Abstract
Autosomal dominant mutations in BTB and Kelch domain containing 13 protein (KBTBD13) are associated with a new type of Nemaline Myopathy (NEM). NEM is a genetically heterogeneous group of muscle disorders. Mutations causing phenotypically distinct NEM variants have previously been identified in components of muscle thin filament. KBTBD13 is a muscle specific protein composed of an N terminal BTB domain and a C terminal Kelch-repeat domain. The function of this newly identified protein in muscle remained unknown. In this study, we show that KBTBD13 interacts with Cullin 3 (Cul3) and the BTB domain mediates this interaction. Using ubiquitination assays, we determined that KBTBD13 participates in the formation of a Cul3 based RING ubiquitin ligase (Cul3-RL) capable of ubiquitin conjugation. Confocal microscopy of transiently expressed KBTBD13 revealed its co-localization with ubiquitin. Taken together, our results demonstrate that KBTBD13 is a putative substrate adaptor for Cul3-RL that functions as a muscle specific ubiquitin ligase, and thereby implicate the ubiquitin proteasome pathway in the pathogenesis of KBTBD13-associated NEM.
Keywords: Nemaline Myopathy, Muscle, BTB and Kelch domain containing 13 protein, Cullin 3, Ubiquitin ligase
1. Introduction
We recently identified dominant mutations in an unknown gene, eventually designated as Kelch-repeat and BTB (POZ) domain containing 13 gene (KBTBD13) that are associated with a new form of Nemaline Myopathy type 6 (NEM6) [1,2]. NEM is a heterogeneous group of inherited myopathies that mostly affect infants and children. NEM is characterized by slowly progressive muscle weakness and the presence of thread- or rod-like so called nemaline bodies in affected muscle [2–4]. Mutations causing phenotypically distinct NEM variants have been identified in genes encoding components of skeletal muscle sarcomeric thin filaments or regulators of their assembly and as such NEM is considered a disease of muscle thin filament [3,4].
KBTBD13 encodes the KBTBD13 protein which is highly expressed in striated muscle [1]; however the role and function of this protein are completely unknown. KBTBD13 contains an N-terminal BTB/POZ domain, named after the Drosophila melanogaster transcription factors, Bric-a-brac, Tramtrack, and Broad-complex [5,6]. The BTB domain is a conserved protein–protein interacting motif that is involved in a variety of cellular functions. Proteins containing the BTB domain often serve as substrate specific adaptors for Cullin 3 (Cul3) based RING ubiquitin ligases (Cul3-RLs) that target proteins for proteasomal degradation [7–9]. The BTB domain is structurally similar to the Cul1 binding domain of Skp1, and forms analogous interactions with Cul3. Skp1 is an integral component of Skp1/Cul1/F-box (SCF) ubiquitin ligases that bridge Cul1 to an F-box substrate adaptor [10]. In contrast to Skp1, BTB proteins contain additional protein–protein interacting domains such as Kelch or MATH [5,6,11] that can bind directly to substrates without the need for an intervening F-box protein.
KBTBD13 contains a C-terminal Kelch domain consisting of five Kelch repeats. The Kelch domain was originally discovered in galactose oxidase in D. melanogaster and is thought to mediate protein–protein interactions [11]. Kelch domain containing proteins are implicated in a broad variety of biological processes, including cytoskeleton modulation, regulation of gene transcription, ubiquitination, cell migration and myofibril assembly [11,12].
The presence of BTB and Kelch domains in KBTBD13 led us to propose that KBTBD13 interacts with Cul3, a core component of Cul3-RL. In this study, we present the first biochemical studies of KBTBD13 protein, its interaction with other proteins and localization in mammalian cells. We show that KBTBD13 directly interacts with Cul3 and forms a functional Cul3-RL complex. Our results suggest that KBTBD13 is a putative adaptor for Cul3-RL and raise the possible involvement of the ubiquitin–proteasome machinery in the pathogenesis of NEM.
2. Materials and methods
2.1. Construction of expression plasmids
The expression construct for C-terminal Myc-Flag tagged human KBTBD13 in pCMV6-vector (KBTBD13-Myc-Flag) has previously been described [1]. A plasmid expressing full length human Cul3 was purchased from OriGene. Vector expressing Myc tagged Ringbox 1 protein (Myc-Rbx1) was obtained from Conaway [13]. Construct, pCMV-(HA-Ub)8, expressing hemagglutinin (HA) tagged octameric ubiquitin was a kind gift from Bohmann [14]. The integrity of all plasmids was confirmed by DNA sequencing.
