Abstract
Purpose:
Radiomics, which is the high-throughput extraction and analysis of quantitative image features, has been shown to have considerable potential to quantify the tumor phenotype. However, at present, a lack of software infrastructure has impeded the development of radiomics and its applications. Therefore, the authors developed the imaging biomarker explorer (ibex), an open infrastructure software platform that flexibly supports common radiomics workflow tasks such as multimodality image data import and review, development of feature extraction algorithms, model validation, and consistent data sharing among multiple institutions.
Methods:
The ibex software package was developed using the matlab and c/c++ programming languages. The software architecture deploys the modern model-view-controller, unit testing, and function handle programming concepts to isolate each quantitative imaging analysis task, to validate if their relevant data and algorithms are fit for use, and to plug in new modules. On one hand, ibex is self-contained and ready to use: it has implemented common data importers, common image filters, and common feature extraction algorithms. On the other hand, ibex provides an integrated development environment on top of matlab and c/c++, so users are not limited to its built-in functions. In the ibex developer studio, users can plug in, debug, and test new algorithms, extending ibex’s functionality. ibex also supports quality assurance for data and feature algorithms: image data, regions of interest, and feature algorithm-related data can be reviewed, validated, and/or modified. More importantly, two key elements in collaborative workflows, the consistency of data sharing and the reproducibility of calculation result, are embedded in the ibex workflow: image data, feature algorithms, and model validation including newly developed ones from different users can be easily and consistently shared so that results can be more easily reproduced between institutions.
Results:
Researchers with a variety of technical skill levels, including radiation oncologists, physicists, and computer scientists, have found the ibex software to be intuitive, powerful, and easy to use. ibex can be run at any computer with the windows operating system and 1GB RAM. The authors fully validated the implementation of all importers, preprocessing algorithms, and feature extraction algorithms. Windows version 1.0 beta of stand-alone ibex and ibex’s source code can be downloaded.
Conclusions:
The authors successfully implemented ibex, an open infrastructure software platform that streamlines common radiomics workflow tasks. Its transparency, flexibility, and portability can greatly accelerate the pace of radiomics research and pave the way toward successful clinical translation.
Keywords: radiomics, quantitative imaging analysis, infrastructure software, collaborative work
1. INTRODUCTION
Patients receive an ever increasing number of multimodality imaging procedures, such as computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET). The use and role of medical images has greatly expanded from primarily as a diagnostic tool to include a more central role in the context of individualized medicine.1–5 At present, the effective utilization of this large amount of medical imaging data is still challenging. Recently, there is an increased interest in the use of quantitative imaging methods to both improve tumor diagnosis and act as proxies of genetics and tumor response. With these improvements, the overall goal is to better inform and enhance clinical decision making.6–19 One important advancement in quantitative imaging analysis is the concept of “radiomics.” Radiomics is the high-throughput extraction and analysis of quantitative imaging features from medical images.20,21 Previous work has shown that radiomics can be used to create improved prediction algorithms for various clinically relevant metrics and endpoints.22–25
The lack of an open infrastructure software platform, however, has made previous radiomics research difficult to share and validate between institutions. Image features with the same name may be implemented differently by different groups. For example, the number of bins used for calculating histograms may vary, as may the use of image interpolation. These differences mean that independent validation of published work is difficult. As a result, the translation of radiomics research findings into improved clinical practices has been notably impeded. There is, therefore, a need for an open infrastructure software platform that is available for all researchers. Currently, no infrastructure software platforms are available to flexibly support common quantitative imaging analysis tasks such as multimodality image data import and review, development and calculation of feature extraction algorithms, model validation, and consistent data sharing among multiple institutions to assess reproducibility. A Computational Environment for Radiotherapy Research (cerr) publication26 states that reproducibility is a key element of the scientific method. This has been difficult to achieve with previous radiomics implementations. Some publicly available software programs do however exist for specific image feature analysis. For example, Chang-Gung Image Texture Analysis (cgtia)27 is an open-source software package for quantifying tumor heterogeneity with PET images. Also, originally designed for MRI texture analysis, MaZda (Ref. 28) is another software package with SDK support. Because of their intended use, both software packages are limited in their functionality or scope. For example, in neither is there a simple way for the user to implement new types of image features. Modifying feature extraction parameters, reviewing, validating intermediate data and results are also challenging using these two software packages. Straightforward multi-institutional reproduction of results is not included in their workflows. Within the field of radiation oncology, cerr demonstrates a successful example of open-source software used for collaborative work, and can be used as a template for the development of similar software geared toward radiomics.
We developed the imaging biomarker explorer (ibex) software package as an open infrastructure software platform to flexibly support common radiomics workflow tasks such as multimodality image data import and review, development of feature extraction algorithms, model validation, and consistent data sharing among multiple institutions. ibex is used for research only. On one hand, ibex is a self-contained and ready-to-use radiomics software, with preimplemented typical data importers, image filters, and feature extraction algorithms. On the other hand, the advanced research developers can extend ibex’s functionality. ibex provides an integrated development environment on top of the matlab (MathWorks, Natick, MA) and c/c++ programming languages. Users are not limited to ibex’s built-in functions: in the ibex developer studio, users can plug in, debug, and test new algorithms, extending the program’s functionality. ibex also supports quality assurance for data and feature extraction algorithms: image data, ROIs, and feature algorithm-related data can be reviewed, validated, and modified. Critically, image data, feature extraction algorithms, and model validation can be anonymized and be easily and consistently shared so that users from different institutions can reproduce the results of radiomics workflows. Finally, windows version 1.0 beta of stand-alone ibex without the requirement of matlab license can be freely downloaded at http://bit.ly/IBEX_MDAnderson. The source-code version of ibex can be downloaded at http://bit.ly/IBEXSrc_MDAnderson for free. Both versions of ibex can be shipped in compact disc form as well.
The alpha version of ibex was developed by Hunter et al.14 for in-house radiomics analysis. The current 1.0 beta version of ibex discussed in this paper was created from scratch in order to increase performance, improve ease of use, and extend functionality. Most importantly, compared to the prior version, the current ibex version has been engineered to have greatly increased modularity and robustness, allowing for it to be used collaboratively across multiple institutions.
2. DESCRIPTION OF ibex
2.A. Software architecture
ibex is written using matlab 2011a, 32-bit programming environment. To overcome poor memory management for large matrices, many three-dimensional (3D) image analysis modules are written in c/c++ and called by matlab via the matlab Executable (MEX) interface. ibex consists of a suite of component-based application and development tools for applying, sharing, and building reliable and reproducible quantitative image analysis algorithms.
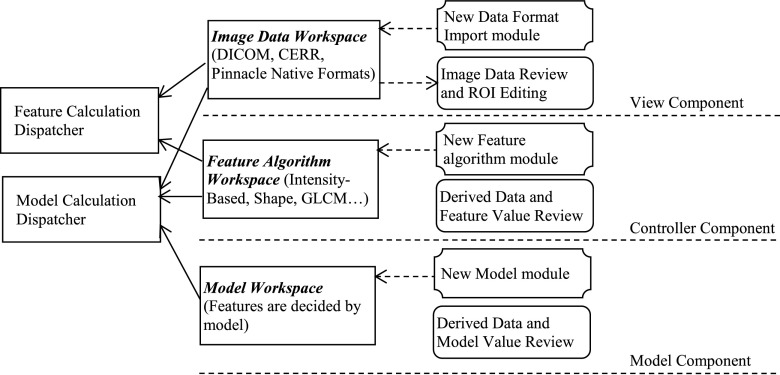
To achieve the goal of being an open infrastructure software platform, three modern programming concepts—model-view-controller (MVC),29 unit testing,30 and function handles—were deployed as shown in Fig. 1.
