terminals of entorhinal layer 2 spiny stellate cells delineate the outer molecular layer in DG and lacunosum-moleculare in CA3. The mossy fiber (MF) axonal terminals of DG granule cells define the hilus in DG and the lucidum layer in CA3. The recurrent and commissural terminals of CA3 pyramidal cells demarcate the oriens and radiatum layers in CA3. Abbreviations: SMo (outer stratum moleculare), SMi (inner stratum moleculare), SG (stratum granulare), H (hilus), SLM (stratum lacunosum-moleculare), SR (stratum radiatum), SL (stratum lucidum), SP (stratum pyramidale), and SO (stratum oriens). C: A periodic table of neuromorphological patterns in the dentate gyrus. The two walls of the box represent the pre-synaptic axonal (red) and post-synaptic dendritic (blue) binary distributions of every neuron type (listed horizontally) across all neuropils (listed vertically). The box floor represents the predicted circuitry, in which black and gray squares correspond to potential excitatory and inhibitory connections, respectively, and white squares indicate absence of synapses. Black dots on the blue wall indicate the location of the somata for the various neuron types. D: A “raindrop cloud” representation of the neuropil tensor, where the ijkth entry represents a potential synapse formed by the axon of the ith neuron onto the dendrite of the jth neuron in the kth neuropil.
This classification recipe provides, to a first approximation, the essential information to predict all possible pre- and post-synaptic partners of every present and future neuron type, as well as the location of the potential synapses. For instance, semilunar granule cells could in principle make synapses onto any target with dendrites in SMi, SG, or H; and they could receive contacts from any neuron with axons in SMo and SMi. More generally, if the axons and dendrites are represented as vectors with binary components corresponding to the neuropils (e.g. the semilunar granule cell axon would be 0111 across SMo, SMi, SG, and H), two neuron types can form a synapse if, and only if, the dot product (·) between the pre-synaptic axon and the post-synaptic dendrite is non-zero. For example, semilunar granule cells can form synapses onto outer molecular layer (OML) cells, whose dendrites span all DG layers [31] (0111·1111≠0), but not with DG neurogliaform cells, whose dendrites are confined to SMo [32] (0111·1000=0).
Notably, unlike the single concept of atomic valence underlying molecular bonds, neuronal connectivity is intrinsically directional. Therefore, the “neuronal valence” must be expressed with distinct pre-synaptic (axonal) and post-synaptic (dendritic) components. Moreover, because the axons and dendrites of individual neurons are often distributed across multiple neuropils, both (pre- and post-synaptic) components of the neuronal valence are multi-dimensional, the dimensionality corresponding to the distinguishable anatomical parcels in which the circuit is embedded.
The boundaries of these parcels are not arbitrary, but are in fact themselves demarcated by the distinct axonal-dendritic neuropils (or by the principal cell layers). In hippocampal area CA3, for instance, the lucidum, radiatum, and lacunosum-moleculare layers correspond to the regions occupied by the apical dendrites of pyramidal neurons that are invaded by the mossy fibers (the axons of DG granule cells), the recurrent commissural and collaterals (the axons of ipsi- and contra-lateral CA3 pyramidal cells), and the perforant pathway (the axons of layer 2 spiny stellate cells in the entorhinal cortex), respectively (Fig. 2B). This conceptual link reveals a fundamentally intertwined relationship between anatomical parceling, neuron types, and circuitry [33].
The proposed definitions of a set of neuron types in a given neural system, such as the rat dentate gyrus, can be compactly summarized, along with the corresponding potential connectivity, in a discrete Cartesian representation (Fig. 2C). The three orthogonal axes correspond to the neuropils (listed vertically) and to the pre- and post-synaptic neuron components (listed in the two horizontal coordinates). The two walls of the resulting box represent the axonal (red) and dendritic (blue) patterns of every neuron type (horizontally) across all neuropils (vertically). The box floor represents the connectivity matrix, in which black and gray squares correspond respectively to potential excitatory and inhibitory connections, and white squares indicate the absence of synapses. The neuropil “raindrop cloud” that conceptually lives inside the box is displayed separately for clarity of illustration (Figure 2D). The ijkth tensor entry (cloud drop) represents a potential synapse formed by the axon of the ith neuron and the dendrite of the jth neuron in the kth neuropil.
