Abstract
Ruminants have the remarkable ability to convert human-indigestible plant biomass into human-digestible food products, due to a complex microbiome residing in the rumen compartment of their upper digestive tract. Here we report the discovery that rumen microbiome components are tightly linked to cows' ability to extract energy from their feed, termed feed efficiency. Feed efficiency was measured in 146 milking cows and analyses of the taxonomic composition, gene content, microbial activity and metabolomic composition was performed on the rumen microbiomes from the 78 most extreme animals. Lower richness of microbiome gene content and taxa was tightly linked to higher feed efficiency. Microbiome genes and species accurately predicted the animals' feed efficiency phenotype. Specific enrichment of microbes and metabolic pathways in each of these microbiome groups resulted in better energy and carbon channeling to the animal, while lowering methane emissions to the atmosphere. This ecological and mechanistic understanding of the rumen microbiome could lead to an increase in available food resources and environmentally friendly livestock agriculture.
Introduction
Ruminants hold enormous significance for man, as they convert the energy stored in plant biomass polymers, which are indigestible for humans, to digestible food products. Humans domesticated these animals for this purpose in the Neolithic era (Ajmone-Marsan et al., 2010) and have been farming them ever since for the production and consumption of animal protein in the form of meat and milk. In today's extensive production regimes, ruminants consume 30% of the crops grown on earth and occupy another 30% of the earth's land mass (Thornton, 2010). These animals also emit methane—a highly potent greenhouse gas—to the atmosphere and are considered to be responsible for a considerable portion of its emission because of anthropogenic activities (McMichael et al., 2007). One way to tackle these problems is to increase the animals' energetic efficiency, that is, the efficiency with which they convert energy from feed, thereby increasing food availability, while lowering the environmental burden, as these animals would produce more and eat less (Bradford, 1999; Thornton, 2010).
Different methods are used to evaluate an animal's energetic efficiency; of these, the residual feed intake (RFI) method (Koch et al., 1963) is highly accepted and widely used (Herd and Arthur, 2009) as it takes into account growth and body size and is thus suitable for comparisons between animals. This parameter is an estimation of the difference between an animal's actual feed intake and its predicted feed intake based on its production level and body weight. The energetic efficiency varies considerably between different individuals from the same breed. Specific genomic regions, such as one that is suggested to be associated with a role in controlling energy metabolism, have been found to correlate to feed efficiency using genome-wide association studies (Pryce et al., 2012). Nevertheless, only a moderate genetic component (heritability ranging from 0.26 to 0.58) affects energy utilization, as has also been demonstrated by elevation of feed efficiency via selection of animals according to their RFI (Archer et al., 1999; Moore et al., 2009).
One important factor that could greatly contribute to variations in these animals' feed efficiency is the rumen microbiome. The ability of these animals to digest plant biomass polymers is attributed to this complex microbiome that resides in their upper digestive tract in a compartment termed the rumen (Mizrahi, 2013). The anaerobic environment in the rumen and the highly complex food webs sustained by the rumen microbiome enable the fermentation of plant material into metabolic end products such as short-chain fatty acids (SCFAs) and methane. Although SCFAs are absorbed through the rumen wall and serve to fulfill the animal's energy needs, methane is not absorbed; it is emitted to the atmosphere together with its retained energy, thereby contributing to energy loss from the feed, as well as global warming (Mizrahi, 2011). Microbial composition of the rumen has been described using various techniques (Brulc et al., 2009; Hess et al., 2011; Jami and Mizrahi, 2012; Henderson et al., 2015). In addition, differences between high and low RFI animals have been reported in terms of methane production, as well as of some differences in microbial composition (Nkrumah et al., 2006; Mizrahi, 2011; Hernandez-Sanabria et al., 2012; Jami et al., 2014; Kittelmann et al., 2014; Shi et al., 2014; Wallace et al., 2015). Nevertheless, a comprehensive and thorough understanding of microbiome structure patterns and how to translate them to functionality at the animal level is still lacking.
Here we determined the feed efficiency phenotype of a cohort of 146 animals; this phenotype was mapped to its underlying microbiome determinants by sampling and analyzing rumen fluid and feces from the animals in the two extreme quartiles (78 animals—40 efficient and 38 inefficient; Supplementary Figure S1 and Supplementary Data 1). We characterized the taxonomic composition, genetic functional potential, metabolomic composition and activity of these rumen microbiomes to explore the hypothesis of a link between the animal's feed efficiency and these microbiome components, and to uncover the potential mechanisms that might explain this link. Our data provide detailed and novel insight into the characteristics and components of the rumen microbiome related to feed efficiency—their ecological context, the underlying mechanisms and their potential as markers to predict the feed efficiency phenotype.
Materials and methods
Trial design
The experimental procedures used in this study were approved by the Faculty Animal Policy and Welfare Committee of the Agricultural Research Organization (ARO), approval number IL-386/12, Volcani Research Center, and were in accordance with the guidelines of the National Council for Animal Experimentation.
A total of 146 Holstein Friesian dairy cows were selected for the experiment and housed at the ARO's experimental dairy farm in Bet Dagan, Israel. Cows with history of diseases, miscarriages and twin pregnancies or that were above first trimester were not included in the experiment. The experimental dairy farm is equipped with a facility that is specially designed to individually monitor all of the animal's functions, feed intake and different physiological parameters (Halachmi et al., 1998). The animals were divided into seven groups according to lactation period such that each cow was between 50 and 150 days of lactation when monitored. Each group contained between 19 and 21 cows that were monitored for 42–49 days. The animals were fed ad libitum a standard lactating cow diet consisting of 30% roughage and 70% concentrate and had free access to water. The cows were habituated with the aforementioned diet for 3 weeks before the start of the experiment so that they would become accustomed to their individual feeding station.
The following parameters were automatically monitored three times a day during the experiment: dry matter (DM) intake (kg), weight (kg), milk yield (kg), milk lactose, fat and protein (g) and somatic cell count using the Afimilk program (Afimilk Ltd, Kibbutz Afikim, Israel). Milk samples were sent to an authorized milk quality lab (National Service for Udder Health and Milk Quality, Caesarea, Israel) three times for each group to verify the Afimilk program analysis. Body conditioning score was measured once a week by the same person throughout the experiment.
Feed efficiency parameters RFI and conversion ratio were calculated according to National Research Council (2001) formulas. In order to increase the statistical power compared with random sampling, extreme phenotypes sampling approach (Li et al., 2011) was applied. Twelve cows with the most extreme and stable RFI values were selected from each group for rumen fluid sampling, six with low and six with high RFI values. Tukey's test was used to verify that the RFI value of each cow was steady throughout the experiment and significantly different from cows in the reciprocal efficiency group. Overall, 78 cows were chosen for sampling and represented the 25% most efficient and 25% most inefficient animals of the whole cohort (P<0.0001; Supplementary Figure S1 and Supplementary Data 1). Our previous work showed that samples from 16 cows are sufficient in order to cover all of the microbial diversity in the bovine rumen on a specific diet (Jami and Mizrahi, 2012). Therefore, this extreme phenotype characterization and sampling approach, together with the large cohort from which the extreme cows were chosen from, ensured increased adequate power to detect microbiome components connected to the host phenotype.
Sample collection
Rumen samples were collected on 3 consecutive days. The cows were sampled 6 h after feeding in which they were not offered feed; 500 ml of rumen contents were collected using a stainless-steel stomach tube with a rumen vacuum sampler, and pH was immediately determined. Samples for DNA and metabolite extraction were snap frozen in liquid nitrogen and stored at −80 °C until analysis. Rumen samples for metabolic assays were filtered through six layers of cheese cloth to remove big feed particles, transferred to CO2-containing bottles and flushed with CO2 to maintain anaerobic conditions. Immediately after collection, the rumen samples were maintained at 39 °C up to 1 h until use, and processed in the laboratory, located 100 m away.
Fresh fecal samples were obtained three times a day for 4 consecutive days. Samples were immediately frozen at −20 °C.