FIG. 1.
The ibex architecture. MVC, unit testing, and function handle programming concepts are deployed to isolate each task, test algorithms, plug in new algorithms, share data, and reproduce data easily and consistently.
The MVC concept is implemented to isolate each task. By implementing a unit testing concept, users are able to validate if their relevant data and algorithms are fit for use. Function handles are widely employed in the supplied developer studio of ibex where users can easily plug in their own algorithms into ibex. The MVC View Component represents the workspace of reviewing multimodality images with delineated structures (if available). The MVC Controller Component represents image preprocessing and feature extraction algorithms. The MVC Model Component represents the predictive model formula and parameters. Because of the unit testing implementation, users have the option of reviewing the corresponding result at each stage to check on the quality of the data and algorithms. Although ibex is self-contained and has standard algorithms and modules for a typical radiomics workflow, it is an open system, so additional algorithms and models can be easily added by defining them in library files in the ibex developer studio. Thanks to the MVC technique, a complete model can be exported or imported easily, including necessary data such as preprocessing algorithms, feature extraction algorithms, model formulas, and model parameters. This greatly helps maintain data consistency and result reproducibility when outside institutions attempt to validate feature extraction algorithms and response models.
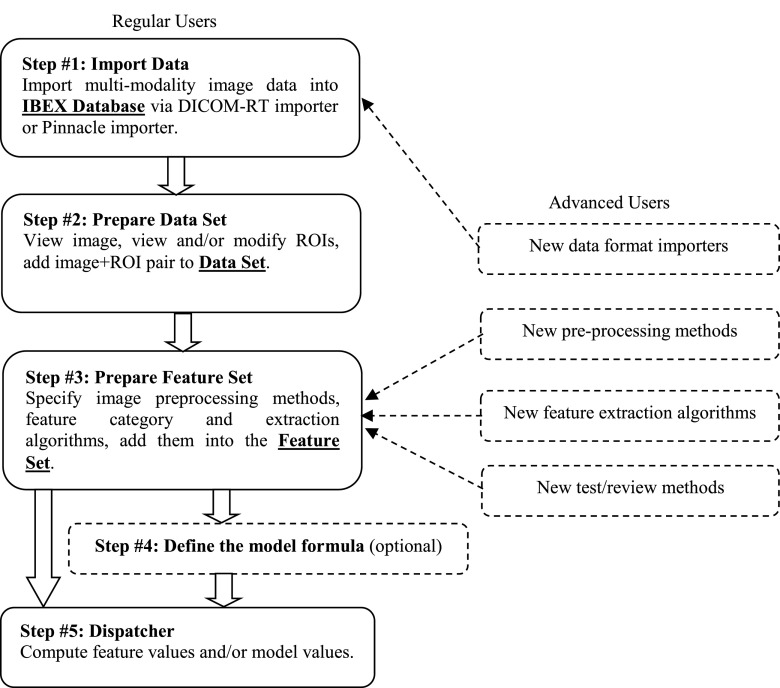
The ibex workflow is shown in Fig. 2. The ibex Database is a local store of patient images with associated data and ROIs. Regular users begin their workflow (Step #1) by importing patient data into the ibex Database using Digital Imaging and Communications in Medicine (DICOM)31 format data importer or pinnacle (Philips Radiation Oncology Systems, Fitchburg, WI) native format data importer. The Data Set is a local store of images that are subportions of images previously added to the ibex Database. To create subimages to populate the Data Set (Step #2), users open an image from the ibex Database; review the image and its associated ROIs; modify or create ROIs if desired; and specify which ROIs to apply to the image to obtain a subimage (multiple subimages can be generated from the same patient image by applying different ROIs). The Feature Set is a local store of the features that the user wishes to have extracted from a subimage. ibex organizes a variety of features into several feature categories based on feature’s nature. For example, all intensity histogram related features belong to the feature category “IntensityHistogram.” Feature category code computes the parent data (the parent data correspond to the histogram data for the feature category IntensityHistogram) and sends the parent data to feature extraction algorithm code to compute the value of each individual feature (features correspond to kurtosis, skewness, etc. for feature category IntensityHistogram). Users add features to the Feature Set (Step #3) by specifying image preprocessing algorithm(s), feature category, and feature extraction algorithm(s). Algorithm results can be reviewed via testing (optional). Users can then specify a model formula (Step #4) if desired. To complete the workflow (Step #5), users specify the Data Set and the Feature Set created in the previous steps and direct ibex to compute the feature values and/or model values. The steps above describe how to use ibex’s built-in functions. Advanced ibex users can use the ibex developer studio to plug in new data format importers, preprocessing methods, feature extraction algorithms, and test/review methods.
FIG. 2.
The ibex workflow. Regular users import data, prepare the data set and feature set, specify the model formula, and compute the feature value and/or model value. Advanced users can plug in new data format importers, preprocessing methods, feature algorithms, and test review methods using the ibex developer studio.
2.B. Image data workspace
The main purpose of the Image Data Workspace in ibex is to create subimages (image/ROI pairs) to add to a Data Set. Each item within a Data Set contains the basic information about an image and ROI pair, such as the imaging modality, medical record number (MRN), ROI statistics, voxel and image information, and item creation time. The Data Set also stores ROI contours, ROI binary masks, and image data in the ROI bounding box.
To prepare each Data Set, ibex includes the functionalities of importing patient data, reviewing images and ROIs, modifying or creating ROIs if necessary, appending Data Set items by adding image and ROI pairs. In compliance with the unit testing philosophy, ibex also supports reviewing and modifying Data Set items in the current workspace.