In what sense is this multi-dimensional table of the neurons periodic? As in Mendeleev’s first table of the elements, horizontal alignment of neurites predicts similar circuit composition. Specifically, neuron types that have axons in the same parcel(s) can contact the same post-synaptic targets. For example, in the dentate gyrus, HIPP and neurogliaform cells align horizontally in the SMo axonal dimension, and they can both in principle target any neuron with dendrites in that region, such as granule cells. Similarly, neuron types that have dendrites in the same parcel(s) can receive inputs from the same sources. For example, MOLAX and dentate basket cells align horizontally in the hilar dendritic dimension, and they can both in principle be synapsed upon by any neuron with axons in that region, such as MOCAP cells.
While these correspondences are useful to guide analysis and interpretation of neural circuits, synaptic selectivity may rule out certain connections despite axonal-dendritic co-localization, as in the cases of interneuron-specific interneurons and Chandelier cells, respectively avoiding glutamatergic and GABAergic targets [34]. Recent experimental evidence [35] suggests that these deviations from the predicted connectivity can themselves be coded by a limited number of rules, refining, rather than undermining, the scaffolding circuitry established by the anatomical distributions of axons and dendrites.
The proposed definition also provides a simple framework to represent the location of potential connections relative to the pre- and post-synaptic somata (black dots in Fig. 2C). For instance, semilunar granule cells of the dentate gyrus (whose somata are in SMi) may receive dendritic inhibition from both HICAP and neurogliaform cells, but the former would synapse proximally, while the latter more distally (in SMo). The synaptic distance along the dendrites can affect signal attenuation and integration depending on passive and active membrane properties [36]. Similarly, the axons of HIPROM cells may contact the dendrites of both mossy cells and granule cells, but in the former case they do so near their somata of origin (in the hilus), while in the latter the contact is farther away (in the stratum moleculare). The synaptic distance along the axon can affect signal delay and signal failure due to intervening branching points [37].
Despite its considerable predictive power, this characterization of neuron types is extremely parsimonious, because it does not require a detailed (and labor-intensive) quantification of axonal and dendritic arbors. Overall arbor morphology is widely adopted in neuroscience as the most common identifier to recognize neuron types [38]. As a consequence, the axonal and dendritic patterns are already known for many neural systems. In addition to the rodent dentate gyrus and all other regions of the hippocampal formation (CA3, CA2, CA1, subiculum, and entorhinal cortex [39]), periodic tables of the neurons can be readily generated for the cat primary visual cortex and the fly brain (Fig. 3). Similar efforts appear viable for cerebellum, spinal cord, olfactory bulb, retina, basal ganglia, amygdala, and thalamus [40].
Figure 3.
Periodic tables and potential circuit connectivity (encoding as in Figure 2, C and D). A: Neuron types from the cat primary visual neocortex: layer 2/3 pyramidal cells (p2/3); layer 2/3 basket cells (b2/3); layer 2/3 double bouquet cells (db2/3); layer 4 spiny stellate cells with axon in layer 4 (ss4 (L4)); layer 4 spiny stellate cells with axon in layer 2/3 (ss4 (L2/3)); layer 4 pyramidal cells (p4); layer 4 basket cells (b4); layer 5 pyramidal cells with axon in layer 2/3 (p5 (L2/3)); layer 5 pyramidal cells with axon in layers 5 and 6 (p5 (L5/6)); layer 5 basket cells (b5); layer 6 pyramidal cells with axon in layer 4 (p6 (L4)); layer 6 pyramidal cells with axon in layers 5 and 6 (p6 (L5/6)). B: Potential connectivity of the neurons represented in panel A. C: Cholinergic (excitatory) and GABAergic (inhibitory) neuron types from the fly brain (see Supplementary Information), with birth day indicated. D: Potential connectivity of the neurons represented in panel A.
While this approach cannot directly compare neuron types across brain areas, useful parallels nonetheless emerge. For example, dentate gyrus HIPP cells have dendrites and somata in the deep layer (hilus), and project their axons superficially (into outer molecular layer). Similarly, CA1 O-LM cells have dendrites and somata in the deep layer (oriens) and project their axons superficially (into lacunosum-moleculare). An identical motif also describes neocortical Martinotti cells, suggesting a shared circuit function for these three neuron types. This speculation is corroborated by analogous molecular (somatostatin-expressing) and electrophysiological (stuttering) characteristics. Likewise, hippocampal and neocortical pyramidal neurons with dendrites spanning all layers may share the same role of universal information recipients. The methodical compilation of periodic tables of neurons in multiple neural systems might thus reveal convergent and divergent architectural modules across species, brain regions, and developmental stages. Parallel morphological patterns across sub-regions could even indicate the existence of to-be-discovered neuron types (e.g. Martinotti-like cells in the subiculum). A parcel containing only axonal or dendritic terminal would also suggest a missing neuron type with complementary neurites.
