Abstract
The evolution of complex traits is hypothesized to occur incrementally. Identifying the transitions that lead to extant complex traits may provide a better understanding of the genetic nature of the observed phenotype. A keystone functional group in wastewater treatment processes are polyphosphate accumulating organisms (PAOs), however the evolution of the PAO phenotype has yet to be explicitly investigated and the specific metabolic traits that discriminate non-PAO from PAO are currently unknown. Here we perform the first comprehensive investigation on the evolution of the PAO phenotype using the model uncultured organism Candidatus Accumulibacter phosphatis (Accumulibacter) through ancestral genome reconstruction, identification of horizontal gene transfer, and a kinetic/stoichiometric characterization of Accumulibacter Clade IIA. The analysis of Accumulibacter's last common ancestor identified 135 laterally derived genes, including genes involved in glycogen, polyhydroxyalkanoate, pyruvate and NADH/NADPH metabolisms, as well as inorganic ion transport and regulatory mechanisms. In contrast, pathways such as the TCA cycle and polyphosphate metabolism displayed minimal horizontal gene transfer. We show that the transition from non-PAO to PAO coincided with horizontal gene transfer within Accumulibacter's core metabolism; likely alleviating key kinetic and stoichiometric bottlenecks, such as anaerobically linking glycogen degradation to polyhydroxyalkanoate synthesis. These results demonstrate the utility of investigating the derived genome of a lineage to identify key transitions leading to an extant complex phenotype.
Introduction
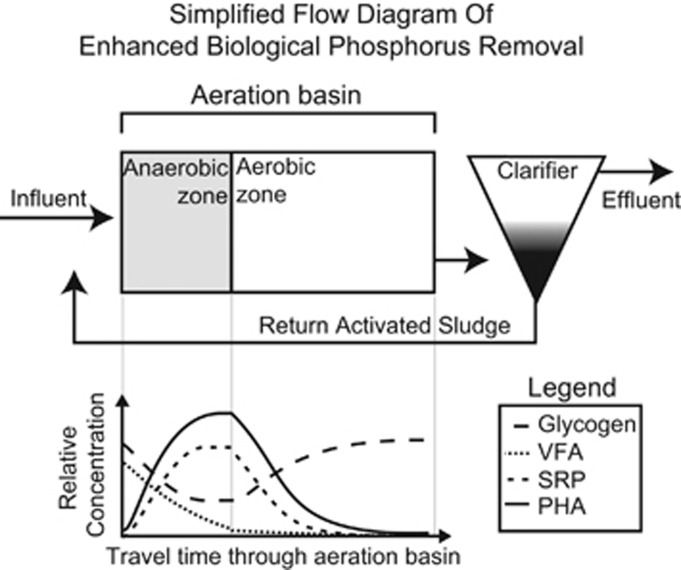
The ability of some microbes to store large quantities of intracellular polyphosphate (polyP) is an important trait that differentiates them from other closely related taxa. Engineers exploit this trait to increase the efficacy of phosphorus (P) removal from wastewater by designing treatment systems that select for a combination of physiological capabilities that comprise a distinct and complex phenotype (Seviour et al., 2003). The term ‘polyphosphate accumulating organism' (PAO) is typically used to distinguish these organisms from others that may not display the complete phenotype needed for successful enrichment in wastewater treatment systems. One of the most abundant PAO in wastewater treatment systems is Candidatus Accumulibacter phosphatis (henceforth Accumulibacter) of the family Rhodocyclaceae (He et al., 2008; Mielczarek et al., 2013). Accumulibacter's ability to sequester P during cyclic ‘anaerobic feast' and ‘aerobic famine' conditions characteristic of enhanced biological P removal (EBPR) wastewater treatment processes (Figure 1) is the result of a complex metabolism that cycles three storage polymers: polyP, glycogen and polyhydroxyalkanoate (PHA) (Seviour et al., 2003).
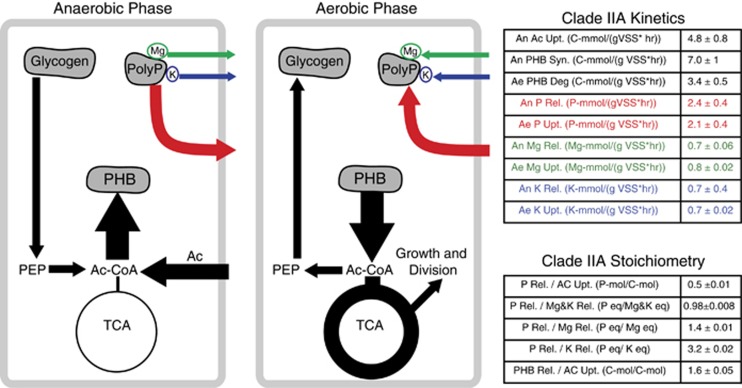
Figure 1.