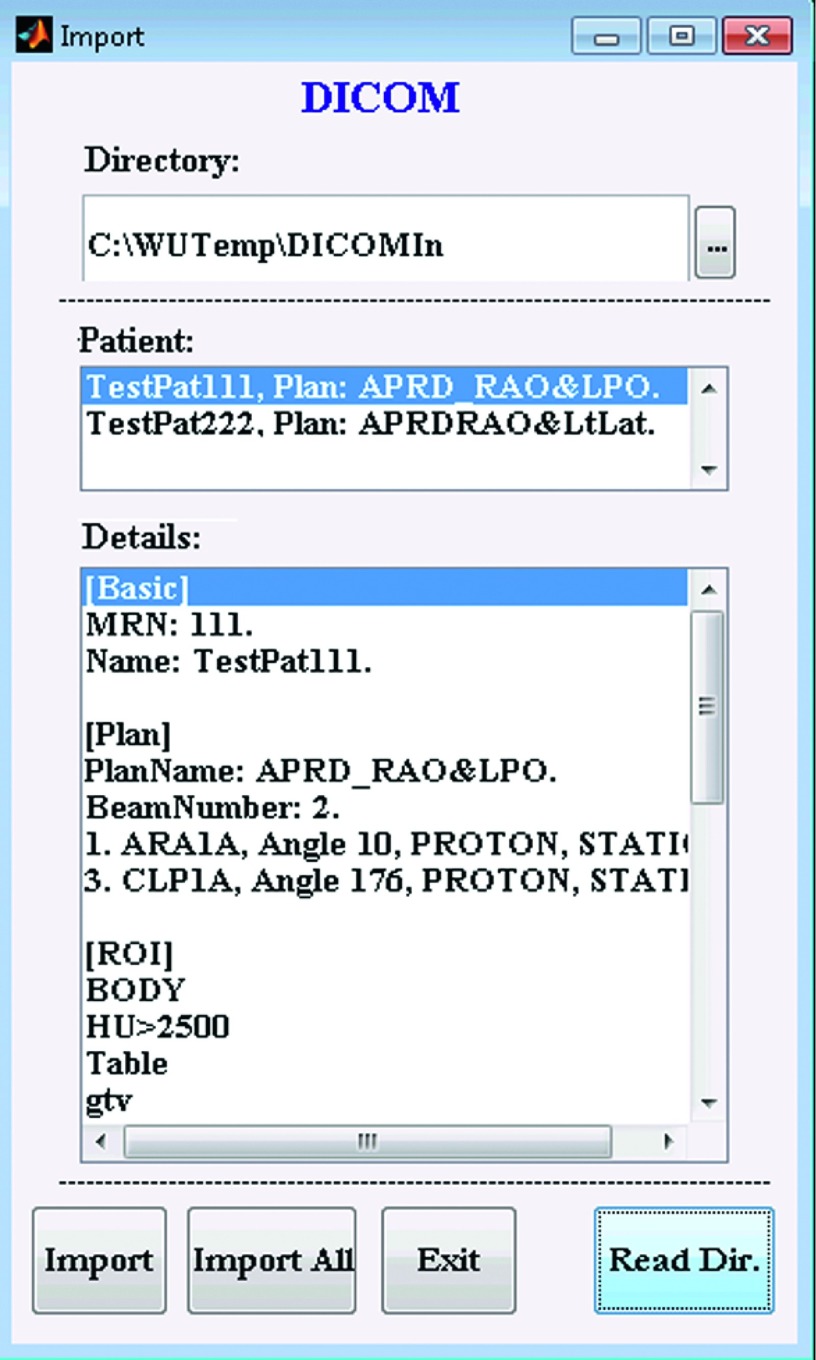
The current version of ibex provides DICOM data and pinnacle native data importers. pinnacle native data are the raw data used by the pinnacle treatment planning system (TPS). DICOM data may originate from numerous sources, including the majority of radiotherapy treatment planning systems, such as Eclipse (Varian Medical Systems, Palo Alto, CA) and pinnacle, many free image viewers/editors, such as 3D slicer (http://www.slicer.org), and the majority of commercial segmentation systems, such as MIMvista (MIM Software, Inc., Cleveland, OH), Velocity (Varian Medical Systems, Palo Alto, CA), and Mirada (Mirada Medical, Oxford, UK). If a computer running ibex has access to a pinnacle postgres database and data storage, ibex can be configured to retrieve data in the pinnacle native format directly from storage. For a DICOM importer, it first reads all the files in a configured DICOM input directory, then sorts and organizes the DICOM data according to the unique identifier (UID), and then lists all the patients available for import. As part of the unit testing implementation, the Details list box in the DICOM data importer describes the patient information and any related plan, ROI, and image information. Figure 3 shows an example of a DICOM data importer. When importing a patient’s DICOM data, ibex converts the data into the pinnacle native format. If DICOM imaging data were obtained using PET, ibex automatically computes the standardized uptake value from the DICOM PET raw uptake value if all the necessary radiopharmaceutical dose information is available. In addition to conversion, ibex also dumps all DICOM file information into the DICOMInfo folder to retain all the information from DICOM files.
FIG. 3.
Example of a DICOM data importer. The importer sorts and organizes DICOM data based on the relationship among MRNs, instance UIDs, study UIDs, series UIDs, and frame UIDs, and then lists all the available patients that could be imported. The Details list box describes the detailed patient information for verification.
ibex cannot connect to any PACS and RIS/HIS at present—the images will first need to be exported from the PACs and then imported into ibex. The current build-in ibex importers cannot import data from non-DICOM or non-pinnacle objects. However, users can plug-in their own customized data importers through ibex Developer Studio to import those non-DICOM or non-pinnacle data.
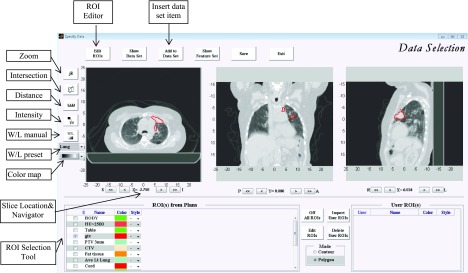
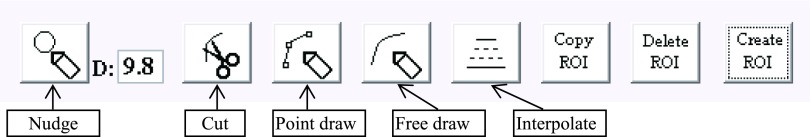
In the ibex Image Data Workspace, users insert Data Set items by specifying image and ROI pairs. Figure 4 is a screenshot of the ibex Image Data Workspace. This workspace supplies the multimodality image viewer for axial, coronal, and sagittal orientations and the ROI editor. Users can navigate to the different image slices, zoom images in and out, quickly go to the corresponding anatomy using the intersection tool, measure the distance, check image intensity values, manually set window/level, select the preset window and/or level setting, and select the preset color map. As part of unit testing implementation, ROIs can be overlaid on images in three orientations to verify contours. If a ROI must be modified, the user can employ the ROI editor to create a new ROI, copy the existing ROI, delete the ROI, nudge contours, delete contours, draw contours by clicking points, freely draw contours, or interpolate contours. Figure 5 is a screenshot of the ROI editor tools in ibex.
FIG. 4.
The ibex image data workspace. The main purpose of this workspace is to insert data set items by specifying image and ROI pairs. Image data can be viewed in axial, coronal, and sagittal orientations. ROIs can be overlaid on images and modified if necessary. Users can navigate to different image slices, zoom images in and out, quickly view the corresponding anatomy using the intersection tool, measure the distance, check the image intensity value, manually set window/level, select the preset window/level setting, and select the preset color map.
FIG. 5.
The ROI editor tools in ibex. Users can use the ROI editor to create new ROIs, copy existing ROIs, delete ROIs, nudge contours, delete contours, draw contours by clicking points, freely draw contours, and interpolate contours.