As more detailed data become available for larger proportions of the brain, quantifying the neuropil distribution of axonal and dendritic length and of the number of pre-synaptic varicosities and post-synaptic densities or spines will provide a tighter correlate of network connectivity [41, 42]. Although simple, however, the binary pattern of axons and dendrites across neuropils is already highly effective in discriminating neuron types. Out of 122 neuron types recently surveyed in the rat entorhinal-hippocampal system (Hippocampome.org [39]), only seven could not be separated by coarse morphology alone (e.g. parvalbumin-expressing fast-spiking vs. CCK-expressing regular-spiking basket cells). The ability to capture most known types into a compact description constitutes an advantage over more complex classification schemes.
More generally, in a neural system with k parcels, (2k-1)2 neurons could be theoretically distinguished just on the basis of binary axonal-dendritic patterns. For example, partitioning a brain region simply into right-left, top-bottom, and front-back could give rise to as many as 65,025 neuron types. In practice, a much smaller number of phenotypes have been observed so far. Dividing the human brain in as few as 40 parcels would already enable more possible binary patterns of axonal and dendritic distributions than the estimated number of neurons it contains. Thus, the recognizably distinct qualitative binary patterns that neurons clearly display are suggestive of discrete categories despite the known cell-to-cell variability in anatomical, biochemical, and physiological variables.
Our representation does not yet consider the molecular and biophysical diversity of neurons [43]. Alternative schemes will be necessary to reveal the complex interaction among transcription factors and circuit specificity [44]. Nevertheless, many studies rely on axonal and dendritic morphology to link intracellular recordings or protein expressions with identified neuron types. Therefore, this proposal is well suited as a core to progressively integrate biochemical, electrophysiological, and synaptic data. For instance, starting from the binary patterns of axons and dendrites, the above-mentioned knowledge base of hippocampal neuron types [39] successfully amassed over 13,000 pieces of molecular and biophysical evidence. This evidence unequivocally delineates the available information regarding the expression of more than 90 biomarkers and the measured ranges of the most important active and passive membrane properties (Hippocampome.org). Additionally, among the features commonly employed to describe neurons, morphology is relatively robust to experimental conditions, hence generally increasing reproducibility across laboratories. Hence, the proposed classification framework offers a practical approach to consolidate and organize data from existing publications as well as forthcoming studies.
Conclusions and outlook
The key to success in biological classification is the judicious selection of pivotal variables with the most discriminant and explanatory power. Luckily, History demonstrates that the initial choice of the organizing criteria has neither to be perfect nor based on a singular set of rules in order to revolutionize the field. Similar to the approximate, yet practical and effective, role of atomic mass in Mendeleev’s original Table, we envision axonal-dendritic patterns to serve as a crucial stepping stone towards neuronal classification. This initial level of description appears particularly amenable to organize the existing data as accessible in the published literature.
Even in terms of binary neurite patterns, knowledge is still sparse with respect to complete axons of projection neurons [45, 46]. However, novel technological advances continue to accelerate the pace of data acquisition [47, 48]. Furthermore, a large influx of new data is widely expected from Big Science endeavors [49] such as the European Union Human Brain Project (humanbrainproject.eu), the United States BRAIN initiative (whitehouse.gov/brain), the Allen Institute Cell Type Project (celltypes.brain-map.org), the Big Neuron consortium (bigneuron.org), and the Howard Hughes Medical Institute Fly Light project (janelia.org/project-team/flylight). Systematic integration of existing and forthcoming information in a periodic table of the neurons organized by axonal and dendritic patterns will have explanatory and predictive power with respect to circuits and could point the way towards more comprehensive neuronal classifications, likely integrating molecular fingerprinting, developmental origins, activity dynamics, and functional plasticity.
Supplementary Material
Acknowledgments
We gratefully acknowledge grants R01NS39600 (NIH), R01NS086082 (NIH), MURI N00014-10-1-0198 (ONR), FA9550-10-1-0385 (AFOSR), DBI-1546335 (NSF), and IIS-1302256 (NSF).
Footnotes
Conflict of Interest
None of the authors has a conflict of interest to declare.