The N-terminal domain (1–383) of Cul3 followed by a hexa -histidine tag was cloned into the NotI and SbfI sites of pMAL-c5X (New England Biolabs) to generate MBP-Cul3(1–383)-(His)6. The BTB domain (6–132) of KBTBD13 followed by a hexa-histidine tag was cloned into the NcoI and NdeI sites of pMAL-c5X to generate MBP-KBTBD13(6–132)-(His)6. The co-expression construct was generated by inserting downstream from the multiple cloning site in pMAL-c5X (MCS-1), a linker, tac promoter, and second multiple cloning site (MCS-2). KBTBD13(6–132) was cloned into NcoI and NdeI sites of MCS-1 and Cul3(1–383)-(His)6 was cloned into the NotI and SbfI sites of MCS-2.
2.2. Expression and purification of recombinant proteins
pMAL-c5X plasmids encoding KBTBD13 and Cul3 proteins were transformed into the NEB Express strain (New England Biolabs) or the BL21 strain (Invitrogen) of Escherichia coli and grown in LB medium. Protein expression was induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside. Bacteria were lysed in buffer containing 20 mM Tris pH 8.0, 500 mM NaCl, 0.05% Tween-20, 10 mM 2-mercaptoethanol, 10% glycerol, and supplemented with Complete Protease Inhibitor Cocktail (Roche Applied Sciences). Fusion proteins were purified using nickel agarose (Qiagen) and amylose resin (New England Biolabs). Cleavage of the MBP tag was performed by overnight digestion at 16 °C with factor Xa. After passage through the amylose resin, eluted proteins were concentrated and loaded onto a Superose 6 size exclusion column (GE Health-care) equilibrated with the lysis buffer. The purified proteins were subjected to SDS–PAGE and Colloidal Blue staining (Invitrogen). Protein samples were used immediately for analysis by sedimentation velocity analytical ultracentrifugation or stored at −80 °C.
The co-expression plasmid encoding MBP-KBTBD13(6–132) and Cul3(1–383)-His6 was used to transform the BL21 strain of E. coli. Bacteria were cultured at 37 °C in 2 × YT medium until the OD reached 0.4. The temperature was decreased to 23 °C and protein expression induced for 4 h by addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside. The bacterial pellet was lysed in a buffer containing 20 mM HEPES pH 8.0, 20 mM NaCl, 200 μM tris-carboxyethyl phosphine, and supplemented with DNase, RNase, and Complete Protease Inhibitor Cocktail. Protein complex was purified as described above except that the MBP tag was not cleaved. Protein samples were used immediately for analysis by sedimentation velocity analytical ultracentrifugation or were frozen at −80 °C.
2.3. Sedimentation velocity analytical ultracentrifugation of the BTB–Cul3 complex
Sedimentation velocity experiments were conducted at 20.0 °C on a Beckman Coulter Proteome XL-A analytical ultracentrifuge using the absorbance optical detection system. Samples were loaded into two-channel, 12 mm path length sector shaped cells (400 μL). Scans were acquired at 4 min intervals and rotor speeds of 45,000 rpm; absorbance data were collected as single absorbance measurements at 280 nm using a radial spacing of 0.003 cm. Data were analyzed in SEDFIT 11.9b [15] in terms of a continuous c(s) distribution. Solution density (ρ) and viscosity (η) were calculated using SEDNTERP 1.2 [16], as was the partial specific volume (v) of the protein complex. The c(s) analyses were carried out using an s range of 0–25 with a linear resolution of 200 and maximum entropy regularization confidence levels (F-ratio) of 0.68. Sedimentation coefficients were corrected to standard conditions at 20.0 °C in water, s20,w.
2.4. Cell cultures, transfection and immunofluorescence
293T, NIH3T3 and C2C12 cells were grown in Dulbecco’s modified eagle medium (DMEM, Invitrogen) containing 10% fetal bovine serum. About 60–70% confluent 293T and NIH3T3 cells were transfected with Lipofectamin 2000 and C2C12 cells were transfected with Lipofectamin LTX according to manufacturer’s protocol. At 48 h post-transfection, cells were fixed in 3.8% paraformaldehyde, permeabilized in PBS with 0.2% TritonX-100. Cells were blocked in 5% goat serum (DAKO) and incubated with a rabbit antibody to the Myc (Abcam) or Flag (Sigma) tag for 1 h followed by a mouse ubiquitin antibody (Chemicon) staining. The secondary antibodies, Alexa 488 and Alexa 594 (Invitrogen) were applied for 30–60 min. Cell nuclei were counterstained using Prolong Gold antifade reagent with DAPI (Invitrogen). Cells were visualized and analyzed with a Zeiss LSM 510 confocal microscope.