2.C. Feature algorithm workspace
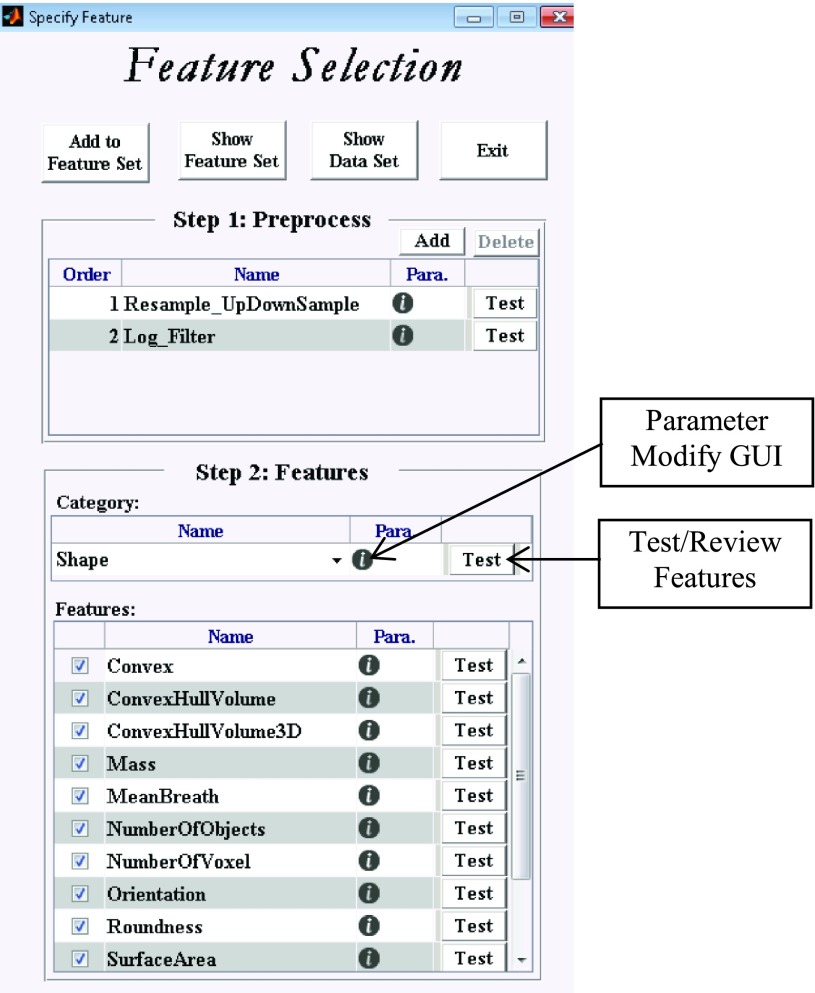
The main purpose of the Feature Algorithm Workspace in ibex is to prepare the Feature Set by specifying image preprocessing algorithms, feature category, and feature extraction algorithms. Each Feature Set item contains preprocessing methods and their parameters, the feature category and its parameters, feature extraction algorithms and their parameters, and the current feature set information (such as comments and its creation date). Figure 6 is a screenshot of the ibex Feature Algorithm Workspace.
FIG. 6.
The feature algorithm workspace in ibex. The main purpose of this workspace is to prepare the feature set by specifying the image preprocessing algorithms, feature category, and feature algorithms.
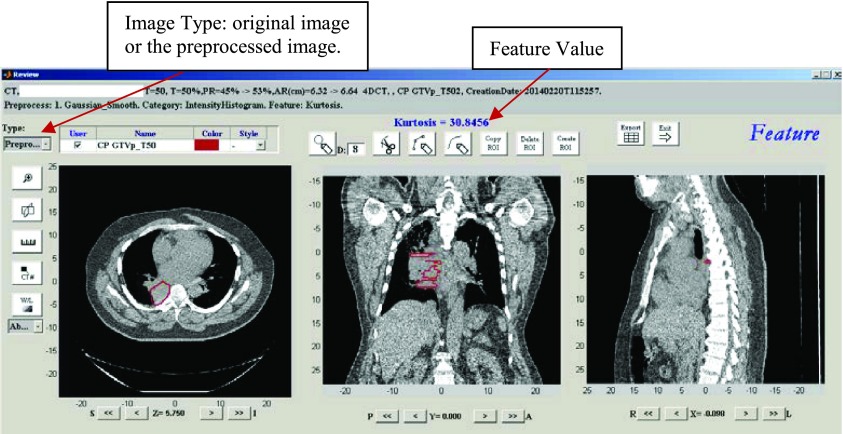
In the Feature Algorithm Workspace, users first specify the image preprocessing algorithms applied to the image. Users can apply multiple preprocessing algorithms in any order. Multiple preprocessing algorithms work in a pipeline style. Table I lists all of the preprocessing algorithms currently available in the current version of ibex. Users then specify the feature category and its feature extraction algorithms. The feature categories and feature extraction algorithms currently available in ibex are listed in Table II. As part of unit testing implementation, users can review and modify algorithm parameters using a parameter modification graphical user interface (GUI). Furthermore, by clicking the test button in the workspace, users can test the algorithm and review the intermediate data and feature calculation result. Figure 7 shows an example of testing the feature “Kurtosis” in the category IntensityHistogram. In the review window, users can check the original and preprocessed images, feature values, and contours.
TABLE I.
The image preprocessing algorithms available in ibex.
| Purpose | Preprocessing name | Comment | References |
|---|---|---|---|
| Image smoothing | Average_Smooth | ||
| EdgePreserve_Smooth3D | |||
| Gaussian_Smooth | 11–14 and 16 | ||
| Gaussian_Smooth3D | |||
| Median_Smooth | |||
| Wiener_Smooth | 11 | ||
| Image enhancement | AdaptHistEqualization_Enhance3D | ||
| HistEqualization_Enhance | |||
| Sharp_Enhance | |||
| Image deblur | Blind_Deblur | ||
| Gaussian_Deblur | |||
| Change enhancement | Laplacian_Filter | 11–14 and 16 | |
| Log_Filter | 11–14 and 16 | ||
| XEdge_Enhance | |||
| YEdge_Enhance | |||
| Resample | Resample_UpDownSample | 9 | |
| Resample_VoxelSize | 9 | ||
| Miscellaneous | Threshold_Image_Mask | 11 and 15 | |
| Threshold_Mask | 11 and 15 | ||
| BitDepthRescale_Range | Change dynamic range | 11 and 15 |
TABLE II.
The feature extraction algorithms available in ibex.