A defining feature of many biological wastewater treatment systems is the recycling of microbial biomass, commonly called activated sludge (AS). Recycling AS provides two features: (1) a mechanism for achieving high densities of microorganisms and (2) a mechanism for the ecological selection of organisms based on their growth characteristics and physiology. A common design is to have an anaerobic basin preceding an aerobic basin. Under these conditions, polyphosphate accumulating organisms (PAO) are selected for, enhancing the phosphorus removal capabilities of the system. This configuration is commonly referred to as Enhanced Biological Phosphorus Removal (EBPR). Anaerobic zone: in the absence of a terminal electron acceptor, volatile fatty acids (VFA) are transported into the cell and stored as polyhydroxyalkanoates (PHA) with a concomitant release of P and degradation of glycogen. Aerobic zone: carbon stored as PHA is used to drive growth, cell division, P-uptake and glycogen synthesis. At the end of the Aerobic zone, the activated sludge is settled in a clarifier and removed from the system to be recycled, further processes or disposed. Figure adapted from McMahon and Read (2013).
Despite the broad phylogenetic distribution of these polymers across all domains of life (Wilkinson, 1963; Kornberg et al., 1999; Jendrossek, 2009), the hallmark anaerobic/aerobic cycling phenotype displayed by Accumulibacter is uncommon, and the specific metabolic traits discriminating the PAO phenotype from non-PAO phenotype have yet to be explicitly defined. Identifying these traits and the boundaries of the PAO phenotype within the Rhodocyclaceae will provide a better understanding of the evolution of the PAO phenotype and hence a metabolic framework for the further identification and monitoring of additional bacterial groups responsible for key EBPR functions. Ultimately, this knowledge may be harnessed for more strategic design of wastewater treatment systems that implement the PAO phenotype.
Several challenges exist in identifying the traits that differentiate PAO from non-PAO. First, the term ‘PAO' is sometimes used indiscriminately, to discuss organisms that store polyP without displaying the full phenotype described above, encompassing many diverse metabolisms that result in polyP synthesis (Grillo, 1979; Rao et al., 1998; García Martín et al., 2006; Kristiansen et al., 2013; Zhang et al., 2015). Second, there are inherent complications in determining which organisms in mixed communities are PAOs because high-throughput techniques to identify organisms that cycle polyP are currently unavailable. Finally, once a putative PAO is identified, the metabolism that allows polyP storage must be determined; an arduous assignment in mixed communities. To date, only two major PAOs have been identified and functionally characterized; organisms belonging to the genera Accumulibacter (Crocetti et al., 2000; Zilles et al., 2002) and Tetrasphaera (Maszenan et al., 2000; Kong et al., 2005).
Although the hunt to identify, characterize and differentiate the phenotypes of PAOs continues, a recent proliferation of available Accumulibacter genomes (García Martín et al., 2006; Flowers et al., 2013; Skennerton et al., 2015) has made it possible to investigate the genomic evolution of the Accumulibacter-type PAO phenotype for the first time. Many simultaneous evolutionary processes such as horizontal gene transfer (HGT), point mutations, re-arrangements, recombination, expansions and contractions contribute to genome evolution (Ochman et al., 2005; Hao and Golding, 2006; Touchon et al., 2009; Zaremba-Niedzwiedzka et al., 2013; Nowell et al., 2014). The result of these concomitant evolutionary processes is that the pan-genome of related bacteria may be categorized into lineage-specific (gene families unique to specific genotypes), flexible (gene families with irregular occurrence across numerous genotypes) and core genomes (gene families present in all genotypes) (Ochman et al., 2000; Hacker and Carniel, 2001). The core genome represents the uniting genomic features, while the flexible and lineage-specific genome provides insight into the evolution of population structure and the speciation process of closely related strains (Ochman et al., 2000; Kettler et al., 2007; Polz et al., 2013; Chan et al., 2015). The categorization of genome content in this way may provide insight on the boundaries of the PAO phenotype within the Rhodocyclaceae and bring us closer to a more high-throughput way to identify other lineages with similar polyP cycling traits.
To investigate ancient evolutionary events, such as the emergence of the PAO phenotype in Accumulibacter, it is common to use ancestral state reconstructions (Schluter et al., 1997; Larsson et al., 2011; Latysheva et al., 2012). Ancestral state reconstructions allow the division of a genome into ancestral and derived traits. Traits present before the last common ancestor (LCA) of a lineage are considered ancestral, whereas those that have transitioned to new states, such as genes acquired through HGT, are considered derived traits. Previous studies on the evolution of bacterial metabolic networks using gene gain and loss analysis have demonstrated the importance of derived traits from HGT in contributing to the expansion of metabolic network capabilities (Pál et al., 2005). Thus, by inferring the laterally acquired derived traits of a lineage using ancestral genome reconstructions, the molecular evolution resulting in the emergence of novel phenotypes may be studied.
Although ancestral reconstructions provide evidence of the changes that occurred in the past, extant phenotypes may be used to deduce the evolutionary pressures which were selected for these changes (Connell, 1980). In this investigation we merged these lines of evidence; using kinetic and stoichiometric values for Accumulibacter Clade IIA, coupled with the reconstructed ancestral states of 26 genomes in the family Rhodocyclaceae (10 Accumulibacter, 4 Dechloromonas, 8 Thauera, 3 Azoarcus, 1 Zooglea,). Using the resulting inferred ancestral states, the Accumulibacter Clade IIA genome, CAP2UW1 (García Martín et al., 2006), was parsed into an ancestral, derived, flexible and lineage-specific genome. A phylogenetic analysis on derived genes within KEGG pathways/COG Inorganic Ion Transport and Metabolism was conducted to determine which were laterally derived. Using these discrete inferred categories, an evolutionary model of Accumulibacter was constructed and integrated with measured phenotypic data, providing the first comprehensive analysis of the molecular evolution of the polyP accumulating phenotype in Accumulibacter. Our results reveal that the laterally derived genes of Accumulibacter's LCA contain numerous adaptations important to the PAO phenotype, demonstrating the utility of investigations into the derived genome of an organism for identifying key adaptations that lead to its present phenotype.