References
- 1.Huang ZJ. Toward a genetic dissection of cortical circuits in the mouse. Neuron. 2014;83:1284–302. doi: 10.1016/j.neuron.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denk W, Briggman KL, Helmstaedter M. Structural neurobiology: missing link to a mechanistic understanding of neural computation. Nat Rev Neurosci. 2012;13:351–8. doi: 10.1038/nrn3169. [DOI] [PubMed] [Google Scholar]
- 3.Lodato S, Arlotta P. Generating neuronal diversity in the mammalian cerebral cortex. Annu Rev Cell Dev Biol. 2015;31:699–720. doi: 10.1146/annurev-cellbio-100814-125353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharpee TO. Toward functional classification of neuronal types. Neuron. 2014;83:1329–34. doi: 10.1016/j.neuron.2014.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–26. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mainetti M, Ascoli GA. A neural mechanism for background information-gated learning based on axonal-dendritic overlaps. PLoS Comput Biol. 2015;11:e1004155. doi: 10.1371/journal.pcbi.1004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton DJ, Wheeler DW, White CM, Rees CL, et al. Name-calling in the hippocampus (and beyond): coming to terms with neuron types and properties. Brain Inform. 2016 doi: 10.1007/s40708-016-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong C, Krook-Magnuson E, Soltesz I. Neurogliaform and Ivy Cells: A Major Family of nNOS Expressing GABAergic Neurons. Front Neural Circuits. 2012;6:23. doi: 10.3389/fncir.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buhl EH, Halasy K, Somogyi P. Diverse sources of hippocampal unitary inhibitory postsynaptic potentials and the number of synaptic release sites. Nature. 1994;368:823–8. doi: 10.1038/368823a0. [DOI] [PubMed] [Google Scholar]
- 10.Daw MI, Tricoire L, Erdelyi F, Szabo G, et al. Asynchronous transmitter release from cholecystokinin-containing inhibitory interneurons is widespread and target-cell independent. J Neurosci. 2009;29:11112–22. doi: 10.1523/JNEUROSCI.5760-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petilla Interneuron Nomenclature Group. Ascoli GA, Alonso-Nanclares L, Anderson SA, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9:557–68. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armañanzas R, Ascoli GA. Towards the automatic classification of neurons. Trends Neurosci. 2015;38:307–18. doi: 10.1016/j.tins.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton DJ, Shepherd GM, Martone ME, Ascoli GA. An ontological approach to describing neurons and their relationships. Front Neuroinformatics. 2012;6:15. doi: 10.3389/fninf.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanes JR, Masland RH. The types of retinal ganglion cells: current status and implications for neuronal classification. Annu Rev Neurosci. 2015;38:221–46. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- 15.Tripathy SJ, Burton SD, Geramita M, Gerkin RC, et al. Brain-wide analysis of electrophysiological diversity yields novel categorization of mammalian neuron types. J Neurophysiol. 2015;113:3474–89. doi: 10.1152/jn.00237.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–42. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 17.Nagayama S, Homma R, Imamura F. Neuronal organization of olfactory bulb circuits. Front Neural Circuits. 2014;8:98. doi: 10.3389/fncir.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.López-Muñoz F, Boya J, Alamo C. Neuron theory, the cornerstone of neuroscience, on the centenary of the Nobel Prize award to Santiago Ramón y Cajal. Brain Res Bull. 2006;70:391–405. doi: 10.1016/j.brainresbull.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Jones EG. Neuroanatomy: Cajal and after Cajal. Brain Res Rev. 2007;55:248–55. doi: 10.1016/j.brainresrev.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Bock O. Cajal, Golgi, Nansen, Schäfer and the neuron doctrine. Endeavour. 2013;37:228–34. doi: 10.1016/j.endeavour.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Senft SL. A brief history of neuronal reconstruction. Neuroinformatics. 2011;9:119–28. doi: 10.1007/s12021-011-9107-0. [DOI] [PubMed] [Google Scholar]
- 22.DeFelipe J, Garrido E, Markram H. The death of Cajal and the end of scientific romanticism and individualism. Trends Neurosci. 2014;37:525–7. doi: 10.1016/j.tins.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 23.DeFelipe J, López-Cruz PL, Benavides-Piccione R, Bielza C, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14:202–16. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ihde AJ. The development of modern chemistry. New York: Dover; 1984. [Google Scholar]