| Category | Feature name | Comment | References |
|---|---|---|---|
| Shape | Compactness1 | 33 | |
| Compactness2 | 33 | ||
| Max3DDiameter | 33 | ||
| SphericalDisproportion | 33 | ||
| Sphericity | 33 | ||
| Volume | 7 and 15 | ||
| SurfaceArea | 7 | ||
| SurfaceAreaDensity | |||
| Mass | Useful for CT only | 7 | |
| Convex | 7 | ||
| ConvexHullVolume | |||
| ConvexHullVolume3D | |||
| MeanBreadth | |||
| Orientation | 7 | ||
| Roundness | 7 | ||
| NumberOfObjects | |||
| NumberOfVoxel | 7 | ||
| VoxelSize | |||
| IntensityDirect | Energy | 33 | |
| RootMeanSquare | 33 | ||
| Variance | 33 | ||
| Kurtosis | 7, 9, 11, and 15 | ||
| Skewness | 7, 9, 11, and 15 | ||
| Range | 9 | ||
| Percentile | 9 and 15 | ||
| Quantile | 9 | ||
| InterQuartileRange | 9 | ||
| GlobalEntropy | 7, 9, 11, 12, and 15, | ||
| GlobalUniformity | 11–14 and 16 | ||
| GlobalMax | 9 and 15 | ||
| GlobalMin | 9 and 15 | ||
| GlobalMean | 7, 9, 11–13, and 15 | ||
| GlobalMedian | 9 and 15 | ||
| GlobalStd | 7, 9, 11–13, and 15, | ||
| MeanAbsoluteDeviation | 9 | ||
| MedianAbsoluteDeviation | |||
| LocalEntropy/Range/StdMax | |||
| LocalEntropy/Range/StdMin | |||
| LocalEntropy/Range/StdMean | |||
| LocalEntropy/Range/StdMedian | |||
| LocalEntropy/Range/StdStd | |||
| IntensityHistogram | Kurtosis | 7, 9, 11, and 15 | |
| Skewness | 7, 9, 11, and 15 | ||
| Range | |||
| Percentile | 9 and 15 | ||
| PercentileArea | |||
| Quantile | 9 | ||
| InterQuartileRange | 9 | ||
| AutoCorrelation | 32 and 33 | ||
| ClusterProminence | 32 and 33 | ||
| ClusterShade | 32 and 33 | ||
| CluseterTendency | 32 and 33 | ||
| DifferenceEntropy | 32 and 33 | ||
| Dissimilarity | 32 and 33 | ||
| Entropy | 32 and 33 | ||
| Homogeneity2 | 32 and 33 | ||
| InformationMeasureCorr1 | 32 and 33 | ||
| InformationMeasureCorr2 | 25:=GLCM is computed from all 2D image slices 3:=GLCM is computed from 3D image matrix | 32 and 33 | |
| GrayLevelCooccurenceMatrix25 GrayLevelCooccurenceMatrix3 | InverseDiffMomentNorm | 32 and 33 | |
| InverseDiffNorm | 32 and 33 | ||
| InverseVariance | 32 and 33 | ||
| MaxProbability | 32 and 33 | ||
| SumAverage | 32 and 33 | ||
| SumEntropy | 32 and 33 | ||
| SumVariance | 32 and 33 | ||
| Variance | 32 and 33 | ||
| Contrast | 7, 9, 11, 15, 28, 32, and 33 | ||
| Correlation | 7, 9, 11, 15, 28, 32, and 33 | ||
| Energy | 7, 9, 11, 15, 28, 32, and 33 | ||
| Homogeneity | 7, 9, 11, 15, 28, 32, and 33 | ||
| NeighborIntensityDifference25 NeighborIntensityDifference3 | Busyness | 25:= neighborhood intensity difference (NID) is computed from all 2D image slices 3:=NID is computed from 3D image matrix | 11, 23, 29, and 34 |
| Coarseness | 11, 23, 29, and 34 | ||
| Complexity | 23, 29, and 34 | ||
| Contrast | 11, 23, 29, and 34 | ||
| TextureStrength | 23, 29, and 34 | ||
| GrayLevelRunLengthMatrix25 | GrayLevelNonuniformity | 25:=run-length matrix (RLM) is computed from all 2D image slices 25:=RLM is computed from all 2D image slices | 7, 15, 30, 31, 35, and 36 |
| HighGrayLevelRunEmpha | 7, 30, 31, 35, and 36 | ||
| LongRunEmphasis | 7, 15, 30, 31, 35, and 36 | ||
| LongRunHighGrayLevelEmpha | 7, 30, 31, 35, and 36 | ||
| LongRunLowGrayLevelEmpha | 7, 30, 31, 35, and 36 | ||
| LowGrayLevelRunEmpha | 7, 30, 31, 35, and 36 | ||
| RunLengthNonuniformity | 7, 15, 30, 31, 35, and 36 | ||
| RunPercentage | 7, 15, 30, 31, 35, and 36 | ||
| ShortRunEmphasis | 7, 15, 30, 31, 35, and 36 | ||
| ShortRunHighGrayLevelEmpha | 7, 30, 31, 35, and 36 | ||
| ShortRunLowGrayLevelEmpha | 7, 30, 31, 35, and 36 | ||
| IntensityHistogramGaussFit | GaussAmplitude | ||
| GaussArea | |||
| GaussMean | |||
| GaussStd | |||
| NumberOfGauss |
FIG. 7.
A testing GUI in ibex. At each stage (import, preprocessing, and feature calculation), users have the option of reviewing the corresponding results and intermediate data.
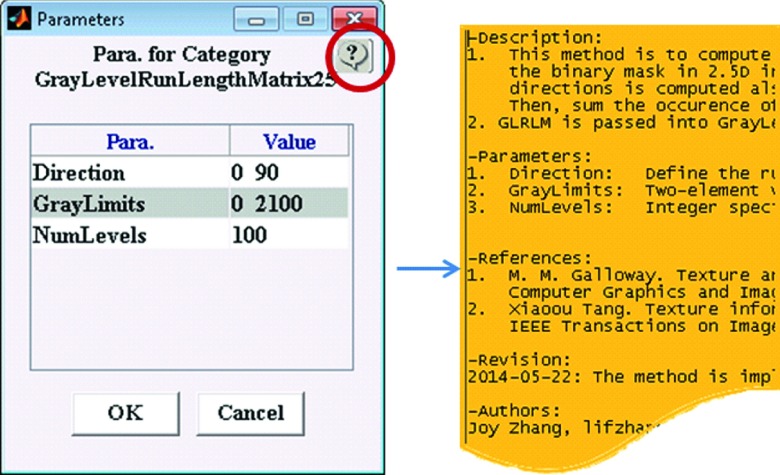
The Feature Algorithm Workspace is also self-documented. Each feature name is self-explanatory, indicating what feature it is. For example, the feature “ConvexHullVolume3D” in the category “Shape” means that the volume of the ROI convex hull is calculated according to the 3D connectivity of adjacent voxels in the binary masks. A detailed description of the algorithm and its parameters is easily accessed by clicking the help button on the parameter modification GUI as shown in Fig. 8.
FIG. 8.
Self-documented algorithm in ibex. The algorithm and feature name are self-explained. The description of the algorithm and its parameters can be easily accessed using the help button on the parameter modification GUI (circled in red).
The feature categories “GrayLevelCoocurrenceMatrix”32,33 and “NeighborhoodInstensityDifference”34 in the Feature Al gorithm Workspace are implemented in both two and a half dimension (2.5D) and 3D versions. This is done in consideration of the fact that most image data are in finer resolution in one orientation than in others. For example, the feature “GrayLevelCoocurrenceMatrix25” computes the co-occurrence of individual intensity pairs in 2D directions in slice by slice manner. Next, the gray-level co-occurrence matrix is the summation of the co-occurrence of individual intensity pairs in all 2D image slices. In contrast, the feature “GrayLevelCoocurrenceMatrix3” directly computes the GLCM as the co-occurrence of individual intensity pairs in the 3D directions. Similarly, in the feature “NeighborhoodInstensityDifference25,” the NID matrix is computed with the voxel’s neighborhood defined in 2D, whereas in the feature “NeighborhoodInstensityDifference3,” the neighborhood is defined in 3D. In the feature “GrayLevelRunLengthMatrix25,”35,36 the RLM is computed in 2D.
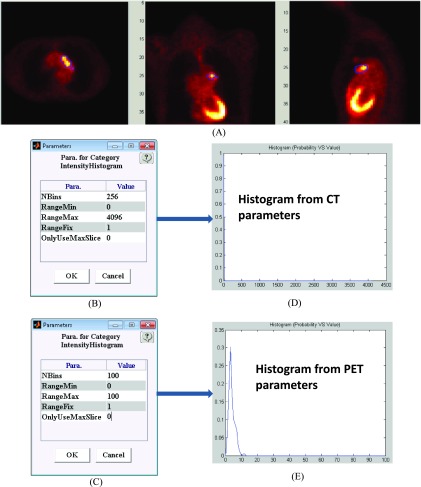
It is important to set the appropriate algorithm parameters for different modality images. The default parameters are set to be suitable for CT modality images. Figure 9 shows histograms created using the different parameters for one PET image set [Fig. 9(A)]. If the CT parameters in Fig. 9(B) are inappropriately applied to the PET data, then the histogram is erroneously compressed into one bin location [Fig. 9(D)]. However, by correctly selected PET-appropriate parameters, the appropriate histogram results are generated [Fig. 9(E)]. Note, consistent with the pinnacle treatment planning system, ibex uses a CT number where water is given a value of 1000.
FIG. 9.