2.5. Immunoprecipitation and immunoblot analysis
At 48 h post-transfection, the cells were lysed in cold lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 or 1% NP40, 1 mM PMSF, and protease inhibitor mixture) and immunoprecipitated using appropriate antibodies followed by protein-A or-G beads (Invitrogen) per manufacturer protocol. Flag-tagged proteins were immunoprecipitated with anti-Flag M2 magnetic beads (Sigma) according to product guidelines. Immunoprecipitates were analyzed by standard immunoblotting. Briefly, membranes were incubated in blocking buffer followed by antibody and anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase (HRP) (GE Healthcare). Detection of the HPR-conjugated antibody was done using a Super Signal WestPico kit (Pierce Biotechnology) and exposure to chemiluminescent film (GE Healthcare).
2.6. Ubiquitination assay
Ubiquitination assays were performed according to published protocols [17,18]. For detection of ubiquitinated protein in vivo, 293T cells were transfected with expression vectors for KBTBD13-Myc-Flag, Cul3, Rbx1-Myc and pCMV-(HA-Ub)8. The octameric ubiquitin is more efficiently conjugated to the protein than monomeric ubiquitin and can be processed in vivo by cellular ubiquitin C-terminal hydrolases [12]. Transfected cells were treated with 50 μM MG132 proteosome inhibitior (Calbiochem) for 3 h prior lysis. The cells were lysed in buffer containing 2% SDS, 50 mM buffer Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM DTT. Lysates were immediately denatured to preserve ubiquitin modification of protein in the cells. The lysates subjected to immunoprecipitation with anti-Flag beads overnight at 4 °C. Ubiquitinated proteins were determined by immunoblotting with anti-HA antibodies (Sigma).
For detection of ubiquitinated protein in vitro, 293T cells were transfected with expression vectors for KBTBD13-Myc-Flag, Cul3 and Rbx1-Myc. Transfected cells were lysed in buffer containing 10 mM buffer Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP40, 1 mM PMSF, 1 mM Na3VO4 and protease inhibitor mixture. The lysates were subjected to immunoprecipitation with anti-Flag beads. Precipitates were washed with lysis and ubiquitin ligase assay buffers. Immobilized protein complex was incubated with 2 μg purified HA-tagged ubiquitin (Ub-HA), E1 (200nM), E2-UbcH5a (1 μM) in 30 μl of ubiquitin reaction buffer with ATP (2 mM) at 37 °C for 1 h. Ub-HA, E1, E2-UbcH5a and ubiquitin reaction buffer were purchased from Boston Biochem.
3. Results
3.1. KBTBD13 interacts with Cullin 3 through its BTB domain