In-vitro digestibility assay
The in-vitro digestibility of plant cell wall fibers, represented by neutral detergent fiber or total feed polymers (in DM), was determined according to the two-stage technique by Tilley and Terry (1963). Briefly, cows' feed was dried for 72 h in an aerated 60 °C oven and then ground to pass a 1-mm screen. The feed was incubated with rumen fluid and artificial rumen buffer, in sealed glass tubes.
Artificial rumen buffer was formulated as described previously (McDougall, 1948; Tilley and Terry, 1963). Briefly, 100 ml of buffer A (98 g NaHCO3, 93 g NaHPO4·12H2O, 5.7 g KCl, 4.7 g NaCl, 1.2 g MgSO4·7H2O, added to double-distilled water (DDW) to final volume of 1 liter) was added to 800 ml DDW. The solution was flushed with CO2 to reduce the pH to 6.8–7.0. Then, 1 ml of buffer B (40 g CaCl2 added to DDW to final volume of 1 liter), 50 ml of buffer C (30 g NH4HCO3 added to DDW to final volume of 1 liter) and 100 μl of buffer D (10 g MnCl2·4H2O, 1 g CoCl2·6H2O, 8 g FeCl3·6H2O added to DDW to final volume of 1 liter) were added and the buffer was brought to a final volume of 1 liter with DDW.
The tubes were flushed with CO2 and closed with a unidirectional valve cap, which only allowed emission of gas from the tube. The tubes were incubated for 24 or 48 h at 39 °C and were shaken five times a day, followed by incubation with acid pepsin. At the end of this procedure, the undigested solids were precipitated by centrifugation at 1000 g for 10 min and dried in an aerated oven at 60 °C for 72 h. The precipitates were used for residual DM determination by weighing or for residual neutral detergent fiber determination by following the procedure of Van Soest et al. (1991). The results are expressed as mean feed digestibility in the rumen from two consecutive sampling days.
In-vivo digestibility
Fecal grab samples were pooled for each cow, dried at 60 °C for 72 h in a forced-air oven and ground to pass a 1-mm screen. The indigestible neutral detergent fiber content was determined in the ration and in the fecal samples according to a previously reported method (Lippke et al., 1986) after incubation with rumen fluid for 72 h and was used as an internal marker for the apparent total-tract DM digestibility analysis. Each cow's in-vivo DM and neutral detergent fiber digestibility of the ration was calculated using its average DM intake and fecal output.
In-vitro methane emission assay
Samples were diluted 1:2 (v/v) with artificial rumen buffer. Duplicates of 5 ml aliquots from each diluted sample were transferred to screw-cap glass tubes (ISI, Israel Scientific Instruments Ltd, Petah-Tikva, Israel) suitable for methane measurement using a gas chromatography (GC) system HP-5890 series II, (Hewlett-Packard, Palo Alto, CA, USA) with a FID detector. The samples were incubated at 39 °C for 24 h with 0.5 g DM feed, and then analyzed by GC for methane emission. Samples of 0.5 ml gas from the tube headspace were injected into a 182.88 cm × 0.3175 cm × 2.1 mm packed Supelco analytical-45/60 Molecular sieve 5 A column (Supelco Inc., Bellefonte, PA, USA) with helium carrier gas set to a flow rate of 10 ml min–1 and an oven temperature of 200 °C. The oven temperature remained steady for a total run time of 5 min. A standard curve was generated using pure methane gas.
Methane production was quantified for 36 rumen microbiome samples of the most extreme animals of the feed efficiency groups (18 efficient and 18 inefficient), with two biological repeats of each animal.
Identification and quantification of rumen fluid metabolites
Frozen rumen fluid samples were thawed at 25 °C and centrifuged at 10 000 g for 15 min. The supernatant was filtered through a sterile 0.45 μm filter (Merck Millipore Ltd., Tullagreen, County Cork, Ireland). Rumen fluid samples were kept on ice during metabolite extraction in the gas chromatography mass spectrometry and GC metabolite identification and quantification pipelines to minimize metabolite degradation.
The rumen samples were analyzed by gas chromatography mass spectrometry for polar metabolites and by GC with a FID detector for SCFAs. The extraction and derivatization protocol for the gas chromatography mass spectrometry analysis was adapted from a previously described method (Saleem et al., 2013). Derivatized extracts were analyzed using an Agilent 5975C GC and an Agilent 7890A MS (Agilent Technologies, Palo Alto, CA, USA) operating in electron impact (EI) ionization mode. Aliquots (1 μl) were injected (splitless) into a 30 m × 0.25 mm × 0.25 μm HP-5MS Ultra Inert column (Agilent Technologies, Berkshire, UK) with helium carrier gas set to a flow rate of 1 ml min–1 and initial oven temperature of 70 °C. The oven temperature was held constant at the initial temperature for 2 min, and thereafter increased at 10 °C min–1 to a final temperature of 310 °C, and a final run time of 45 min. Samples were run using full scan in a mass range of 50–500 m/z (1.7 scan s–1) with a detection delay of 4 min. Retention indices were calculated using a C8-C20 alkane standard mixture solution (Sigma-Aldrich, Buchs, Switzerland) as the external standard. Quantification and identification of trimethylsilylated metabolites were performed using the NIST database and high performance liquid chromatography grade standards.
For SCFA identification and quantification, 400 μl of filtered rumen fluid were mixed with 100 μl of 25% metaphosphoric acid solution (w/v in DDW) and vortexed for 1 min. The samples were incubated at 4 °C for 30 min and subsequently centrifuged for 15 min at 10 600 g. The supernatant was decanted into new tubes, then 250 μl methyl tert-butyl ether (Sigma-Aldrich) was added and the tubes were vortexed for 30 s. Another cycle of centrifugation was performed for 1 min at 10 600 g. The upper phase, which contained methyl tert-butyl ether+SCFAs, was analyzed using an Agilent 7890B GC system (Agilent Technologies, Santa Clara, CA, USA) with a FID detector. The temperatures at the inlet and detector were 250 °C and 300 °C, respectively. Aliquots (1 μl) were injected with a split ratio of 1 : 25 into a 30 m × 0.32 mm × 0.25 μm ZEBRON ZB-FFAP column (Phenomenex, Torrance, CA, USA) with helium carrier gas set to a flow rate of 2.4 ml min–1 and initial oven temperature of 100 °C. The oven temperature was held constant at the initial temperature for 5 min, and thereafter increased at 10 °C min–1 to a final temperature 125 °C, and a final run time of 12.5 min.
Quantification and identification of metabolites were performed using high performance liquid chromatography-grade standards. All metabolites were normalized to the organic matter content of the rumen fluid they were extracted from. Rumen samples were filtered through a sterile 0.45 μm Supor Membrane filter (PALL Life Sciences Ann Arbor, MI, USA). The organic C in the rumen samples was analyzed with a Formacs, combustion total organic carbon analyzer (Skalar, De Breda, The Netherlands).
Microbial DNA extraction
The rumen microbial fraction was separated according to Stevenson and Weimer (2007), with minor modifications to suit the needs of these experiments as described in Jami et al. (2013). The DNA extraction was performed as described by Stevenson and Weimer (2007).
Shotgun DNA sequencing and analysis
Metagenomic DNA libraries were constructed with the TruSeq DNA Sample Prep kit (Illumina, San Diego, CA, USA). Libraries were pooled and sequenced on two lanes for 151 cycles from each end on a HiSeq2500 (Illumina) and processed with Casava 1.8.2 (Illumina). On average, 35 581 041±6 899 269 paired end reads were obtained from each sample and 2 775 321 186 paired end reads were obtained overall. In all, 18.6% of the reads did not pass artifact filtering and trimming using MOCAT pipeline (Kultima et al., 2012).