Materials and methods
Accession numbers
The genomes used for the ancestral genome reconstruction were downloaded from version 4.0 of the Integrated Microbial Genomes (IMG) (http://img.jgi.doe.gov) database (IMG genome ID, January 20th 2015) (Markowitz et al., 2012) and included all 10 publically available Accumulibacter genomes at that time (2556921090, 2100351004, 2556921089, 644736333, 2556921085, 2556921086, 2556921084, 2556921087, 2556921083, 2556921088), and 16 out-group genomes including Azoarcus (2563366569, 2518645585, 639633007), Dechloromonas (2506520024, 2528768215, 2506520023, 637000088) Thauera (2579778713, 2531839537, 2556921623, 2531839206, 2531839276, 2531839205, 2537561694, 643692051) and Zooglea (2596583626), all within the Rhodocyclaceae family.
Orthologous gene clusters
Reconstruction of ancestral states inferred by gene gain/loss analysis requires the assignment of orthologous gene clusters. Initial all vs all BLAST of each Rhodocyclaceae genome was conducted using BLASTP 2.2.28+ (Altschul et al., 1990) with parameters -seg yes, -soft_masking true, -use_sw_tback, -evalue 1e-5, which have been shown to be sensitive for identifying orthologs (Moreno-Hagelsieb and Latimer, 2008). BLAST results were then filtered to query coverage of 75% and percent identity of 70%. Finally, orthologous genes clusters were identified using MCL version 14–137 with an inflation value of 1.1 (van Dongen, 2000).
Phylogenetic analysis of pan orthologs
Seventy-four pan single-copy genes were identified to construct a robust phylogeny for gene gain/loss analysis. These pan single-copy genes were aligned using the linsi option in MAFFT version 7.215 (Katoh and Standley, 2013) and masked in Gblocks version 0.91b (Castresana, 2000) permitting gaps in up to half of the taxa. A phylogenetic analysis was then conducted on the concatenated 74 aligned, masked pan orthologs using RAxML version 8.0.14 with a protein-specific amino-acid substitution model identified using RAxML (PROTGAMMAAUTO) with 100 bootstraps (Zaremba-Niedzwiedzka et al., 2013).
Gene flux analysis
Gene flux analysis was conducted using Count (Csurös, 2010) based on the matrix of orthologous gene family abundance obtained from MCL, as well as the phylogeny obtained from the concatenated 74 orthologous single-copy genes. For each gene family, Wagner parsimony with a gene gain/loss penalty of 2 (Pál et al., 2005; Zaremba-Niedzwiedzka et al., 2013) was used to infer the most parsimonious ancestral states. Inferred patterns of gene gain and loss were mapped onto the orthologous single-copy gene tree. Ancestral genes were defined as those inferred gained before the Accumulibacter LCA node. Derived genes were defined as those inferred gained at the Accumulibacter LCA node. Laterally derived genes were defined as derived genes with phylogenetic evidence of HGT. Lineage-specific genes were defined as those that were unique to a single Accumulibacter genome. Flexible genes were defined as those represented in more than a single Accumulibacter genome but not core. Pseudogenes were omitted from the analysis.
Core genome determination
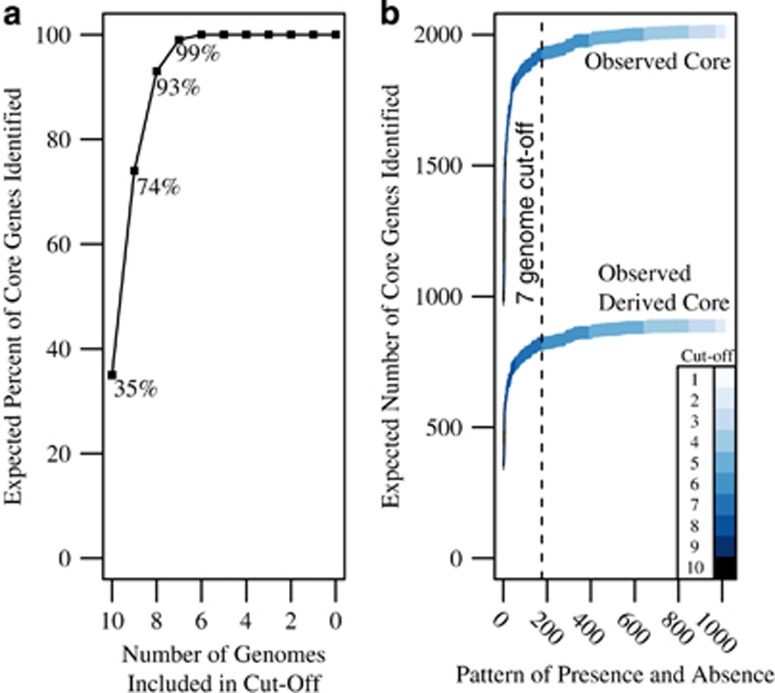
The core of a set of genomes is dependent on the quantity and phylogenetic distance of the genomes included in an analysis such that as these values increase, the number of core genes identified decreases (Lefébure and Stanhope, 2007; Ozer et al., 2014). When genomes are incomplete, it is common to determine a threshold number of genomes in which a gene must be observed in order to call it ‘core'. This cut-off may be based on the average estimated completeness of the genomes within the analysis (Ghylin et al., 2014). Genome completeness estimates may also be used to calculate the probability of observing a pattern of presence and absence given that a gene family is core. The probability of each pattern is simply the product of the estimated genome completeness of all genomes a gene family is present in, multiplied by 1 minus the completeness estimate if a gene family is absent (Table 1). Summing these products for a given abundance thus provides information on the percentage of core genes one would expect to identify at each cut-off (Table 1).