The appropriate algorithm parameters for different modality images. (A) PET image. (B) CT-type parameters. (C) PET-type parameters. (D) Histogram from CT-type parameters that is meaningless and squeezed into one bin. (E) Histogram from PET-type parameters. The PET-type parameters zoom in on a CT-type histogram and can provide meaningful results for a PET image.
2.D. Model workspace
The main purpose of the Model Workspace in ibex is to prepare the model formula by specifying the expression, features, and parameters of each item. In the current version of ibex, the model formula (i.e., the formula that adds different features with different weights to give an outcome prediction) is simply defined in an ASCII text file to make it readable and easily shared. Defining the model formula naturally indicates which features are used in the model. Modeling is the informatics analysis of features. Models can be generated for different applications, such as tumor diagnosis, tumor staging, gene prediction, and outcome prediction. Developing a good model and selecting appropriate model features are beyond the scope of this report. Examples of model development have been described by several authors.7,11 That is, the predictive models must be developed outside of ibex. The image features (including all necessary parameters) and model coefficients can then be added to ibex for the purpose of internal or independent validation, etc.
2.E. Computation dispatcher
The Computation dispatcher (see Fig. 3, Step #5) in ibex is used to compute the feature or model values. Users first specify the data set, feature set, and/or model. The dispatcher engine then computes the feature and/or model value. Last, the dispatcher writes the result along with the information of data set, feature set, and model into one Excel spreadsheet (Microsoft Corporation, Redmond, WA). Users can then use this common format to import the result into the statistical program (SPSS, R, SAS, etc.) that they prefer. To comply with unit testing implementation, the information in the Excel file contains the data set item description, features’ names and parameters, and model formula so that users can reproduce the results and determine what was used to generate the results. Thanks to MVC concept implementation, the data sets, feature sets, and models are relatively independent of one another. Thus, one data set can be applied to different feature sets and models, one feature set can be applied to different data sets and models, and one model can be applied to different feature and data sets. This independence in the ibex workspaces enables ibex to serve as the infrastructure platform for testing and developing feature algorithms and models for quantitative imaging analysis.
2.F. Developer studio/extensibility
The ibex developer studio enables users to extend the functionality of the software. In the developer studio, advanced users can plug in new data importers for any data format, new preprocessing algorithms, new feature algorithms, and new test/review functions. The ibex plug-in feature is based heavily on the matlab function handle technique. The ibex developer studio works in the same way as Visual Studio (Microsoft Corporation). Depending on the type of plug-in, the developer studio generates the skeleton code with simple functions and puts this code under the designated directory for the ibex platform to recognize. The skeleton code itself is ready to use. Advanced users can first run the skeleton code to get an idea of plug in input arguments, and then modify and enrich the skeleton code to meet their purposes.
2.G. Reproducibility
Because of the MVC architecture, interinstitutional comparison and reproducibility can be easily provided by ibex. Data and feature sets are stored in the individual matlab MAT files, and models are stored in readable ASCII files. Data sets, feature sets, and models are all self-contained, including all information necessary for the ibex computation dispatcher to calculate the result of feature and/or model. The ibex users anonymize and export their own data set, feature set, and/or model files. An ibex user from a different institution can then import these files into the ibex database and then compute the result of feature and/or model. The result can be reproduced, as all the data are shared consistently among institutions. The second user can double-check the first user’s algorithms by examining the parameters and reviewing the intermediate data. Data anonymization can be done in several scenarios: Users have the option to anonymize their data when data are imported into ibex; users can anonymize patients in the ibex database; data sets created in ibex can be anonymized; in the data workspace, the user has a tool to anonymize the ROI data.
2.H. Quality assurance/reliability
Thanks to the implementation of unit testing philosophy, ibex users can review the relevant data at each stage involved in the feature calculation. Specifically, ibex provides the GUIs for users to review image data, review and modify ROIs and algorithm parameters, read algorithm descriptions, test algorithms, review intermediate and final results for algorithms, and review model formulas. ibex itself supports reviewing 3D and 2D matrices, single values, gray-level co-occurrence matrices, curves, meshes, and layers along with the image display. Furthermore, users can even plug in their own review callback functions to customize the review requirements. All of these capabilities enable users to perform quality assurance for their image data, ibex’s built-in algorithms, users’ plug-ins, and models.
2.I. Testing
The implementation of data importers, preprocessing algorithms, and feature extraction algorithms in ibex was validated using commercial and free software. Specifically, the ibex DICOM importer was compared with DICOM importers in the eclipse and pinnacle TPSs. Also, the ibex pinnacle importer was compared with the pinnacle TPS database. ibex preprocessing algorithms were validated against matlab’s built-in functions and cerr implementation qualitatively by visually reviewing the preprocessed images. Software developers and physics users visually reviewed the preprocessed images for each modality (5+ images for CT, MRI, and PET modalities). This qualitative comparison was subjective, with the users visually searching the differences in the preprocessed images created by ibex, matlab, and cerr. For the purpose of the quantitative validation, we created four digital sphere phantoms with one known volume size (volume = 65.3 cm3), one known mean intensity value (mean = 1025), and four different intensity standard deviations (SD = 25, 47, 50, and 75). By comparing with the known value, the average volume differences were 0.15, 0.03, 0.69, and 1.28 cm3 from ibex, pinnacle TPS, eclipse TPS, and cgita; the average intensity mean difference was 0.20, 0.16, and 0.21 from ibex, pinnacle TPS, and cgita, respectively; the average intensity standard deviation difference was 0.23, 0.22, and 0.24 from ibex, pinnacle TPS, and cgita, respectively. Feature values of Kurtosis and skewness from ibex on these four digital phantoms were compared with those from cgita. The average Kurtosis difference and skewness difference are 0.02 and 0.00. We qualitatively validated feature algorithm implementation for categories GLCM, NID, and IntensityHistogramCurveFit by validating the intermediated data such as GLCM matrix, NID matrix, and the fitted Gaussian curves. It is impossible to quantitatively validate them against cgita because cgita implementation is mainly for PET images and its ROI boundary handling is different from ibex, pinnacle TPS, and eclipse.
At the time of this writing, ibex has been used for two substantial projects11,15 and is currently being used by around 35 researchers from different countries with CT (including contrast-enhanced CT, noncontrast-enhanced CT, cone beam CT, and 4D CT), PET, and MRI images. ROIs have been successfully imported from commercial and research software such as eclipse, pinnacle, MIMvista, velocity, slicer, and mirada. Researchers were able to use ROI editors to create new ROIs and modify the existing ROIs in ibex ROI editor. Several researchers reported that ibex is intuitive, powerful, and easy to use.
2.J. Distribution
Windows version 1.0β of ibex is freely distributed. About 35 researchers around the world are using it and have contributed to the development of new preprocessing and feature extraction algorithms and review callback functions. The stand-alone version of ibex without the requirement of a matlab license can be downloaded at http://bit.ly/IBEX_MDAnderson. The source-code ibex version requires installation of matlab and can be downloaded at http://bit.ly/IBEXSrc_MDAnderson for free. Both versions of ibex can be shipped via compact disc. ibex-related documents can be found at http://bit.ly/IBEX_Documentation.
An ibex discussion group is available for users to post and answer any ibex-related questions. Users can review the discussion threads at https://groups.google.com/forum/#!forum/IBEX_users. Individuals can subscribe to the group by e-mailing ibex_users+subscribe@googlegroups.com to obtain posting rights.