To obtain a more comprehensive metagenome, a joint assembly of all data from the 78 cows was created. This compensated for the lower sequencing depth of each individual sample and any bias caused by assembly of individual samples. Reads from all samples were pooled and assembled into one metagenome using CLC Bio, package CLC Assembly Cell version 3.2.2 (Qiagen, Redwood, CA, USA) with K-mer=21 and default parameters; 16 784 830 contigs were obtained. A QC pipeline of dereplication and screening for Bos taurus reads was performed using the MG-RAST pipeline. No redundancies were found and 0.43% of the contigs were discarded after removing Bos taurus contaminants. The phylogenetic origin of each contig was annotated with RefSeq database (Pruitt et al., 2007) (E⩽10−5) using the MG-RAST pipeline (Meyer et al., 2008).
Gene calling was performed on the contigs using FragGeneScan (Rho et al., 2010); 21 531 511 genes were identified over all. Each sample's reads were recruited against the overall genes using burrows-wheeler alignment tool (Li and Durbin, 2009) with 98% identity and default parameters; a threshold of one read for gene identification was chosen to include rare genes in the analysis. On average, 52.4% of the reads from each sample were mapped to the obtained genes, without differences between the efficiency groups (Supplementary Figure S2). An average of 4 079 212 genes were identified in each sample. The abundance of a specific gene was calculated by the number of reads uniquely recruited, normalized to the length of the gene and total reads obtained from the sample. The number of genes detected had no dependence on the number of mapped reads (Supplementary Figure S3).
16S ribosomal DNA sequencing and analysis
The 16S V3 region was amplified using the primers 357F 5′-CCTACGGGAGGCAGCAG-3′ and 926R 5′-CCGTCAATTCMTTTRAGT-3′ (Peterson et al., 2009). The libraries were pooled and sequenced on one MiSeq flowcell (Illumina) for 251 cycles from each end of the fragments and analyzed with Casava 1.8. Overall, 49 760 478 paired end reads were obtained for all 3 sampling days, with an average of 212 652 reads per sample per day.
Data quality control and analyses were performed using the QIIME pipeline version 1.7.0 (Caporaso et al., 2010). Species were defined at 97% identity using UCLUST (Edgar, 2010). Taxonomy assignment of species was performed using BLAST against the 16S rRNA reference database RDP (version 10) (Cole et al., 2003). All singletons and doubletons were removed from the data set, resulting in 85 225 species with an average of 5039 per sample. Species were binned at different taxonomic levels to receive taxon abundances for each phylogenetic level (Supplementary Figure S4 and Supplementary Data 2).
Biodiversity analysis
Within-sample (alpha) diversity was calculated using Shannon index and dominance was determined according to 1–Simpson index (Harper, 1999). The indices of 16S rRNA gene profiles were calculated using bootstrapping with 9999 replicates. Richness of genes and taxa are presented as simple counts of genes and taxa.
Identifying differential species and genes
The statistical significance of differences in species and gene abundance between the efficiency groups was tested by Wilcoxon rank-sum test coupled with a bootstrapping approach adopted from Le Chatelier et al. (2013): 70% of the whole sampled cohort was randomly chosen 30 times and significance was determined at P<0.05 with bootstrap=0.8 as a threshold. This process was repeated with another 30 iterations on the 48 most extreme cows (24 efficient and 24 inefficient). Overall, 18 significantly different species and 34 166 significantly different genes common to all 60 tests were further analyzed. Species and genes that were significantly different were correlated to the RFI parameter using Spearman correlation. Functional annotation of significant genes was achieved using BLASTP with E⩽10−6 against KEGG PATHWAY, MODULE, BRITE, GENES and ORTHOLOGY databases (2014; 25% annotation) (Kanehisa et al., 2011). These genes were also blasted against the NR database (Pruitt et al., 2007), and their phylogenetic annotation was determined according to the best hit (BLASTP with E⩽10−6; 89% annotation).
Statistical tests and estimation of false discovery rate
Tukey's, Student's t and Wilcoxon rank-sum tests were conducted depending on the normality of distribution of the input data. All tests were corrected for false discovery rate using the method described by Benjamini and Hochberg (1995) unless otherwise noted. In permutation t-test, significance of the difference between means was inferred by performing t-test between the two groups and comparing the resulting t-statistic to the t-statistics resulting from 9999 permutations of random group assignments (two-tailed, P<0.05) (Davis, 1986). For multiple hypothesis correction, the distribution of t-test P-values was compared with the lowest P-values distribution resulting from 9999 permutations of random group assignments according to Westfall and Young (1993). This procedure was performed using the R bioconductor package multtest (Pollard et al., 2005), function mt.maxT, individually for each metabolic or activity test, namely polymers, SCFAs, methane and all other measured metabolites. Variance similarity was tested where required by the statistical test.
Predictions of different physiological parameters
Feature selection of microbial species and genes was conducted by choosing species or genes that were significantly different in their presence/absence using the Fisher's exact test. Species and genes were sorted separately according to their P-value in ascending order and grouped into bins of 100 features. Each bin was used as predictive features for the feed efficiency phenotype using the k-nearest neighbors algorithm (Aha, 1997) with k=3. The mean accuracy of the prediction was calculated using cross-validation of 1000 iterations for each bin, in which 70% of the samples were used as a training set and the remaining 30% were used as a test set to measure the accuracy of the prediction. Changing the bin size (bins ranging in size from 50 to 1000 features per bin) did not affect the accuracy of the prediction (Supplementary Data 3). To check the significance of the classifications accuracy, a permutations technique was used. The classification procedure was repeated 100 times, each time after randomly shuffling (permutating) the sample labels. The P-value for each classification accuracy was then obtained by the percentage of permutation runs in which the accuracy achieved was greater than the classification accuracy achieved with the original non-permutated data. The same prediction methodology, accuracy and P-value determination were applied to several other metabolic parameters—conversion ratio, milk yield, milk energy, milk lactose, milk fat, milk protein, body conditioning score, pH and DM intake. For each metabolic parameter prediction test, the cows were separated into two groups, by the physiological parameter's mean value (Supplementary Data 4).
For each physiological index, receiver operation characteristics curves and area under curve (AUC) measures were obtained based on the average of 1000 k-nearest neighbors cross-validation iterations. The analysis was performed with the Metrics class that is part of the SKLEARN python machine-learning framework.
Recruitment to microbial genomes and metabolic pathways
Reads from each sample were subsampled according to the sample with lowest number of reads (21 486 100). The reads from each sample were aligned using burrows-wheeler alignment tool to a data set of 59 microbial genomes downloaded from NCBI using burrows-wheeler alignment tool with 98% identity and default parameters. Reads were also recruited to metabolic pathways of the significantly different metabolites (P<0.05) using the same method. Our database consisted of all possible KEGG enzymes for each metabolic pathway. The EC numbers used for each metabolic pathway are described in Supplementary Table S1.
The existence of the propionate production acrylate pathway in the genomes of the examined lactate utilizers Selenomonas ruminantium and Anaerovibrio lipolyticus was additionally tested by blasting them against all possible KEGG enzymes belonging to the acrylate pathway (EC 1.3.8.7, 2.8.3.1 and 4.2.1.54) using a threshold of above 70% identity, 70% alignment length of the subject gene and E⩽10−5.
Results
Construction of a rumen metagenome reference data set
To determine whether there are microbiome features that are associated with the cow's energetic efficiency, the individual feed efficiency of 146 Holstein Friesian cows was first determined. Each animal was automatically monitored for multiple parameters used to calculate feed efficiency (using the RFI approach). For further analyses, the upper and lower 25% of the animals that exhibited extreme feed efficiency values were chosen, for a total of 78 animals—40 efficient and 38 inefficient (Supplementary Figure S1 and Supplementary Data 1). Metagenomic DNA samples of these animals' rumen microbiomes were subjected to 16S rRNA gene sequencing and whole-genome shotgun sequencing. The metagenomics reads of all samples were pooled and assembled, and the predicted genes served as a reference data set (see Materials and methods section). The metagenome contained 96.72% bacterial sequences, 1.73% archaeal sequences and 1.34% eukaryotic sequences, similar to what was previously described for rumen microbiome metagenomes (Brulc et al., 2009). None of the eukaryotic sequences showed significance in the analyses.