Table 1. (A) The estimated completeness for the 10 Accumulibacter genomes in this study. (B) The expected probability of observing pattern of presence and absence across the 10 Accumulibacter genome set.
|
(A) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genome | AW09 | AW06 | CAPSK01 | AW08 | AW07 | AW12 | CAP2UW1 | CAP1UW1 | AW11 | AW10 |
| Completeness | 0.92 | 0.92 | 0.87 | 0.91 | 0.89 | 0.88 | 1 | 0.85 | 0.89 | 0.88 |
|
(B) | |||
|---|---|---|---|
| Patterns | Calculation | Expected probability | Sum |
| PPPPPPPPPP | 0.92 × 0.92 × 0.87 × 0.91 × 0.89 × 0.88 × 1 × 0.85 × 0.89 × 0.88 | 0.3494 | 0.349 |
| APPPPPPPPP | 0.08 × 0.92 × 0.87 × 0.91 × 0.89 × 0.88 × 1 × 0.85 × 0.89 × 0.88 | 0.0304 | |
| PAPPPPPPPP | 0.92 × 0.08 × 0.87 × 0.91 × 0.89 × 0.88 × 1 × 0.85 × 0.89 × 0.88 | 0.0304 | |
| PPAPPPPPPP | 0.92 × 0.92 × 0.13 × 0.91 × 0.89 × 0.88 × 1 × 0.85 × 0.89 × 0.88 | 0.0522 | |
| PPPAPPPPPP | 0.92 × 0.92 × 0.87 × 0.09 × 0.89 × 0.88 × 1 × 0.85 × 0.89 × 0.88 | 0.0346 | |
| PPPPAPPPPP | 0.92 × 0.92 × 0.87 × 0.91 × 0.11 × 0.88 × 1 × 0.85 × 0.89 × 0.88 | 0.0432 | 0.391 |
| PPPPPAPPPP | 0.92 × 0.92 × 0.87 × 0.91 × 0.89 × 0.12 × 1 × 0.85 × 0.89 × 0.88 | 0.0476 | |
| PPPPPPAPPP | 0.92 × 0.92 × 0.87 × 0.91 × 0.89 × 0.88 × 0 × 0.85 × 0.89 × 0.88 | 0.0000 | |
| PPPPPPPAPP | 0.92 × 0.92 × 0.87 × 0.91 × 0.89 × 0.88 × 1 × 0.15 × 0.89 × 0.88 | 0.0617 | |
| PPPPPPPPAP | 0.92 × 0.92 × 0.87 × 0.91 × 0.89 × 0.88 × 1 × 0.85 × 0.11 × 0.88 | 0.0432 | |
| PPPPPPPPPA | 0.92 × 0.92 × 0.87 × 0.91 × 0.89 × 0.88 × 1 × 0.85 × 0.89 × 0.12 | 0.0476 | |
Given the completeness estimates, it is possible to calculate the expected probability of observing pattern of presence and absence across the 10 Accumulibacter genome set. For example, here we present 11 patterns of presence and absences and demonstrate how the probability of each pattern was calculated. The first pattern represents a gene that is present in all genomes. The 10 patterns below represent the possibilities for a single absence. Presence is indicated by a 'P', and absence is indicated by an 'A' or in bold for the calculation. For each pattern, if a gene family was present in a genome, the product of the completeness estimates for those genome was calculated. This was then multiplied by the product of 1 minus the completeness estimate of genomes in which the gene family was absent. The sum of these probabilities within a particular number of genomes may then be calculated. Presence and absence is binomial, therefore, there are 210 (1024) possible patterns.