Studies demonstrate that the proteins containing BTB and Kelch domains function as substrate-specific adaptors for Cul3-RL [7–10]. However, whether KBTBD13 forms a complex with Cul3 to target substrate(s) for ubiquitination is unknown. To determine if KBTBD13 interacts with Cul3, we set out to identify stable constructs for both proteins. A fragment of human Cul3 containing residues 1–383 was chosen for expression in bacteria considering its homology with the Cullin domain of Cullin proteins [19]. MBP-Cul3(1–383)-(His)6 was expressed as a soluble protein. After cleavage and removal of MBP, Cul3(1–383)-(His)6 was analyzed by size exclusion chromatography and sedimentation velocity analytical ultracentrifugation (AUC), which revealed that the protein behaves as a monomer in solution (Supplementary Fig. S1). The KBTBD13 protein was analyzed using Conserved Domain Search algorithm provided by the National Center for Biotechnology Information (Fig. 1A). Based on this analysis, a fragment (6–132) of KBTBD13 encompassing the BTB domain was chosen for the construction of MBP-KBTBD13(6–132)-(His)6. Most of the protein formed inclusion bodies when expressed in bacteria. A small amount of the protein was soluble and was purified, but still aggregated as determined by size exclusion chromatography (data not shown). Due to poor solubility of the KBTBD13(6–132) fragment, co-expression was used for isolation of a stable complex between KBTBD13 and Cul3. For this purpose, a vector was constructed to co-express KBTBD13(6–132) and Cul3(1–383). After a two-step affinity purification, the protein complex was injected onto a Superose 6 size-exclusion column. Protein eluted from the column in two main peaks with an intervening shoulder peak (Fig. 1B). Proteins in the peak eluting at 9-mL (labeled H) as well as the peak eluting at 17-mL (labeled L) were separated by SDS–PAGE and stained with Colloidal Blue (Fig. 1C). The data reveal that peak H (near the void volume of 8 mL) contained MBP-KBTBD13(6–132) in a high-molecular-weight aggregate while peak L contained the MBP-KBTBD13(6–132):Cul3(1–383)-(His)6 complex. Comparison of band intensities for MBP-KBTBD13(6–132) and Cul3(1–383)-(His)6 suggests that the subunit stoichiometry of the complex is 1:1 or possibly 2:2. Sedimentation velocity AUC was employed to determine the molecular mass and subunit stoichiometry of the MBP-KBTBD13(6–132):Cul3(1–383)-(His)6 complex. Peak L collected from the size exclusion column was concentrated, and filtered immediately before analysis by sedimentation velocity AUC. Protein concentration was determined to be ~0.8 mg/mL (8 μM, based on the calculated protomer masses of 57 kDa for MBP-KBTBD13(6–132) and 46 kDa for Cul3(1–383)-His6). C(s) data analysis using SEDFIT revealed a major sediment species at 5-S (~90% of total loaded protein) with a molecular mass of 90 kDa (Fig. 1D), consistent with the 1:1 MBP-KBTBD13(6–132):Cul3(1–383)-(His)6 complex (Mcalc = 103 kDa). Minor species with s20,w values of 3.4-S (2% of loaded protein) and 6.1-S (8% of loaded protein) were