Microbiome features differ and can predict feed efficiency phenotype
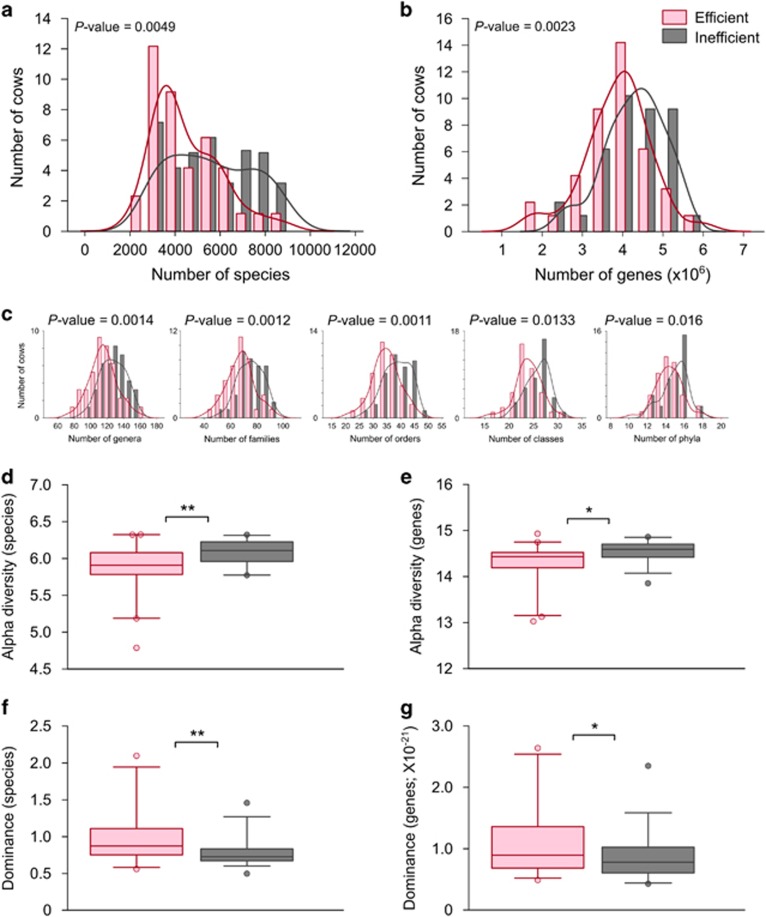
A comparison of microbiome richness across the animals revealed significantly lower richness in the efficient cows' microbiomes in both species (P=0.0049) and gene content (P=0.0023; Figures 1a and b). The differences in taxon richness were apparent up to the phylum level (Figure 1c), further stressing the intensity of this phenomenon. Taxon composition and gene content were derived from two different procedures of sequencing and analysis, and therefore the agreement between these findings highlights the robustness of the observation. The differences in richness were also accompanied by significantly lower diversity and higher dominance in the efficient animals' microbiomes at the species and gene levels (P<0.01 and P<0.05, respectively, for both diversity and dominance; Figures 1d–g and Supplementary Table S2). These differences were apparent up to the family level (Supplementary Figure S5). These differences in microbiomes of efficient and inefficient cows begged the question of whether microbiome features could be used as markers for the feed efficiency trait.
Figure 1.
Community parameters of efficient and inefficient cows' microbiomes. (a, b) Microbiome richness. Species (based on 16S amplicon sequencing) (a) and gene (based on metagenomics sequencing) (b) counts were calculated and expressed as simple richness. Kernel density of the efficient and inefficient histograms emphasizes the different distribution of counts in each microbiome group. P-values of the difference in richness between efficient and inefficient cows are shown. (c) Microbiome richness at different phylogenetic levels. (d, e) Alpha diversity (Shannon index) measurements according to species (d) and genes (e). (f, g) Dominance of the microbiome according to species (f) and genes (g). Data are expressed as mean±s.e.m. Wilcoxon rank-sum, *P<0.05, **P<0.01.
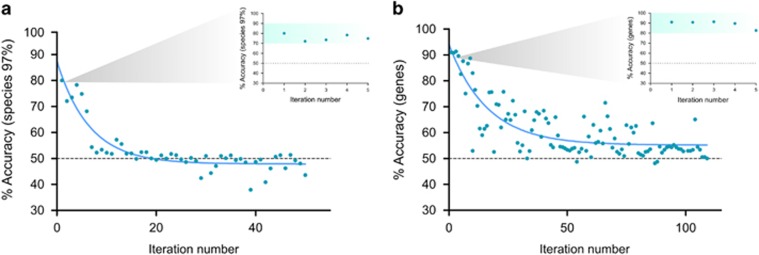
Thereupon, the species and gene composition of the rumen microbiomes were used to successfully predict the animals' feed efficiency phenotypes with up to 91% accuracy using the k-nearest neighbors algorithm (Aha, 1997). For the feature selection process, a Fisher's exact test was used to measure differences in presence/absence between microbiomes of efficient and inefficient animals. The species and genes were ranked separately according to their P-values in ascending order and divided into bins of 100 features to be used for prediction. Each bin was tested for its ability to predict high or low feed efficiency. The mean prediction accuracy was calculated using cross-validation for each bin (1000 iterations). The first species bin's prediction accuracy was 80%, while the first gene bin reached an accuracy of 91% (Figure 2). The species prediction accuracy declined to 50% (accuracy of a random guess) after the fifth bin, whereas the decline in prediction accuracy for the genes followed a much more moderate slope, with the first four predictive bins at above 90% accuracy with highly significant P-values. These differences in the slope of prediction accuracy could stem from the fact that each species represents a single genome containing thousands of genes therefore declining more rapidly compared to bins composed of hundreds of single genes. Supplementary Data 5 and 6 contain the identity of species and genes of the first five bins.
Figure 2.
Feed efficiency predictions according to species and genes. Species (a) and genes (b) that differed in presence/absence between efficient and inefficient cows were ranked according to their P-values and grouped into bins of 100. The bins were used as predictive features for the RFI feed efficiency parameter using the k-Nearest Neighbors (KNN) algorithm with k=3. Each iteration used a different bin as predictive features, in ascending P-value order. Inset in both graphs represents the first five prediction accuracy values (permutations of random classes shuffling, P-value=0.009).
The microbiome features were also highly predictive of other physiological parameters, such as milk lactose content and milk yield (Supplementary Figures S6 and S7). The sensitivity and specificity of the predictive bins was further assessed by performing receiver operating characteristic analysis for the first five bins, for both the species and genes data of each physiological parameter (Supplementary Figures S8 and S9). This analysis showed high sensitivity and specificity of the predictions of the host physiological traits based on these microbiome features, as the AUC index had high values that are considered to be good for the species data, and excellent for the genes data (AUC>0.8, AUC>0.9 respectively). This high prediction accuracy indicated that the differences in microbiome gene content and taxonomic composition could be used to classify and predict the cow's energetic efficiency.
Microbiome metabolic activity varies in cows with different feed efficiencies
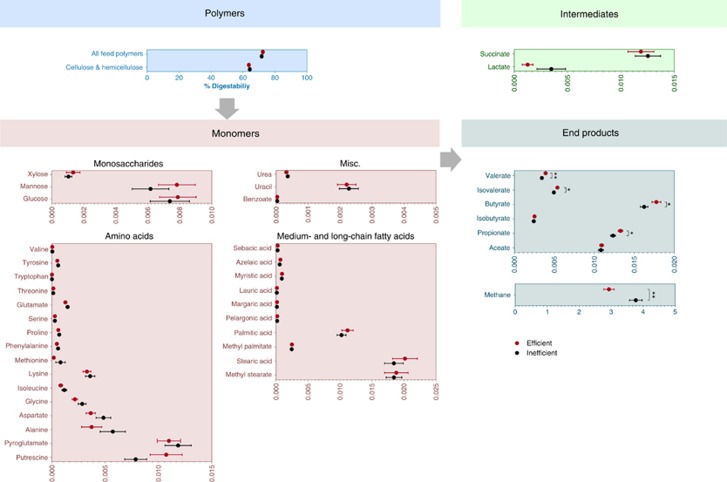
Diversity, richness and dominance are key ecological determinants that, when altered in a given ecosystem, are expected to have a marked effect on its functionality (Hooper et al., 2005). Hence, following the findings of evident differences in these parameters (Figure 1, Supplementary Figure S5 and Supplementary Table S2), the functionality of the rumen ecosystem was further investigated. Several microbial activity assays, as well as a series of 41 metabolites, were targeted and measured, representing the processes and products of different trophic levels of the rumen microbiome from efficient and inefficient cows, starting from degradation of the ingested plant fiber to the end products (Figure 3).