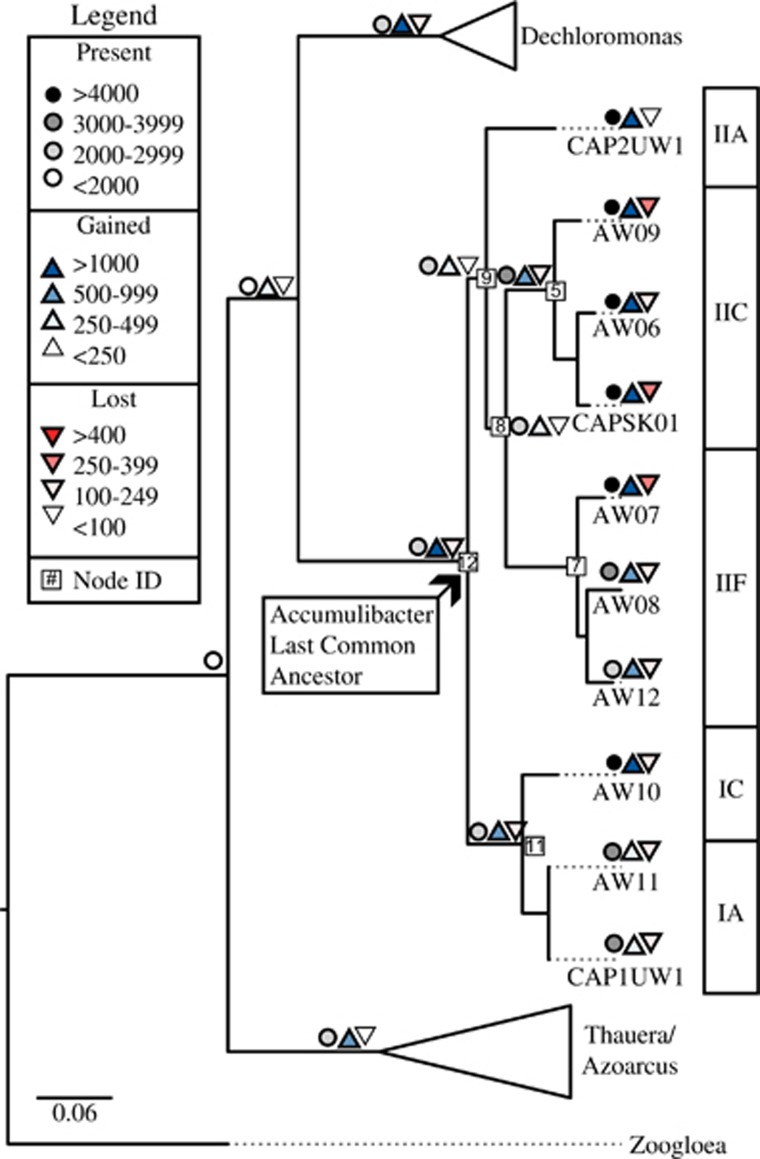
Using these probabilities, we would expect genome-number cutoffs of 10, 9, 8 and 7 to identify approximately 35%, 74%, 93% and 99% of core genes in this analysis. Therefore, a cutoff value of seven genomes was chosen (Figure 2, Supplementary Spreadsheet 6). This cutoff does not take into account phylogenetic relationships; a gene absent in all three Clade IIC genomes would be permissible within this cutoff. Therefore, we developed a second criterion that a core gene must have been inferred at the LCA of Accumulibacter (node 12, Figure 3) and retained at each internal Accumulibacter node (nodes, 11, 9, 5, 7 and 8, Figure 3). As the focus of this analysis was on the flux of new genetic content, gene duplications and reduction events were not included in downstream analysis.
Figure 2.
(a) The expected percentage of core gene families identified for each pattern of presence and absence was calculated using the genome completeness estimates. Using these probabilities, a cutoff of seven genomes is expected to identify 99% of all core genes. This cutoff was used in conjunction with ancestral state reconstructions to determine the core genome of the Accumulibacter lineage. Only gene families that were inferred at the LCA of Accumulibacter and all internal nodes (for example, not lost until a terminal node) and were present in seven or more genomes were considered core in this analysis. (b) The observed number of core and derived core gene families using variable cutoffs. Each potential core gene family was sorted based on the number of genomes they were present in and then on the expected frequency of the pattern. Next, the cumulative sum of each additional pattern was calculated as patterns of increasing likelihood were added. The cutoff at seven genes is demarcated with a dotted line.
Figure 3.
Gene gain (blue triangle), loss (red triangle) and presence (circle) at each node in the Accumulibacter lineage with other Rhodocyclaceae branches collapsed. The gains and losses were inferred using Count implementing Wagner parsimony with a cost of 2 for gain and 1 for loss.
Metabolic function analysis
To determine which pathways had undergone the greatest evolutionary change during the transition between non-PAO to PAO hypothesized to have occurred at the Accumulibacter LCA, the relative proportion of ancestral, derived, flexible and lineage-specific genes within the CAP2UW1 genome in each metabolic pathway annotated within KEGG (Kanehisa et al., 2014), and those annotated as Inorganic Ion Transport and Metabolism from the COG database (Tatusov et al., 2000), were determined by dividing the number of genes parsed into each category by the total number of genes in a respective metabolic pathway.
Identifying laterally derived genes with KEGG annotations
Two approaches exist to infer HGT, parametric (nucleotide composition, structural features, genomic context) and phylogenetic (test of topologies, top sequence matches) (Ravenhall et al., 2015). Here a phylogenetic approach was used to determine whether the new gene families were laterally acquired. First, each derived gene was queried against the NR database (Release 7 May 2015) (Pruitt et al., 2007) using the following BLASTP parameters [-max_target_seqs 100 -evalue 1E-6] and the top 100 BLAST results were retained. For each set of BLAST results, the number of classes, orders, families and the number of non-Accumulibacter Rhodocyclaceae represented in the top 100 BLAST results was calculated. A gene was considered to be a putative HGT if less than 10% of the top 100 BLAST hits were assigned to Rhodocyclaceae, a cutoff based on the average number of non-Accumulibacter Rhodocyclaceae hits in the top 100 BLAST hits of all 238 derived genes with KEGG annotations (Supplementary Spreadsheet 5, Sheets 1 and 2). In addition, a sensitivity analysis was conducted to compare results obtained using thresholds of 5% and 0% of top 100 BLAST hits (Supplementary Spreadsheet 5, Sheets 4 and 5). Derived genes that were classified as HGT are henceforth referred to as laterally derived genes.