also observed. The 3.4-S peak returned a molecular mass of 62 kDa, consistent with MBP-KBTBD13(6–132) monomer (Mcalc = 57 kDa). The 6.1-S peak returned a molecular mass of 130 kDa, which is close to the mass of the MBP-KBTBD13(6–132) dimer (Mcalc = 114 kDa) or possibly the 2:1 MBP-KBTBD13(6–132):Cul3(1–383)-(His)6 complex (Mcalc = 160 kDa). Taken together, the data in Fig. 1B–D provide strong evidence for a direct interaction between the BTB domain of KBTBD13 and the Cullin domain of Cul3.
Fig. 1.
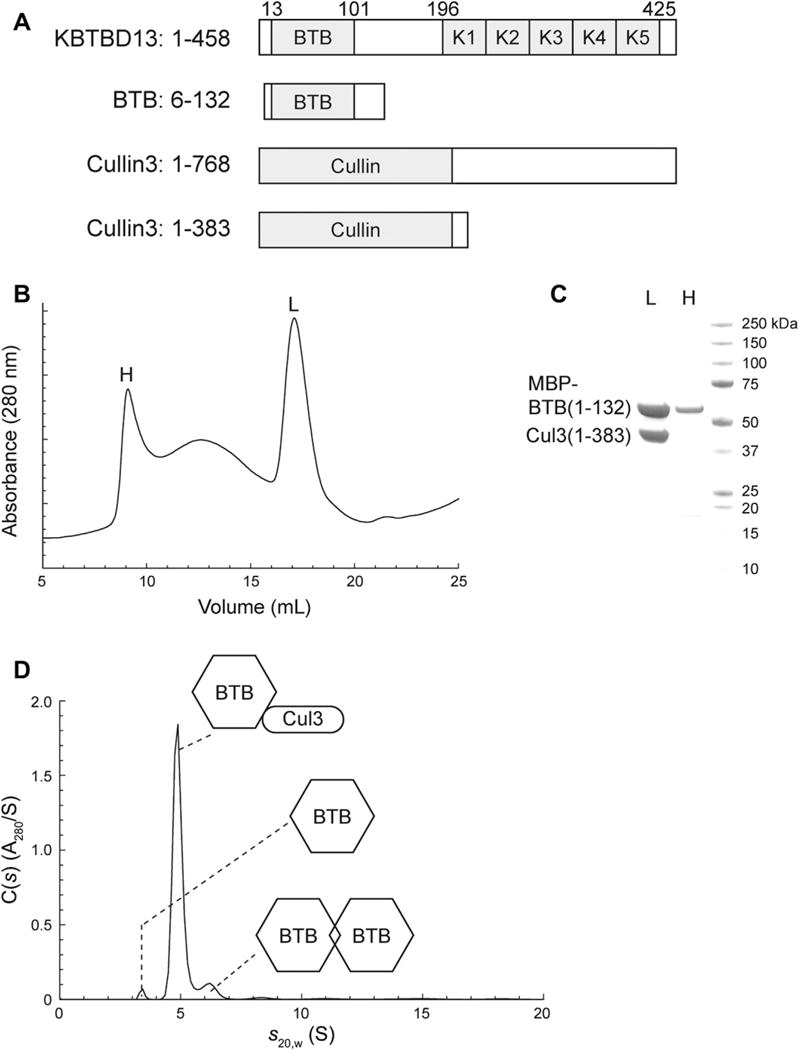
KBTBD13 interacts with Cullin 3 through its BTB domain. (A) Domain structure of KBTBD13 protein. Domain predictions are based on results from NCBI’s Conserved Domain Search algorithm. (B) Size-exclusion chromatography of the MBP-KBTBD13(6–132):Cul3(1–383)-(His)6 complex. About 5 mg of protein was loaded onto a Superose 6 10/300 column equilibrated at 0.5 mL/min in buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, and 200 μM tris(2-carboxyethyl)phosphine. (C) SDS–PAGE analysis of fractions from size-exclusion purification of the MBP-KBTBD13(6–132):Cul3(1–383)-(His)6 complex. Lane “L”: fraction from the peak that eluted at 17 mL from the Superose 6 column. Lane “H”: fraction from the peak that eluted at 9 mL from the Superose 6 column. (D) Sedimentation velocity analytical ultracentrifugation of the MBP-KBTBD13(6–132):Cul3(1–383)-(His)6 complex. The complex was purified as described in Section 2. The fit was judged to be excellent with a root mean square deviation of 0.0070 absorbance units.
We further examined interaction between full length KBTBD13 and Cul3 by ectopic expression of both proteins in mammalian cells. 293T cells were co-transfected with plasmids expressing KBTBD13-Myc-Flag and Cul3. Immunoblot analysis of cell lysates with anti-Flag and anti-Cul3 antibodies revealed that KBTBD13 and Cul3 were expressed as 50 kDa and 90 kDa proteins, respectively (Fig. 2A). Interaction of expressed proteins was then assessed by immunoprecipitation followed by immunoblotting. As shown in Fig. 2B, KBTBD13 and Cul3 were detected in immunoprecipitates with anti-Cul3 and anti-Flag, respectively. These results along with data from size exclusion chromatography and AUC demonstrate a direct interaction of two proteins and show that BTB domain of KBTBD13 is sufficient and necessary for binding to Cul3.
Fig. 2.
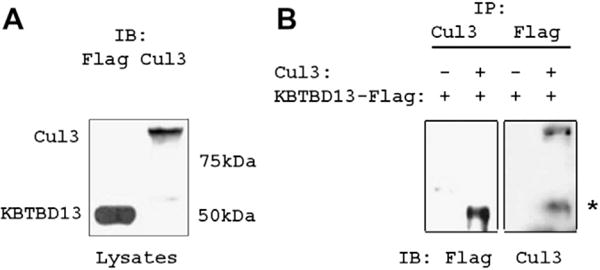
Interaction of full-length KBTBD13 and Cullin 3 in mammalian cells. (A) Lysates from 293T cells transfected with vectors for KBTBD13-Flag and Cul3 were immnoblotted with anti-Flag and anti-Cul3 antibodies. (B) Cell lysates were immunoprecipitated with anti-Flag and anti-Cul3 Abs and immunoblotted with anti-Cul3 and anti-Flag antibodies, respectively. *Indicates nonspecific binding of immunoglobulin.