Figure 3.
Metabolome and microbial activity of rumen microbiomes of efficient and inefficient cows. In-vivo and in-vitro digestibility methods were performed on rumen fluid of efficient and inefficient cows in addition to extraction, identification and quantification of 41 different metabolites by GC and gas chromatography mass spectrometry. These metabolites were normalized to the organic matter content of the rumen fluid from which they were extracted. Metabolites are organized according to trophic levels. Multiple hypothesis correction with 9999 permutations was performed individually for each metabolic or activity test using the t-statistic (Materials and methods section). Data are expressed as mean±s.e.m. *P<0.05, **P<0.01.
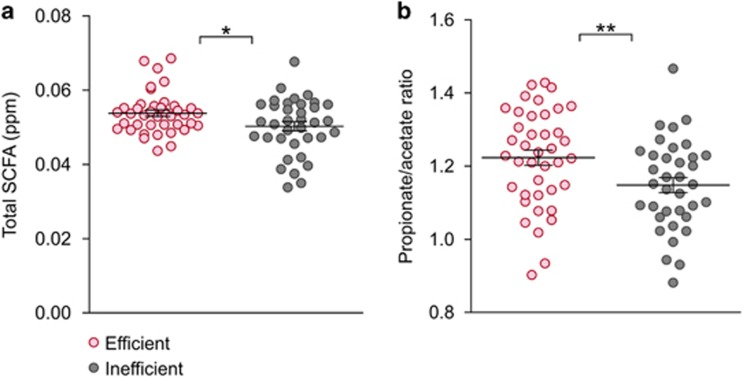
Significant differences were discovered in most SCFAs. Out of the six SCFAs measured, four—propionate, butyrate, valerate and isovalerate—were at higher concentrations in the rumen of efficient cows (Figure 3, metabolic end products and Supplementary Table S3). In addition, the total concentration of SCFAs was higher in the efficient animals showing an increase of 10% between the two efficiency groups (P<0.01; Figure 4a). These differences are considered to have a marked effect on animal productivity, given that approximately 70% of the net energy requirements of the animal are supplied by SCFAs (Seymour et al., 2005).
Figure 4.
SCFA concentration in rumen fluids of efficient and inefficient cows. (a) Total SCFA concentrations in efficient and inefficient rumen samples. (b) Propionate/acetate ratio in the efficient and inefficient rumen samples. Data are expressed as mean±s.e.m. *P<0.05, **P<0.01.
Interestingly, the propionate-to-acetate ratio in the efficient animals was also significantly higher than in the inefficient ones (P<0.05; Figure 4b); an increase in this ratio is associated with a decline in methane production and increased energy retention by cattle (Russell, 1998). This finding was congruent with the measurements of the microbiomes' methanogenesis potential, where it was evident that the efficient cows' microbiomes produce significantly less methane than their inefficient counterparts (P<0.01; Figure 3, metabolic end products). The finding of higher concentrations of SCFAs and lower methane emission from the efficient rumen microbiomes is consistent with the notion that propionate and butyrate production competes with methanogenesis for hydrogen and presents an alternative mechanism that serves as an electron sink (Ungerfeld, 2015). The production of more SCFAs and less methane by the efficient cows' microbiomes is in agreement with the higher energetic efficiency.
Our analysis did not reveal any significant differences in the microbiomes' ability to degrade the plant cell wall in the diet, in vitro or in vivo (Figure 3, polymers and Supplementary Figure S10).
Differential abundance of rumen microbes and metabolic pathways
The lower diversity and higher dominance in gene content and taxonomic composition apparent in the microbiomes of efficient cows, together with changes in metabolite assortments, suggested that the flux through collective metabolic pathways is different in microbiomes of this efficiency group. This raised the hypothesis that this may be due to changes in the occupancy of specific rumen microbial niches, defined by metabolic and physical characteristics, by functional groups that differ in their resource demands or output products.
To explore this hypothesis, a permutative Wilcoxon rank-sum test was conducted in which gene and taxonomic profiles were compared between the microbiomes of efficient and inefficient animals (see Materials and methods section). Overall, 18 species and 34 166 genes differentiated the microbiomes of efficient and inefficient cows (Supplementary Figures S11 and S12, and Supplementary Data 7 and 8); of these, 2 species and 227 genes were more abundant in efficient cows. These species and genes were not only differentially abundant in cows with different RFI values, but were also significantly correlated to the intensity of the phenotype (Figure 5a and Supplementary Data 7). The lower numbers of species and genes that were more abundant in the efficient cows' microbiomes are compatible with the higher dominance and lower richness in species and gene composition of these microbiomes. The annotation and analysis of the differentiating genes against the KEGG database (Kanehisa et al., 2011) were also in agreement with these findings, as well as with the metabolomic analysis. Among the KEGG pathways and resultant metabolites that were enriched in the inefficient cows' microbiomes were enzymes from the protein digestion and absorption category, amino-acid biosynthesis and the methane metabolism category (ko numbers of these pathways are detailed in Supplementary Table S4). Furthermore, a significantly lower number of KEGG pathways were enriched in the efficient cows' microbiomes, resulting in a significantly lower number of potential products (Supplementary Figures S13 and S14, Supplementary Data 9).
Figure 5.
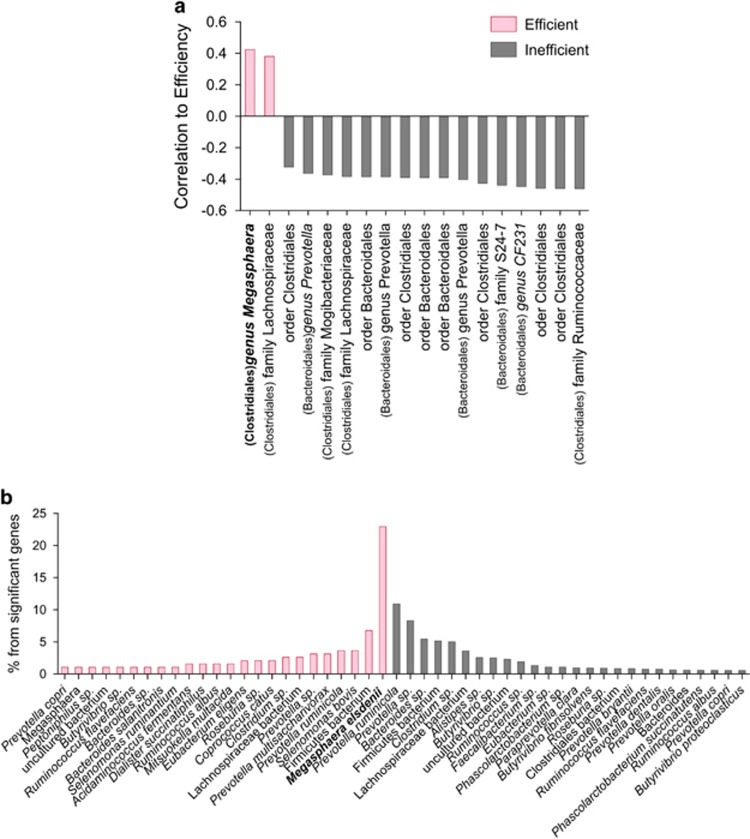
Taxonomic annotations of species and genes enriched in each microbiome group. (a) Spearman's correlation of significantly enriched species to the feed efficiency parameter. The annotations are presented at the lowest phylogenetic level obtained, as well as at the order level in parentheses. (b) The distribution of the phylogenetic annotations of genes enriched in each of the microbiome groups. Phylogenetic annotations above a threshold of 2% are presented.