Reactor operation, population characterization, kinetics and stoichiometry
In order to obtain phenotypic measurements for Accumulibacter Clade IIA, sequencing batch reactors were operated under standard anaerobic feast/aerobic famine conditions (García Martín et al., 2006) and monitored for Accumulibacter population dynamics using fluorescent in situ hybridization (FISH) and quantitative PCR (He et al., 2007; Flowers et al., 2009). Total Accumulibacter was monitored with PAOMIX probes (Crocetti et al., 2000), and Clade I and II were monitored with Acc-I-444 and Acc-II-444, respectively, as previously described (Flowers et al., 2009). Due to the cross hybridization potential of Acc-II-444 with Clade IIA, IIC and IID (Flowers et al., 2009), qPCR using specific primers (He et al., 2007) was conducted to confirm Clade IIA enrichment. Counterstaining of cells was achieved with 4′,6-diamidino-2-phenylindole (DAPI). During periods of high enrichment (>80% Clade IIA Accumulibacter abundance), soluble phosphate, total suspended solids, volatile suspended solids and acetate were measured using previously described methods (Flowers et al., 2009). In addition, PHA analysis was performed using a GC-MS as outlined previously (Comeau et al., 1988). Calcium, magnesium and potassium were analyzed using a VISTA-MPX CCD Simultaneous ICP-OES (Varian Ibérica SL, Madrid, Spain). Kinetic rates for acetate, soluble P, polyhydroxybutryate (PHB), polyhydroxyvalerate (PHV), magnesium, potassium and calcium, over an anaerobic/aerobic cycle were calculated based on linear rates of change observed for each analyte and were normalized to the VSS and Accumulibacter Clade IIA relative abundance.
Results
Identification of orthologous gene clusters
The pan Rhodocyclaceae genome contains 40 263 orthologous gene families and the pan Accumulibacter genome contains 14 702 orthologous gene clusters (Supplementary Spreadsheet 1, Sheets 1 and 3). The largest portions of the pan Rhodocyclaceae and pan Accumulibacter genome are gene families present in only a single genome (65% and 57%, respectively). Only a small fraction (2%) of gene families identified were present in 23 or more of the 26 genomes included in this study to define the pan Rhodocyclaceae genome. A larger fraction of the pan Accumulibacter genome (7%) was present in 8 or more of the 10 Accumulibacter genomes. Within each genome, genes families tended to be present in a single copy with very few paralogs. Non-paralogous genes represented 92% and 87% of the pan Rhodocyclaceae and Accumulibacter genomes, respectively. A more detailed breakdown of the pan Rhodocyclaceae and Accumulibacter genomes is provided in Supplementary Spreadsheet 1 (Sheets 2 and 4).
Gene flux analysis
To determine the flux (that is, loss and gain) of gene families in the Accumulibacter pan genome, a phylogenetic tree was constructed using concatenated pan orthologs and gene gain and loss was inferred using Wagner parsimony implemented in Count (Csurös, 2010). Figure 3 depicts the flux of the 14 702 orthologous gene clusters represented in the pan Accumulibacter genome. Supplementary Figure 1 provides bootstrap values and Supplementary Figure 2 provides specific gain/loss/presence values at each node. Approximately 17% (2459 orthologous gene clusters) of the pan Accumulibacter genome orthologous gene clusters were inferred present in the Accumulibacter LCA; 1477 of these orthologous gene clusters were ancestral (present before the Accumulibacter LCA), whereas 1106 were derived (gained at the Accumulibacter LCA) (Figure 3). In order to filter out non-core genes, a genome-number cutoff of 7, which is expected to include 99% of core genes, was determined (Figure 2). After filtering non-core orthologous gene clusters based on this definition, a total of 1918 core genes clusters were present at the Accumulibacter LCA, of these 1090 were ancestral and 828 were derived. More stringent cutoffs for the number of genomes needing to contain a gene cluster in order to declare it ‘core' led to lower estimates: cutoffs of 8, 9 and 10 would have identified 95%, 81% and 51% of these core gene families, respectively (Supplementary Spreadsheet 6).