3.2. KBTBD13 forms a functional complex with Cul3 RING ubiquitin ligase
Stable interaction between KBTBD13 and Cul3 suggests that KBTBD13 may form a functional complex with Cul3-RL promoting a substrate ubiquitination. Given a lack of data on substrates specific for KBTBD13, we sought to investigate whether KBTBD13 is ubiquitinated by Cul3 and Rbx1 using in vivo and in vitro ubiquitination assays. For in vivo ubiquitination, 293T cells were co-transfected with expression vectors for KBTBD13-Myc-Flag and pCMV-(HA-Ub)8. Transfected cells treated with MG132 were lysed under denaturing conditions and subjected to immunoprecipitation with anti-Flag beads. The presence of ubiquitin conjugated KBTBD13 proteins promoted by cellular ubiquitination machinery were readily seen as determined by immunoblot analysis with anti-HA antibodies (Fig. 3A). To determine whether KBTBD13 is specifically ubiquitinated by Cul3-RL, we next performed in vitro ubiquitination assay. 293T cells were co-transfected with expression vectors for KBTBD13-Myc-Flag, Cul3 and Rbx1-Myc, and the immunocomplex of proteins was immobilized to anti-Flag beads. Proteins bound to beads were subjected to ubiquitin ligation reaction with purified E1, E2-UbcH5a, and HA-Ub in the presence of ATP. Immunoblot analysis using anti-HA antibodies identified ubiquitinated KBTBD13 only in the presence of Cul3 and Rbx1 (Fig. 3B). Ubiquitination of KBTBD13 by Cul3-RL was further confirmed in vivo. 293T cells were co-transfected with vectors for KBTBD13-Myc-Flag and pCMV-(HA-Ub)8 in the presence of Cul3 and or Rbx1-Myc. Cells treated with MG132 were lysed under denaturing conditions and subjected to purification using anti-Flag beads. Immunocomplexes precipitated from the cells were assessed by immunoblotting with anti-HA antibodies (Supplemental Fig. S2). Ubiquitination of KBTBD13 occurred in the presence of Cul3 and was enhanced by co-expression with Cul3 and Rbx1. The results of ubiquitination assays demonstrate that KBTBD13 is able to form a functional Cul3-RL complex capable of conjugating ubiquitin both in vivo and in vitro.
Fig. 3.
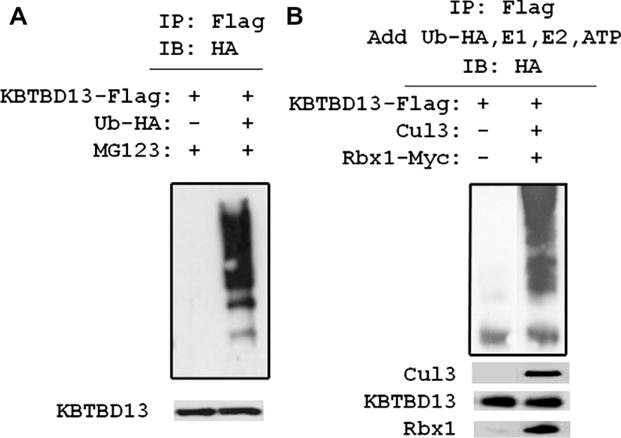
KBTBD13 forms a functional Cullin 3 based RING ubiquitin ligase complex. (A) 293T cells were transfected with KBTBD13-Flag and pCMV-(HA-Ub)8 vectors. Anti-Flag immunoprecipitates were analyzed with HA antibodies. (B) 293T cells were co-transfected with indicated vectors. Lysates precipitated with Flag beads incubated with E1, E2, HA-ubiquitin in ubiquitin ligase assay buffer in the presence of 1 mM ATP. Precipitated complex analyzed with anti-HA antibodies. Bottom panels, cell lysates analyzed by immunoblotting with anti-Flag, anti-Cul3 and anti-Myc antibodies.
3.3. KBTBD13 is a cytoplasmic protein that co-localizes with ubiquitin
To determine whether KBTBD13 and components of proteasome machinery are expressed in the same subcellular compartments, Myc-Flag and GFP tagged KBTBD13 were expressed in C2C12 and 3T3 cells. The cells were stained for anti-Myc or anti-Flag and anti-ubiquitin antibodies and analyzed by confocal microscopy. KBTBD13 located in cytoplasm primarily around the nucleus in muscle and non-muscle cells (Fig. 4). Notably, KBTBD13 does not co-localize with alpha-actin, the major protein component of muscle thing filament (Fig. 4A) This may indicate that the disease mechanisms in KBTBD13 associated myopathy are fundamentally different from other types of NEMs. KBTBD13 often formed cytoplasmic inclusions and formations of these inclusion bodies were dependent on duration and the expression level of the protein (Fig. 4B). These inclusions appear to coalesce to form larger aggregates that were more prominent in cells expressing higher amount of KBTBD13. Cytoplasmic inclusions of KBTBD13 were immunoreactive for ubiquitin (Fig. 4B). Untransfected cells stained with anti-ubiquitin antibodies showed a diffuse staining pattern of ubiquitin (Fig. 4B, middle). Upon transfection with vector expressing KBTBD13, a punctate staining pattern indicative of regions enriched in ubiquitin was detected (Fig. 4B, bottom). Confocal microscopy results further confirm the formation of active Cul3-RL that potentially can recruit unidentified substrates for ubiquitination through its adaptor KBTBD13.
Fig. 4.