3. DISCUSSION
ibex implemented the underlying modules and framework for radiomics and quantitative imaging analysis. ibex serves as an open infrastructure software platform to accelerate collaborative work. Using ibex, researchers can focus on their application and development of radiomics workflows without worrying about data consistency and review, algorithm reliability, and result reproducibility. The ibex plug-in mechanism facilitates contribution of creative algorithms and implementation of customized requirements by users around the world.
Model development in quantitative imaging analysis is a major topic involving how to analyze and/or classify features. Many approaches to model development can be used, such as regression, principal component analysis, artificial neural networks, Bayesian networks, and support vector machines. Model development techniques can differ greatly depending on individual model applications. At this point, establishing a universal workflow for model development is difficult, so the current version of ibex does not provide a tool for model development.
The ibex developer studio is only available in the source-code version. This is because the stand-alone matlab program does not run unencrypted M-files. In other words, matlab does not allow mixing an encrypted M-file from the stand-alone version of ibex with an unencrypted M-file from the ibex developer studio. Developing the source code within the matlab environment and on the ibex platform is always a good practice, as it enables advanced users to use the debugging and testing functionalities of both.
The ibex database has a file-based structure and is organized in the same way in which pinnacle native data storage is organized. Also, the pinnacle native data format is used as the ibex data format. As a result, ibex data can be imported directly into the pinnacle system. The pinnacle data format basically has two parts: (1) the readable and modifiable ASCII header file describing the data and (2) the corresponding raw binary data. pinnacle format data can be read quickly and efficiently, as a series of DICOM images is stored in one large portion of binary data. Users can use any text editor to open the ASCII pinnacle header file to explore the data and modify the information as needed.
Although version 1.0β of ibex has a radiomics infrastructure platform, we have been diligently working on the next version of ibex, mainly focusing on improving the convenience and robustness of multi-institution, multidisciplinary collaborative research. Our near-term development goals include adding functions to do the following:
-
•
Export intermediate data from ibex for users to be able to check and use it for other research purposes.
-
•
Archive completed projects including all necessary information so that project-related data can be restored or shared if reproduction or repeating of any analyses is needed.
-
•
Add additional data importers (e.g., cerr format) as identified by the user network. Although use of DICOM is fairly standard for importing images and ROIs, many other formats can be used for images and/or delineated structures.
-
•
Develop an extension for 3D slicer to bridge 3D slicer and ibex.
4. SUMMARY
We successfully implemented ibex, an open infrastructure software platform that streamlines common radiomics workflow tasks. Its transparency, flexibility, and portability can greatly accelerate the pace of radiomics and its collaborative research and pave the way toward successful clinical translation. ibex flexibly supports common radiomics workflow tasks such as multimodality imaging data import and review, development of feature extraction algorithms, model validation, and consistent data sharing among multiple institutions. On one hand, ibex is self-contained and ready to use, with preimplemented typical data importers, image filters, and feature extraction algorithms. On the other hand, users can extend ibex’s functionality by plugging in new algorithms. ibex also supports quality assurance for data and feature extraction algorithms. Image data, feature algorithms, and model formulas can be easily and consistently shared using ibex for reproducibility purposes.
ACKNOWLEDGMENT
Conflicts of interest and sources of funding: Supported in part by a grant from the NCI (R03CA178495-01).
REFERENCES
- 1.Chen H. Y., Yu S. L., Chen C. H., Chang G. C., Chen C. Y., Yuan A., Cheng C. L., Wang C. H., Terng H. J., Kao S. F., Chan W. K., Li H. N., Liu C. C., Singh S., Chen W. J., Chen J. J., and Yang P. C., “A five-gene signature and clinical outcome in non-small-cell lung cancer,” N. Engl. J. Med. 356, 11–20 (2007). 10.1056/NEJMoa060096 [DOI] [PubMed] [Google Scholar]
- 2.Eisenhauer E. A., Therasse P., Bogaerts J., Schwartz L. H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., Rubinstein L., Shankar L., Dodd L., Kaplan R., Lacombe D., and Verweij J., “New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1),” Eur. J. Cancer 45, 228–247 (2009). 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 3.Fass L., “Imaging and cancer: A review,” Mol. Oncol. 2, 115–152 (2008). 10.1016/j.molonc.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machtay M., Duan F., Siegel B. A., Snyder B. S., Gorelick J. J., Reddin J. S., Munden R., Johnson D. W., Wilf L. H., DeNittis A., Sherwin N., Cho K. H., Kim S. K., Videtic G., Neumann D. R., Komaki R., Macapinlac H., Bradley J. D., and Alavi A., “Prediction of survival by [18F]fluorodeoxyglucose positron emission tomography in patients with locally advanced non-small-cell lung cancer undergoing definitive chemoradiation therapy: Results of the ACRIN 6668/RTOG 0235 trial,” J. Clin. Oncol. 31, 3823–3830 (2013). 10.1200/JCO.2012.47.5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raz D. J., Ray M. R., Kim J. Y., He B., Taron M., Skrzypski M., Segal M., Gandara D. R., Rosell R., and Jablons D. M., “A multigene assay is prognostic of survival in patients with early-stage lung adenocarcinoma,” Clin. Cancer Res. 14, 5565–5570 (2008). 10.1158/1078-0432.CCR-08-0544 [DOI] [PubMed] [Google Scholar]
- 6.Al-Kadi O. S. and Watson D., “Texture analysis of aggressive and nonaggressive lung tumor CE CT images,” IEEE Trans. Biomed. Eng. 55, 1822–1830 (2008). 10.1109/TBME.2008.919735 [DOI] [PubMed] [Google Scholar]
- 7.Basu S., “Developing predictive models for lung tumor analysis,” M.S. thesis, University of South Florida, 2012. [Google Scholar]
- 8.Cunliffe A. R., Al-Hallaq H. A., Labby Z. E., Pelizzari C. A., Straus C., Sensakovic W. F., Ludwig M., and Armato S. G., “Lung texture in serial thoracic CT scans: Assessment of change introduced by image registration,” Med. Phys. 39, 4679–4690 (2012). 10.1118/1.4730505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunliffe A. R., Armato III S. G., Fei X. M., Tuohy R. E., and Al-Hallaq H. A., “Lung texture in serial thoracic CT scans: Registration-based methods to compare anatomically matched regions,” Med. Phys. 40, 061906(9pp.) (2013). 10.1118/1.4805110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunliffe A. R., Armato S. G., Straus C., Malik R., and Al-Hallaq H. A., “Lung texture in serial thoracic CT scans: Correlation with radiologist-defined severity of acute changes following radiation therapy,” Phys. Med. Biol. 59, 5387–5398 (2014). 10.1088/0031-9155/59/18/5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganeshan B., Abaleke S., Young R. C., Chatwin C. R., and Miles K. A., “Texture analysis of non-small cell lung cancer on unenhanced computed tomography: Initial evidence for a relationship with tumour glucose metabolism and stage,” Cancer Imaging 10, 137–143 (2010). 10.1102/1470-7330.2010.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganeshan B., Goh V., Mandeville H. C., Ng Q. S., Hoskin P. J., and Miles K. A., “Non-small cell lung cancer: Histopathologic correlates for texture parameters at CT,” Radiology 266, 326–336 (2013). 10.1148/radiol.12112428 [DOI] [PubMed] [Google Scholar]