These findings suggest that there is more diverse use of resource compounds, such as dietary proteins, pyruvate, acetyl-CoA and hydrogen, in the inefficient cows' microbiomes, resulting in a more diverse array of produced metabolites, some of which affect the animal's energy harvest in a negative manner or cannot be utilized by the animal for its energy requirements. In the efficient cows' microbiomes, the use of these compounds is dominated by a limited number of metabolic pathways that are more relevant and valuable for the energy needs of the animal.
The phylogenetic annotations of genes that were enriched in the efficient cows' microbiomes were dominated by the rumen bacterial species Megasphaera elsdenii, a highly potent utilizer of lactate for the production of butyrate and propionate (Figure 5b). This annotation, or any other closely related annotation, did not appear in the inefficient cow microbiomes' enriched genes. Overall, the inefficient cows' microbiomes were less dominated by a specific taxon unique to that microbiome group (Figure 5b), further supporting the hypothesis of higher dominance of specific functional groups in the microbiomes of efficient cows. This was also reinforced by the annotation of the two species that were significantly more abundant in the efficient cows' microbiomes in the 16S rRNA gene analysis. One annotation that appeared exclusively in this group was of the genus Megasphaera. The other abundant species belonged to the family Lachnospiraceae, which also had a representative in the species that were more abundant in the inefficient cows' microbiomes (Figure 5a).
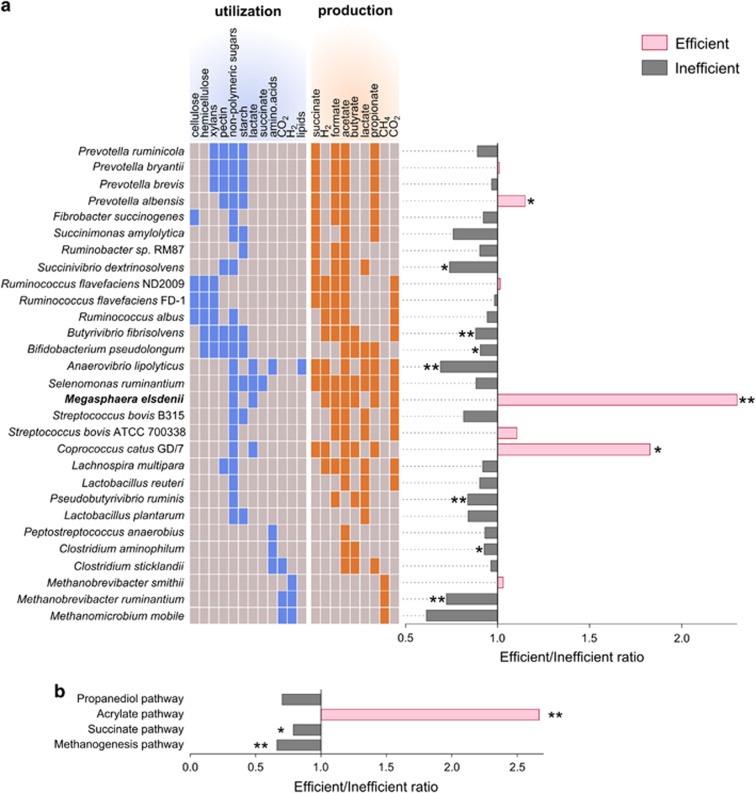
M. elsdenii was also highly enriched in the efficient cows' microbiomes using a different genomic analysis, in which reads from all samples were aligned to a database of 59 sequenced rumen and gut microbial genomes that are known to be involved in various metabolic processes and were also identified in the previous analysis. Here again, inefficient microbiomes were significantly enriched in several microbial genomes, among them Methanobrevibacter ruminantium (P<0.01), a methanogenic archaeon of the most abundant genus in the rumen (Figure 6a and Supplementary Figure S15). This exploration was further expanded by asking whether these observations are true not only for genomes of specific microbes but for all possible KEGG enzymes belonging to rumen end product metabolic pathways by using the same read-alignment approach (see Materials and methods section). In agreement with the previous results, the methanogenesis pathway was significantly enriched in the inefficient cows' microbiomes (P<0.01). Out of all examined pathways for propionate production, only the acrylate pathway that utilizes lactate to propionate was enriched in the efficient cows' microbiomes (P<0.01; Figure 6b). It should be noted that this pathway is encoded in the genome of M. elsdenii (Prabhu et al., 2012) and Coprococcus catus (Reichardt et al., 2014), which were both found by the analyses to be significantly enriched in efficient animals' microbiomes (Figures 5 and 6a, Supplementary Figure S15), and not in the other examined lactate utilizing microbial genomes (S. ruminantium and A. lipolyticus). Furthermore, reads aligned to this pathway are predominantly annotated as M. elsdenii and C. catus, however, annotations of Clostridium propionicum and Clostridium botulinum were also detected (Supplementary Figure S16). This highlights the acrylate pathway as the main contributor to the increase in propionate and decrease in lactate observed in the metabolomic analysis of the efficient cows' microbiome group (Figure 3).
Figure 6.
Microbiome features enriched in each microbiome group. (a) Reads from each sample were aligned to sequenced genomes of known rumen microorganisms using the burrows-wheeler alignment tool. The ratios between alignments of efficient/inefficient samples to each genome are presented. The utilization and production of metabolites for each microorganism based on the known growth characteristics (Holdman and Moore, 1974; Russell and Rychlik, 2001; Duncan et al., 2009) are colored in blue and orange, respectively. (b) Reads from each sample were aligned to KEGG enzymes of different metabolic pathways using the burrows-wheeler alignment tool. Propanediol, acrylate and succinate pathways are different propionate production pathways. The ratios between alignments of efficient/inefficient samples to each pathway are presented. Data are expressed as ratio of means. Permutations t-test,*P<0.05, **P<0.01.
Discussion
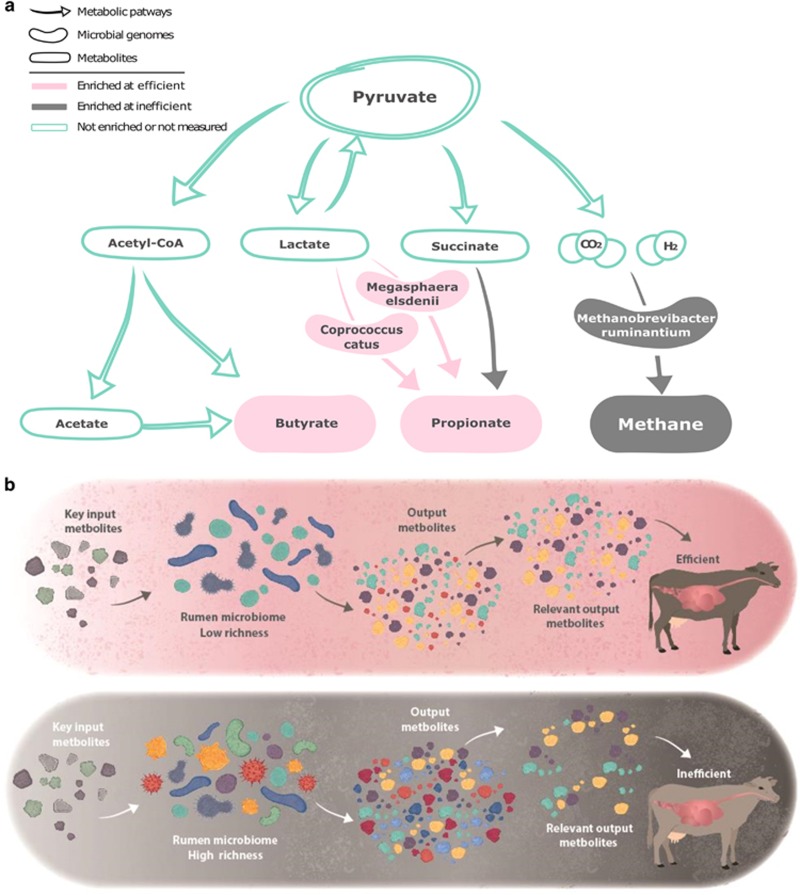
Our analyses of multiple animals feeding on the exact same diet and kept under the same conditions showed that there are large variations in the individual animals' ability to extract energy from their feed. These variations are tightly linked to several microbiome features that include a decrease in richness and increase in dominance of taxonomic and coding capacity in the efficient cow's microbiome. They are reflected as changes in this ecosystem's functionality, where changes in the dominance of specific functional components affect the overall availability of ecosystem goods that are of high value to the hosting animal. Higher microbiome richness and changes in specific functional groups have been recently described to affect host productivity in plants (Wagg et al., 2014) as well as humans, where lower diversity and richness have been associated with higher energy harvest from feed in obese humans (Turnbaugh et al., 2009; Le Chatelier et al., 2013). A possible explanation for this phenomenon could stem from a more diverse use of resource compounds in the inefficient cow's microbiomes that are enriched in species, genes and KEGG pathways resulting in a wider array of output metabolites (Figures 3, 6 and 7); this was also confirmed by significantly more KEGG output metabolites (Supplementary Figures S13 and S14). On the other hand, in the efficient cow's microbiome, simpler metabolic pathway networks result in increased dominance of specific functional components, which leads to higher concentrations of ecosystem goods that are relevant to the host (Figure 7b). Therefore, the efficient microbiomes are less complex but more specialized to support the host's energy requirements.