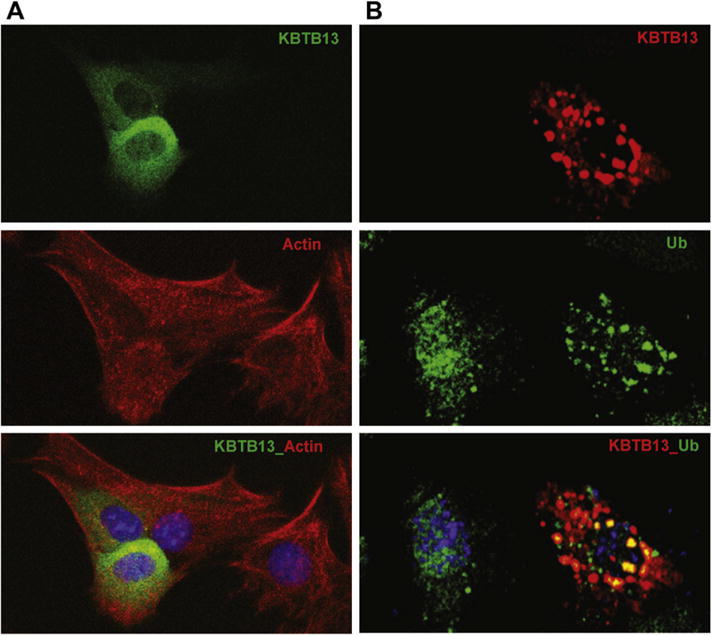
Localization of KBTBD13 in perinuclear areas. (A) C2C12 cells expressing KBTBD13-GFP (top) stained with muscle actin (middle). (B) 3T3 cells expressing KBTBD13-Myc-Flag. The co-localization between ubiquitin and KBTBD13 is indicated by the yellow color. The nucleus is stained in blue with DAPI.
4. Discussion
KBTBD13 was originally identified as a muscle protein mutated in a new subtype of NEM [1]. Its expression exclusively in skeletal and cardiac muscles and association with congenital myopathy suggest an important role of this protein for the integrity of muscle structure and function. To understand the function of KBTBD13 protein, we performed experiments aimed to identify KBTBD13 interacting partners. Based on the domain structure of the protein, we hypothesized that KBTBD13 may serve as substrate specific adaptor for Cul3-RL. Using biochemical analysis of purified recombinant proteins and transient expression of both proteins in mammalian cells, we provide several lines of evidence that KBTBD13 interacts with Cul3 and this interaction is mediated by the BTB domain of KBTBD13 and the Cullin domain of Cul3. We further show that KBTBD13 forms a functional Cul3-Rbx1 ubiquitin ligase complex capable of conjugating ubiquitin both in vivo and in vitro. Taken together, our data demonstrate that KBTBD13–Cul3–Rbx1 is a muscle specific ubiquitin ligase.
Based on size exclusion chromatography and sedimentation velocity AUC, we concluded that the BTB domain of KBTBD13 binds to Cul3 with 1:1 stoichiometry. Most BTB domains form dimers. Dimerization is mediated by two elements located in the N-terminal to the BTB domain, a beta strand and an alpha helix, which are conserved in the “long form” of the BTB domain [5,6]. Analysis of the sedimentation velocity AUC data revealed the presence of minor species consistent with monomeric and dimeric forms of MBP-KBTBD13(6–132). The size exclusion data and SDS–PAGE indicate that a fraction of MBP-KBTBD13(6–132), which was not bound to Cul3, formed a high-molecular-weight aggregate. The reasons for the poor solubility of MBP-KBTBD13(6–132) are not clear. The MBP fusion partner may prevent normal dimerization of the BTB domain and favor the formation of high-molecular-weight species. Further work is required to characterize the BTB domain in KBTBD13 and its interaction with Cul3.
Our study further demonstrates that KBTBD13 is able to form a complex with Cul3 and Rbx1 to reconstitute a functional ubiquitin ligase capable of conjugating ubiquitin onto KBTBD13. Auto-ubiquitination of substrate adaptors in the absence of substrates is achieved through the same chemical reaction as substrate ubiquitination and has also been reported for other BTB and Kelch domain containing proteins [7,20]. Our results support the common theme that BTB and Kelch domain proteins are able to assemble with Cul3-RL into a functional ubiquitin ligase complex and ubiquitinate themselves within the assembled complex. Although the functional role of this shared feature of BTB and Kelch domain proteins are unknown, studies of SCF complexes demonstrate that auto-ubiquitination of F-box proteins is responsible for their dynamic turnover during cell cycle [21,22]. Given functional similarity between these adaptor proteins and their capability for auto-ubiquitination within assembled complexes, it is conceivable that ubiquitination of KBTBD13 and other BTB and Kelch domain proteins allows cells to control a dynamic equilibrium among multiple Cul3-RL complexes and likely between substrates and substrate adaptors.