- 13.Ganeshan B., Panayiotou E., Burnand K., Dizdarevic S., and Miles K., “Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: A potential marker of survival,” Eur. Radiol. 22, 796–802 (2012). 10.1007/s00330-011-2319-8 [DOI] [PubMed] [Google Scholar]
- 14.Hunter L. A., Krafft S., Stingo F., Choi H., Martel M. K., Kry S. F., and Court L. E., “High quality machine-robust image features: Identification in nonsmall cell lung cancer computed tomography images,” Med. Phys. 40, 121916(12pp.) (2013). 10.1118/1.4829514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravanelli M., Farina D., Morassi M., Roca E., Cavalleri G., Tassi G., and Maroldi R., “Texture analysis of advanced non-small cell lung cancer (NSCLC) on contrast-enhanced computed tomography: Prediction of the response to the first-line chemotherapy,” Eur. Radiol. 23, 3450–3455 (2013). 10.1007/s00330-013-2965-0 [DOI] [PubMed] [Google Scholar]
- 16.Segal E., Sirlin C. B., Ooi C., Adler A. S., Gollub J., Chen X., Chan B. K., Matcuk G. R., Barry C. T., Chang H. Y., and Kuo M. D., “Decoding global gene expression programs in liver cancer by noninvasive imaging,” Nat. Biotechnol. 25, 675–680 (2007). 10.1038/nbt1306 [DOI] [PubMed] [Google Scholar]
- 17.Tan H., Liu T., Wu Y., Thacker J., Shenkar R., Mikati A. G., Shi C., Dykstra C., Wang Y., Prasad P. V., Edelman R. R., and Awad I. A., “Evaluation of iron content in human cerebral cavernous malformation using quantitative susceptibility mapping,” Invest. Radiol. 49, 498–504 (2014). 10.1097/RLI.0000000000000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Win T., Miles K. A., Janes S. M., Ganeshan B., Shastry M., Endozo R., Meagher M., Shortman R. I., Wan S., Kayani I., Ell P. J., and Groves A. M., “Tumor heterogeneity and permeability as measured on the CT component of PET/CT predict survival in patients with non-small cell lung cancer,” Clin. Cancer Res. 19, 3591–3599 (2013). 10.1158/1078-0432.CCR-12-1307 [DOI] [PubMed] [Google Scholar]
- 19.Fried D. V., Tucker S. L., Zhou S., Liao Z., Mawlawi O., Ibbott G., and Court L. E., “Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung Cancer,” Int. J. Radiat. Oncol., Biol., Phys. 90, 834–842 (2014). 10.1016/j.ijrobp.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambin P., Rios-Velazquez E., Leijenaar R., Carvalho S., van Stiphout R. G., Granton P., Zegers C. M., Gillies R., Boellard R., Dekker A., and Aerts H. J., “Radiomics: Extracting more information from medical images using advanced feature analysis,” Eur. J. Cancer 48, 441–446 (2012). 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar V., Gu Y., Basu S., Berglund A., Eschrich S. A., Schabath M. B., Forster K., Aerts H. J., Dekker A., Fenstermacher D., Goldgof D. B., Hall L. O., Lambin P., Balagurunathan Y., Gatenby R. A., and Gillies R. J., “Radiomics: The process and the challenges,” Magn. Reson. Imaging 30, 1234–1248 (2012). 10.1016/j.mri.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson A., O’Connor J. P., Parker G. J., and Jayson G. C., “Imaging tumor vascular heterogeneity and angiogenesis using dynamic contrast-enhanced magnetic resonance imaging,” Clin. Cancer Res. 13, 3449–3459 (2007). 10.1158/1078-0432.CCR-07-0238 [DOI] [PubMed] [Google Scholar]
- 23.Rose C. J., Mills S., O’Connor J. P., Buonaccorsi G. A., Roberts C., Watson Y., Whitcher B., Jayson G., Jackson A., and Parker G. J., “Quantifying heterogeneity in dynamic contrast-enhanced MRI parameter maps,” International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) (Springer, Verlag/Berlin/Heidelberg (NY), 2007), Vol. 10, pp. 376–384. [DOI] [PubMed] [Google Scholar]
- 24.Gibbs P. and Turnbull L. W., “Textural analysis of contrast-enhanced MR images of the breast,” Magn. Reson. Med. 50, 92–98 (2003). 10.1002/mrm.10496 [DOI] [PubMed] [Google Scholar]
- 25.Canuto H. C., McLachlan C., Kettunen M. I., Velic M., Krishnan A. S., Neves A. A., Backer M. de, Hu D. E., Hobson M. P., and Brindle K. M., “Characterization of image heterogeneity using 2D Minkowski functionals increases the sensitivity of detection of a targeted MRI contrast agent,” Magn. Reson. Med. 61, 1218–1224 (2009). 10.1002/mrm.21946 [DOI] [PubMed] [Google Scholar]
- 26.Deasy J. O., Blanco A. I., and Clark V. H., “CERR: A computational environment for radiotherapy research,” Med. Phys. 30, 979–985 (2003). 10.1118/1.1568978 [DOI] [PubMed] [Google Scholar]
- 27.Fang Y. H., Lin C. Y., Shih M. J., Wang H. M., Ho T. Y., Liao C. T., and Yen T. C., “Development and evaluation of an open-source software package ‘CGITA’ for quantifying tumor heterogeneity with molecular images,” BioMed Res. Int. 2014, 248505 (2014). 10.1155/2014/248505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szczypinski P. M., Strzelecki M., Materka A., and Klepaczko A., “MaZda–A software package for image texture analysis,” Comput. Methods Programs Biomed. 94, 66–76 (2009). 10.1016/j.cmpb.2008.08.005 [DOI] [PubMed] [Google Scholar]
- 29.Krasner G. E. and Pope S. T., “A cookbook for using the model-view controller user interface paradigm in Smalltalk-80,” J. Object Oriented Program. 1, 26–49 (1988). [Google Scholar]
- 30.Xie T., Taneja K., Kale S., and Marinov D., “Towards a framework for differential unit testing of object-oriented programs,” in Proceedings of the Second International Workshop on Automation of Software Test (IEEE Computer Society, Washington, DC, 2007), p. 5. [Google Scholar]
- 31.Mildenberger P., Eichelberg M., and Martin E., “Introduction to the DICOM standard,” Eur. Radiol. 12, 920–927 (2002). 10.1007/s003300101100 [DOI] [PubMed] [Google Scholar]
- 32.Haralick R. M., Shanmuga K., and Dinstein I., “Textural features for image classification,” IEEE Trans. Syst., Man, Cybern. 3, 610–621 (1973). 10.1109/tsmc.1973.4309314 [DOI] [Google Scholar]
- 33.Aerts H. J., Velazquez E. R., Leijenaar R. T., Parmar C., Grossmann P., Cavalho S., Bussink J., Monshouwer R., Haibe-Kains B., Rietveld D., Hoebers F., Rietbergen M. M., Leemans C. R., Dekker A., Quackenbush J., Gillies R. J., and Lambin P., “Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach,” Nat. Commun. 5, 4006 (2014). 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amadasun M. and King R., “Textural features corresponding to textural properties,” IEEE Trans. Syst., Man, Cybern. 19, 1264–1274 (1989). 10.1109/21.44046 [DOI] [Google Scholar]
- 35.Tang X. O., “Texture information in run-length matrices,” IEEE Trans. Image Process. 7, 1602–1609 (1998). 10.1109/83.725367 [DOI] [PubMed] [Google Scholar]
- 36.Galloway M. M., “Texture analysis using gray level run lengths,” Comput. Graphics Image Process. 4, 172–179 (1975). 10.1016/S0146-664X(75)80008-6 [DOI] [Google Scholar]