Figure 7.
Consolidated results and model. (a) Consolidation of results from the metabolomics, genome and pathway recruitment analyses. Green: pathways and metabolites that were not significantly different or that were not assessed. Pink: enriched in efficient microbiomes. Grey: enriched in inefficient microbiomes. (b) Proposed model. From left to right: identical key input metabolites are ingested by the cow and presented to either an efficient microbiome (top panel) with lower richness and diversity, or an inefficient microbiome (bottom panel) with higher richness and diversity. Differences in richness result in the production of different metabolites. The efficient microbiome produces a smaller range of output metabolites than the inefficient microbiome, however, with larger amounts of relevant output metabolites, which are available for the animal's energetic needs.
This notion is exemplified by the finding of higher concentrations of SCFAs, which are valuable to the hosting animal. SCFAs are absorbed through the rumen wall to serve the energetic needs of the animal; propionate, for example, is the main precursor for gluconeogenesis in animals (Russell and Wilson, 1996; Mizrahi, 2011; Mizrahi, 2013). This is not the case with methane as the energy retained in it cannot be absorbed by the animals, and is lost to the atmosphere. Such metabolic changes are usually achieved via the use of antibiotic growth promoters that increase the animal's feed efficiency (Duffield et al., 2012). Such is the case with monensin, a carboxylic polyether ionophore that selectively affects some of the rumen microbes, therefore changing the structure of the rumen microbiome and subsequently the ratio of SCFAs in the rumen, increasing propionic acid and decreasing methane production (Thornton and Owens, 1981; Callaway et al., 2003; Weimer et al., 2008; Duffield et al., 2012). It has been shown that when administered orally, monensin improves feed efficiency in cattle in a dose-dependent manner. Therefore, it has been used for this purpose extensively since its approval for cattle agriculture in the mid-1970s (Duffield et al., 2012). This effect of rumen microbiome manipulation achieved via antibiotics further supports the connection of the rumen microbiome with the feed efficiency of the animal.
Here we show that these metabolomic changes are the outcome of microbiome structures that are naturally occurring and are highly correlated with, and predictive of the feed efficiency phenotype. Therefore, these findings could be harnessed to reduce the use of antibiotic growth promoters in agriculture. Such prospects, together with examination of the true causal nature of the rumen microbiome on its host, would be made possible when non-antibiotic rumen microbiome manipulation techniques, as well as adult cow germ-free facilities, will be better established.
From an ecological perspective, the lower abundance of methanogenesis pathways and methanogenic archaea in the efficient cow's low-richness microbiome concurs with the notion that processes that are performed by small taxonomic groups, such as the methanogenic archaea that occupy only small percentages of the rumen microbiome, are more sensitive to changes in diversity and richness (Hooper et al., 1995). These changes are usually accompanied by occupation and dominance of the available niche by different species using the same resources (Grime, 1998). Such is the case with M. elsdenii and C. catus, independently found to be enriched in the efficient animals' microbiomes in different analyses (Figures 5 and 6a, Supplementary Figure S15), which use electrons for the production of the valuable SCFAs propionate and butyrate, thereby diverting them from reducing CO2 to methane (Prabhu et al., 2012; Ungerfeld, 2015). A similar principle was shown to apply in Tammar wallabies, where Succinivibrio bacteria were suggested to utilize hydrogen for the production of succinate, therefore lowering its availability for methanogenesis (Pope et al., 2011). It is also possible that the Lachnospiraceae detected in the efficient animals' microbiomes are butyrate producers (Figure 5) and are contributing further to this effect (Louis and Flint, 2009; Meehan and Beiko, 2014). Nevertheless, as other SCFAs are enriched in this microbiome group and most of the carbon flux in the system goes to acetyl-CoA, formate or hydrogen and carbon dioxide, it is likely that more genes and pathways are involved in this effect.
A cardinal point that emerges from our findings is that the functional characteristics of a small number of species can have a large impact on community structure and ecosystem functioning. This, in turn, can change the productivity of the supraorganism—the host and its residing rumen microbiome.
These findings could potentially be harnessed to increase the production of food resources for mankind in a more sustainable manner, as well as to understand the underlying ecological mechanisms that govern complex microbial communities and their interactions with their hosts.
Acknowledgments
This research was supported by Binational Agricultural Research and Development fund project number 362032713, the Israel Science Foundation project number 1313/13 and the European Research Council under the European Union's Horizon 2020 research and innovation program, project number 640384.