- 25.Bota M, Swanson LW. The neuron classification problem. Brain Res Rev. 2007;56:79–88. doi: 10.1016/j.brainresrev.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ascoli GA. On synaptic circuits, memory, and kumquats. N Engl J Med. 2015;373:1170–2. doi: 10.1056/NEJMcibr1509692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuste R. From the neuron doctrine to neural networks. Nat Rev Neurosci. 2015;16:487–97. doi: 10.1038/nrn3962. [DOI] [PubMed] [Google Scholar]
- 28.Yu J, Proddutur A, Elgammal FS, Ito T, et al. Status epilepticus enhances tonic GABA currents and depolarizes GABA reversal potential in dentate fast-spiking basket cells. J Neurophysiol. 2013;109:1746–63. doi: 10.1152/jn.00891.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Binzegger T, Douglas R, Martin K. A quantitative map of the circuit of cat primary visual cortex. J Neurosci. 2004;24:8441–53. doi: 10.1523/JNEUROSCI.1400-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang A-S, Lin C-Y, Chuang C-C, Chang H-M, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 31.Mott DD, Turner DA, Okazaki MM, Lewis DV. Interneurons of the dentate-hilus border of the rat dentate gyrus: morphological and electrophysiological heterogeneity. J Neurosci. 1997;17:3990–4005. doi: 10.1523/JNEUROSCI.17-11-03990.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armstrong C, Szabadics J, Tamás G, Soltesz I. Neurogliaform cells in the molecular layer of the dentate gyrus as feed-forward γ-aminobutyric acidergic modulators of entorhinal-hippocampal interplay. J Comp Neurol. 2011;519:1476–91. doi: 10.1002/cne.22577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ascoli GA. Trees of the brain, roots of the mind. Cambridge, Massachusetts; London, England: The MIT Press; 2015. [Google Scholar]
- 34.Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–7. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang X, Shen S, Cadwell CR, Berens P, et al. Principles of connectivity among morphologically defined cell types in adult neocortex. Science. 2015;350:aac9462. doi: 10.1126/science.aac9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komendantov A, Ascoli G. Dendritic excitability and neuronal morphology as determinants of synaptic efficacy. J Neurophysiol. 2009;101:1847–66. doi: 10.1152/jn.01235.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debanne D. Information processing in the axon. Nat Rev Neurosci. 2004;5:304–16. doi: 10.1038/nrn1397. [DOI] [PubMed] [Google Scholar]
- 38.Parekh R, Ascoli GA. Neuronal morphology goes digital: a research hub for cellular and system neuroscience. Neuron. 2013;77:1017–38. doi: 10.1016/j.neuron.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler DW, White CM, Rees CL, Komendantov AO, et al. Hippocampome.org: a knowledge base of neuron types in the rodent hippocampus. eLife. 2015;4:e09960. doi: 10.7554/eLife.09960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Handbook of brain microcircuits. New York: Oxford University Press; 2010. [Google Scholar]
- 41.Kubota Y. Untangling GABAergic wiring in the cortical microcircuit. Curr Opin Neurobiol. 2014;26:7–14. doi: 10.1016/j.conb.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Jonas E, Kording K. Automatic discovery of cell types and microcircuitry from neural connectomics. eLife. 2015;4:e04250. doi: 10.7554/eLife.04250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seung HS, Sümbül U. Neuronal cell types and connectivity: lessons from the retina. Neuron. 2014;83:1262–72. doi: 10.1016/j.neuron.2014.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bikoff JB, Gabitto MI, Rivard AF, Drobac E, et al. Spinal inhibitory interneuron diversity delineates variant motor microcircuits. Cell. 2016;165:207–19. doi: 10.1016/j.cell.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao T, Kusefoglu D, Hooks BM, Huber D, et al. Long-range neuronal circuits underlying the interaction between sensory and motor cortex. Neuron. 2011;72:111–23. doi: 10.1016/j.neuron.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Economo MN, Clack NG, Lavis LD, Gerfen CR, et al. A platform for brain-wide imaging and reconstruction of individual neurons. eLife. 2016;5:e10566. doi: 10.7554/eLife.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, et al. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–69. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng H, Hawrylycz M, Roskams J, Hill S, et al. BigNeuron: Large-Scale 3D neuron reconstruction from optical microscopy images. Neuron. 2015;87:252–6. doi: 10.1016/j.neuron.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kandel ER, Markram H, Matthews PM, Yuste R, et al. Neuroscience thinks big (and collaboratively) Nat Rev Neurosci. 2013;14:659–64. doi: 10.1038/nrn3578. [DOI] [PubMed] [Google Scholar]
- 50.Ascoli GA. Mobilizing the base of neuroscience data: the case of neuronal morphologies. Nat Rev Neurosci. 2006;7:318–24. doi: 10.1038/nrn1885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.