KBTBD13 localized in cytoplasm, predominantly around the nucleus in all cells assessed in this study. KBTBD13 forms punctate structures that resemble inclusion bodies. Inclusion bodies often contain misfolded proteins that are usually degraded by the ubiquitin proteasome pathway [23,24]. Keeping with this, the colocalization of KBTBD13 with ubiquitin suggests that proteins ubiquitinated by KBTBD13–Cul3–Rbx1 will most likely be degraded by the ubiquitin proteasome system. Dominant mutations in KBTBD13 lead to childhood onset NEM characterized by slowly progressive muscle weakness and hypotrophy in the neck and proximal limb muscles [1,2]. Affected muscles show numerous rods or nemaline bodies clustering into cytoplasmic aggregates and resulting in sarcomere disorganization. In contrast to the thin filament proteins mutated in other subtypes of NEM, the role of KBTBD13 as a muscle specific ubiquitin ligase suggests an involvement of ubiquitin proteasome pathway in pathogenesis of KBTBD13 associated NEM. Identification of substrate proteins targeted by KBTBD13–Cul3–Rbx1 complex will provide further understanding of the molecular mechanisms of this myopathy.
Supplementary Material
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bbrc.2012.04.074.
Contributor Information
Nyamkhishig Sambuughin, Email: nyamkhishig.sambuughin@usuhs.edu.
Wieslaw Swietnicki, Email: wieslaw_swietnicki@hotmail.com.
Stephen Techtmann, Email: stephen.techtmann@usuhs.edu.
Vera Matrosova, Email: vera.matrosova@usuhs.edu.
Tarina Wallace, Email: tarina.wallace@usuhs.edu.
Lev Goldfarb, Email: goldfarbl@ninds.nih.gov.
Ernest Maynard, Email: ernest.maynard@usuhs.edu.
References
- 1.Sambuughin N, Yau KS, Olive M, et al. Dominant mutations in KBTBD13, a member of the BTB/Kelch family, cause nemaline myopathy with cores. Am J Hum Genet. 2010;87:842–847. doi: 10.1016/j.ajhg.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olive M, Goldfarb L, Lee HS, et al. Nemaline myopathy type 6: clinical and myopathological features. Muscle Nerve. 2010;42:901–907. doi: 10.1002/mus.21788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallgren-Pettersson C, Sewry CA, Nowak C, Laing NG. Nemaline myopathies. Semin Pediatr Neurol. 2011;18:230–238. doi: 10.1016/j.spen.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Nance JR, Dowling JJ, Gibbs EM, Bonnemann CG. Congential myopathies: an update. Curr Neurol Neurosci Rep. 2012 doi: 10.1007/s11910-012-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein–protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 6.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- 8.Xu L, Wei Y, Reboul J, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB–Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 10.Petroski MD, Deshaies RJ. Function and regulation of cullin-ring ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 11.Prag S, Adams JC. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinform. 2003;4:42. doi: 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paxton CW, Cosgrove RA, Drozd AC, et al. BTB–Kelch protein Krp1 regulates proliferation and differentiation of myoblasts. Am J Physiol Cell Physiol. 2011;300:C1345–C1355. doi: 10.1152/ajpcell.00321.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamura T, Koepp DM, Conrad MN, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Sceince. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 14.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 15.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole JL, Lary JW, Moody TP, Laue TM. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008;84:143–179. doi: 10.1016/S0091-679X(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furukawa M, Andrews PS, Xiong Y. Assays for RING family ubiquitin ligases. Methods Mol Biol. 2005;301:37–46. doi: 10.1385/1-59259-895-1:037. [DOI] [PubMed] [Google Scholar]
- 18.Bloom J, Pagano M. Experimental tests of definitively determine ubiquitilation of a substrate. Methods Enzymol. 2005;399:249–265. doi: 10.1016/S0076-6879(05)99017-4. [DOI] [PubMed] [Google Scholar]
- 19.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. BMC Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zang DD, Lo SC, Sun Z, Habib GM, Leiberman MW, Hannik M. Ubiquitination of Keap1, a BTB–Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 21.Zhou P, Howley PM. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin–protein ligases. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- 22.Galan JM, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 24.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