Data deposition
Assembled Contigs were deposited in the MG-RAST server under IDs 4547164.3 and 4548996.3.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aha DW. (1997) Lazy Learning. Kluwer Academic Publishers: Norwell, MA, USA. [Google Scholar]
- Ajmone-Marsan P, Garcia JF, Lenstra JA. (2010). On the origin of cattle: how aurochs became cattle and colonized the world. Evol Anthropol Issues News Rev 19: 148–157. [Google Scholar]
- Archer JA, Richardson EC, Herd RM, Arthur PF. (1999). Potential for selection to improve efficiency of feed use in beef cattle. Aust J Agric Res 50: 147–162. [Google Scholar]
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodological) 57: 289–300. [Google Scholar]
- Bradford GE. (1999). Contributions of animal agriculture to meeting global human food demand. Livest Prod Sci 59: 95–112. [Google Scholar]
- Brulc JM, Antonopoulos DA, Miller ME, Wilson MK, Yannarell AC, Dinsdale EA et al. (2009). Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA 106: 1948–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway TR, Edrington TS, Rychlik JL, Genovese KJ, Poole TL, Jung YS et al. (2003). Ionophores: their use as ruminant growth promotants and impact on food safety. Curr Issues Intest Microbiol 4: 43–51. [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA et al. (2003). The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31: 442–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JC. (1986) Statistics and Data Analysis in Geology. Wiley: New York. [Google Scholar]
- Duffield TF, Merrill JK, Bagg RN. (2012). Meta-analysis of the effects of monensin in beef cattle on feed efficiency, body weight gain, and dry matter intake. J Anim Sci 90: 4583–4592. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Thomson JM, Flint HJ. (2009). The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol 11: 2112–2122. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Grime J. (1998). Benefits of plant diversity to ecosystems: immediate, filter and founder effects. J Ecol 86: 902–910. [Google Scholar]
- Halachmi I, Edan Y, Maltz E, Peiper U, Moallem U, Brukental I. (1998). A real-time control system for individual dairy cow food intake. Comput Electron Agr 20: 131–144. [Google Scholar]
- Harper D. (1999) Numerical Palaeobiology. Wiley: London. [Google Scholar]
- Henderson G, Cox F, Ganesh S, Jonker A, Young W et al Collaborators GRC. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep 5: 14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herd RM, Arthur PF. (2009). Physiological basis for residual feed intake. J Anim Sci 87: E64–E71. [DOI] [PubMed] [Google Scholar]
- Hernandez-Sanabria E, Goonewardene LA, Wang Z, Durunna ON, Moore SS. (2012). Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl Environ Microb 78: 1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G et al. (2011). Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331: 463–467. [DOI] [PubMed] [Google Scholar]
- Holdman LV, Moore WEC. (1974). New genus, Coprococcus, twelve new species, and emended descriptions of four previously described species of bacteria from human feces. Int J Syst Evol Micr 24: 260–277. [Google Scholar]
- Hooper DU, Hawksworth D, Dhillion S. (1995) Microbial diversity and ecosystem processes. Global biodiversity assessment UNEP. Cambridge University Press: Cambridge, pp 433–443. [Google Scholar]
- Hooper DU, Chapin FS, Ewel JJ, Hector A, Inchausti P, Lavorel S et al. (2005). Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol Monogr 75: 3–35. [Google Scholar]
- Jami E, Israel A, Kotser A, Mizrahi I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J 7: 1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami E, Mizrahi I. (2012). Composition and similarity of bovine rumen microbiota across individual animals. PLoS One 7: e33306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami E, White BA, Mizrahi I. (2014). Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One 9: e85423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Dudoit S, van der Laan MJ. (2005). Multiple testing procedures: the multtest package and applications to genomics. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S (eds), Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer: New York, USA, pp 249–271. [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. (2011). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittelmann S, Pinares-Patiño CS, Seedorf H, Kirk MR, Ganesh S, Mcewan JC et al. (2014). Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS One 9: e103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch RM, Swiger LA, Chambers D, Gregory KE. (1963). Efficiency of feed use in beef cattle. J Anim Sci 22: 486–494. [Google Scholar]
- Kultima JR, Sunagawa S, Li J, Chen W, Chen H, Mende DR et al. (2012). MOCAT: a metagenomics assembly and gene prediction toolkit. PLoS One 7: e47656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541–546. [DOI] [PubMed] [Google Scholar]
- Li D, Lewinger JP, Gauderman WJ, Murcray CE, Conti D. (2011). Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genet Epidemiol 35: 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippke H, Ellis WC, Jacobs BF. (1986). Recovery of indigestible fiber from feces of sheep and cattle on forage diets. J Dairy Sci 69: 403–412. [Google Scholar]
- Louis P, Flint HJ. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 294: 1–8. [DOI] [PubMed] [Google Scholar]
- McDougall E. (1948). Studies on ruminant saliva. 1. The composition and output of sheep's saliva. Biochem J 43: 99. [PMC free article] [PubMed] [Google Scholar]
- McMichael AJ, Powles JW, Butler CD, Uauy R. (2007). Food, livestock production, energy, climate change, and health. Lancet 370: 1253–1263. [DOI] [PubMed] [Google Scholar]
- Meehan CJ, Beiko RG. (2014). A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol 6: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'souza M, Olson R, Glass E, Kubal M et al. (2008). The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform 9: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi I. (2011) The role of the Rumen microbiota in determining the feed efficiency of dairy cows. In: Rosenberg E, Gophna U (eds), Beneficial Microorganisms in Multicellular Life Forms. Springer Science & Business Media: Berlin Heidelberg, pp 203–210. [Google Scholar]
- Mizrahi I. (2013) Rumen symbioses. In: Rosenberg E, DeLong F, Lory S, Stackebrandt E, Thompson F (eds), The Prokaryotes. Springer Science & Business Media: Berlin Heidelberg, pp 533–544. [Google Scholar]
- Moore SS, Mujibi FD, Sherman EL. (2009). Molecular basis for residual feed intake in beef cattle. J Anim Sci 87: E41–E47. [DOI] [PubMed] [Google Scholar]
- National Research Council. (2001) Nutrient Requirements of Dairy Cattle. National Academy Press: Washington: DC, USA. [Google Scholar]
- Nkrumah JD, Okine EK, Mathison GW, Schmid K, Li C, Basarab JA et al. (2006). Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J Anim Sci 84: 145–153. [DOI] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, Mcinnes P, Wang L, Schloss JA et al. (2009). The NIH human microbiome project. Genome Res 19: 2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope PB, Smith W, Denman SE, Tringe SG, Barry K, Hugenholtz P et al. (2011). Isolation of Succinivibrionaceae implicated in low methane emissions from Tammar wallabies. Science 333: 646–648. [DOI] [PubMed] [Google Scholar]
- Prabhu R, Altman E, Eiteman MA. (2012). Lactate and acrylate metabolism by Megasphaera elsdenii under batch and steady-state conditions. Appl Environ Microbiol 78: 8564–8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. (2007). NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce J, Arias J, Bowman P, Davis S, Macdonald K, Waghorn G et al. (2012). Accuracy of genomic predictions of residual feed intake and 250-day body weight in growing heifers using 625,000 single nucleotide polymorphism markers. J Dairy Sci 95: 2108–2119. [DOI] [PubMed] [Google Scholar]
- Reichardt N, Duncan SH, Young P, Belenguer A, Mcwilliam Leitch C, Scott KP et al. (2014). Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J 8: 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho M, Tang H, Ye Y. (2010). FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res 38: e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JB. (1998). The importance of pH in the regulation of ruminal acetate to propionate ratio and methane production in vitro. J Dairy Sci 81: 3222–3230. [DOI] [PubMed] [Google Scholar]
- Russell JB, Rychlik JL. (2001). Factors that alter rumen microbial ecology. Science 292: 1119–1122. [DOI] [PubMed] [Google Scholar]
- Russell JB, Wilson DB. (1996). Why are ruminal cellulolytic bacteria unable to digest cellulose at low pH? J Dairy Sci 79: 1503–1509. [DOI] [PubMed] [Google Scholar]
- Saleem F, Bouatra S, Guo AC, Psychogios N, Mandal R, Dunn SM et al. (2013). The bovine ruminal fluid metabolome. Metabolomics 9: 360–378. [Google Scholar]
- Seymour WM, Campbell DR, Johnson ZB. (2005). Relationships between rumen volatile fatty acid concentrations and milk production in dairy cows: a literature study. Anim Feed Sci Tech 119: 155–169. [Google Scholar]
- Shi W, Moon CD, Leahy SC, Kang D, Froula J, Kittelmann S et al. (2014). Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res 24: 1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson DM, Weimer PJ. (2007). Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol 75: 165–174. [DOI] [PubMed] [Google Scholar]
- Thornton JH, Owens FN. (1981). Monensin supplementation and in vivo methane production by steers. J Anim Sci 52: 628–634. [DOI] [PubMed] [Google Scholar]
- Thornton PK. (2010). Livestock production: recent trends, future prospects. Philos Trans R Soc Lond B Biol Sci 365: 2853–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley J, Terry R. (1963). A two-stage technique for the in vitro digestion of forage crops. Grass Forage Sci 18: 104–111. [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al. (2009). A core gut microbiome in obese and lean twins. Nature 457: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerfeld EM. (2015). Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: a meta-analysis. Front Microbiol 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest PJ, Robertson JB, Lewis BA. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci 74: 3583–3597. [DOI] [PubMed] [Google Scholar]
- Wagg C, Bender SF, Widmer F, van der Heijden MG. (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc Natl Acad Sci USA 111: 5266–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RJ, Rooke JA, Mckain N, Duthie CA, Hyslop JJ, Ross DW et al. (2015). The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer PJ, Stevenson DM, Mertens DR, Thomas EE. (2008). Effect of monensin feeding and withdrawal on populations of individual bacterial species in the rumen of lactating dairy cows fed high-starch rations. Appl Microbiol Biotechnol 80: 135–145. [DOI] [PubMed] [Google Scholar]
- Westfall PH, Young SS. (1993) Resampling-Based Multiple Testing: Examples and Methods for P-Value Adjustment. John Wiley & Sons: New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.