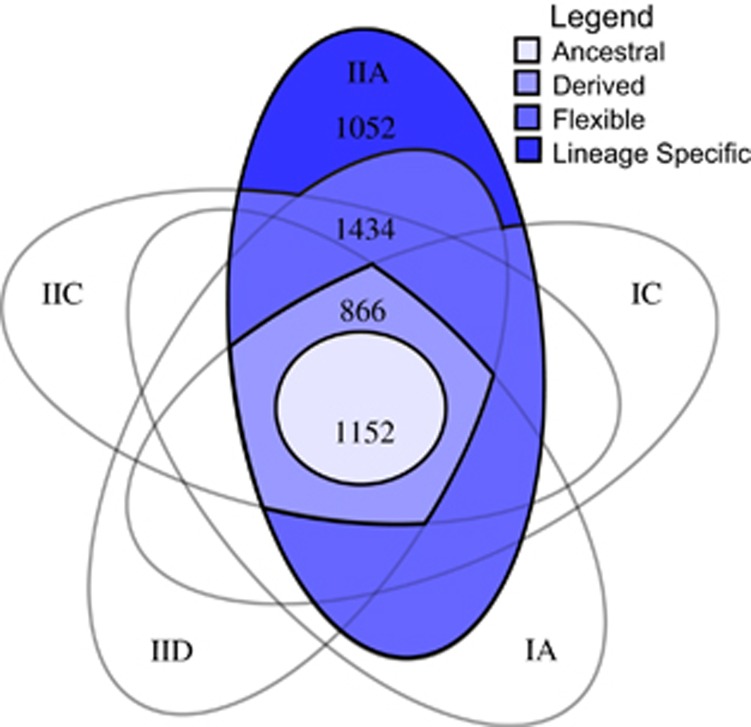
Based on the gene clusters identified within the pan Accumulibacter genome, each gene within the CAP2UW1 genome was coded as ancestral, derived, lineage-specific or flexible. After correcting for non-core genes, 45% (2018) of genes in CAP2UW1 belonged to gene clusters that were inferred present at the Accumulibacter LCA. Of these, ancestral genes represented ~25% (1152) and derived genes represented ~19% (866) of the CAP2UW1 genome. Flexible genes represented 31% (1434) and lineage-specific genes represented nearly 23% (1052) of the CAP2UW1 genome (Figure 4). Supplementary Spreadsheet 2 provides additional details about the presence, gains and losses of genes, and the discrete categories to which they were assigned.
Figure 4.
Five-way Venn diagram depicting the number of ancestral, derived, flexible and lineage-specific genes within the CAP2UW1 Accumulibacter genome. Although many comparative genomic studies use similar plots, they generally do not highlight the shared derived genome, which we have shown to be important in understanding both the ecology and evolution of the lineage.
Substrate uptake and internal flux kinetics and stoichiometry
The phenotype of an organism is important in deducing the selective pressures that shaped an organism's evolutionary history. However, defining phenotypic traits for uncultivated organisms is difficult partially due to the unknown contributions of flanking community members to overall ecosystem function. To minimize the contribution of the flanking community to measured parameters, chemical analysis was only conducted on days of high enrichment of a single Accumulibacter clade as demonstrated by FISH and qPCR. On these four dates (17 July 2013 to 19 July 2013 and 23 July 2013), Accumulibacter relative abundance was on average 84% of total DAPI-stained cells. FISH results showed that Clade IA and IIA accounted for 0.1–0.2% and 95–99% of total Accumulibacter, respectively. The presence of Clade IIC and IID, as measured by qPCR, was negligible, with Clade IIA representing >99% of Clade II sequences (Supplementary Spreadsheet 7). Thus, Clade IIA dominated the community, accounting for 95–99% of total PAOs. Supplementary Table 1 shows the relative abundance during the dates of kinetic and stoichiometric investigation.
Numerous previous investigations have measured key parameters of bulk PAOs, but these investigations have only sporadically included specific molecular identification of the dominant Accumulibacter clade being investigated (Welles et al., 2015), and only in very recent studies. Here the average kinetic parameters and stoichiometric values of Clade IIA for acetate, PHB, P, magnesium and potassium were estimated based on results from high enrichment cultures (Figure 5, Supplementary Spreadsheet 3). The calcium uptake/release and PHV synthesis/degradation measurements were negligible and are not reported. Anaerobic acetate uptake was measured at a rate of 4.8±0.8 C-mmol (gVSS-h)−1. Anaerobic PHB synthesis was higher than acetate uptake rates and aerobic PHB degradation (7.0±1 and 3.4±0.5 C-mmol (gVSS-h)−1, respectively). In contrast, the uptake and release of P (2.4±0.4 and 2.1±0.4 P-mmol (gVSS-h)−1, respectively), Mg (0.7±0.06 and 0.8±0.02 Mg-mmol (gVSS-h)−1, respectively), K (0.7±0.4 and 0.7±0.02 K-mmol (gVSS-h)−1, respectively) were relatively stable across both anaerobic and aerobic phases. Mg and K were the dominant counter cations for PolyP with sum molar equivalents of ~1 (0.98±0.008 P eq./Mg and K eq.).
Figure 5.
A simplified biochemical model and the measured kinetic and stoichiometric parameters for phosphorus, magnesium, potassium, acetate and polyhydroxybutyrate (PHB) of Accumulibacter Clade IIA. Calcium and polyhydroxyvalerate (PHV) were measured but showed negligible changes over an anaerobic/aerobic cycle.
Evolution of Accumulibacter metabolic pathways
To determine the influence of genetic flux on metabolic pathways at the Accumulibacter LCA, genes annotated within KEGG pathways (Kanehisa et al., 2014) were parsed into ancestral, derived, flexible and lineage-specific portions (Supplementary Spreadsheet 4). Similar delineation was conducted for and all inorganic ion transporters identified in the COG database (Tatusov et al., 2000). The KEGG categories of Carbohydrate metabolism, Lipid metabolism, Metabolism of other Amino Acids, and the COG category Inorganic Ion Transport and Metabolism showed the highest proportions of derived genes (Figure 6a). In contrast, the KEGG categories of Translation, Amino Acid Metabolism and Nucleotide Metabolism showed high proportions of ancestral genes (Figure 6a). Specific pathways also showed differential contributions from ancestral and derived genes. Within the broad KEGG category of Carbohydrate metabolism, the starch and sucrose, glycolysis/gluconeogenesis and pyruvate metabolic pathways had a high proportion of derived genes, whereas ancestral genes dominated the citric acid cycle (TCA cycle) and glyoxylate/dicarboyxlate pathways (Figure 6b). In Lipid Metabolism, the glycerophospholipids sub-category contained a high abundance of derived genes, whereas fatty acid degradation had a higher ancestral composition. For Inorganic Ion Transport and Metabolism, P, K, Mg and Fe all showed high proportions of derived genes, especially P (Figure 6b).
Figure 6.
(a) The contribution of ancestral and derived genes to broad KEGG maps and the COG categories involved in Inorganic ion transport and metabolism. (b) The contribution of ancestral and derived genes to specific KEGG pathways and COG categories involved in specific inorganic ion transporters.
Phylogenetic analysis of derived genes
Determination of orthologous gene clusters is conditional on the BLAST and MCL parameters chosen. Strict parameters (for example, high percent identity and coverage requirements) will increase the number of clusters identified, potentially splitting true clusters that have diverged sufficiently through mutation. In contrast, loose parameters will result in grouping of potentially non-orthologous clusters. To address these concerns, we used relatively strict parameters and then manually differentiated between derived genes that likely arose through sufficient accumulation of mutations and those that arose through HGT. To do so, we conducted a phylogenetic analysis for each of the 238 derived genes involved in KEGG pathways or in the COG category Inorganic Ion Transport and Metabolism. The average number of non-Accumulibacter Rhodocyclaceae BLAST hits per gene was ~10%, with 135 genes being identified with fewer than this average (Supplementary Spreadsheet 5). Based on these results, a separate classification within the derived portion of the CAP2UW1 genome was distinguished as ‘laterally derived' genes. Figure 7 depicts an evolutionary model of Accumulibacter with color-coded ancestral, derived, laterally derived, flexible and lineage-specific genes. A sensitivity analysis demonstrated that if the threshold were lowered to 5% of the top 100 BLAST hits, 106 (79%) of these genes would be identified as having arisen by HGT, including 82% of the genes in the evolutionary model (Supplementary Spreadsheet 5).
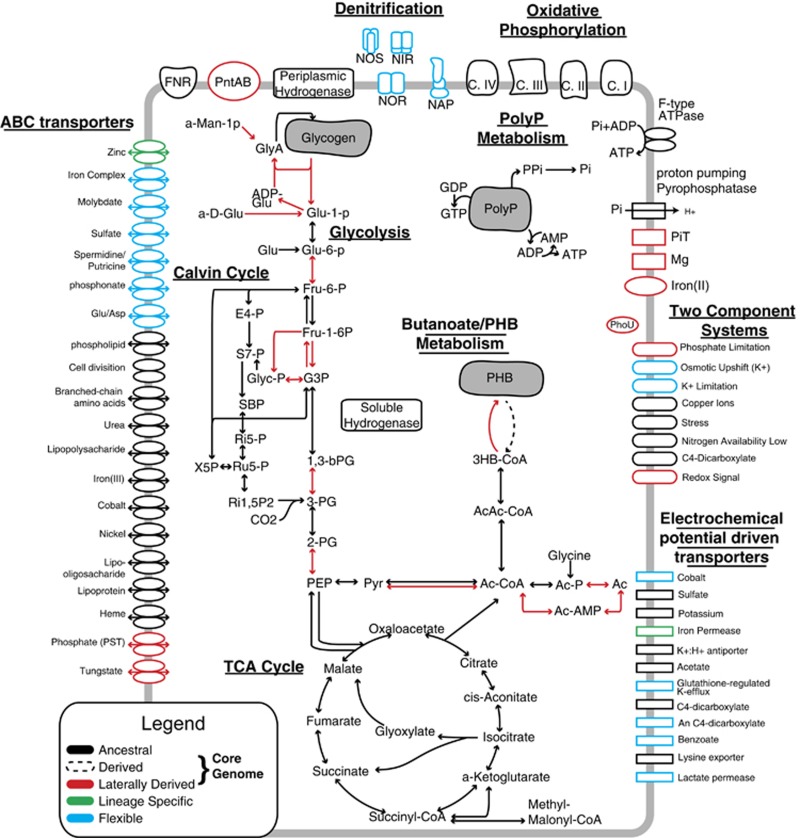
Figure 7.
An evolutionary model of CAP2UW1 depicting ancestral, laterally derived, flexible and lineage-specific genes. Ac, acetate; AcAc-CoA, acetoacetyl-CoA; Ac-CoA, acyl-CoA; Ac-AMP, acetyl AMP; Ac-P, acetyl-P; ADP-Glu, adenosine 5-diphosphoglucose; CDPD, cytidine diphosphate diacylglycerol; C.I, complex I oxidative phosphorylation; C.II, complex II oxidative phosphorylation; C.III, complex III oxidative phosphorylation; C.IV, complex IV oxidative phosphorylation; E4-P, erythrose 4-phosphate; FNR, NADPH-ferredoxin reductase; Fru-1-6P, fructose 1,6-bisphosphate; Fru-6-P, fructose 6-phosphate; G3P, glyceraldehyde 3-phosphate; Glu, glucose; Glu-1-p, glucose 1-phosphate; Glu-6-P, glucose 6-phosphate; Gly, glycogen; GlyA, glycogen amylose; Glyc-P, glycerone-P; Long Chain FA, long chain fatty acid; PE, phosphatidylethanolamine; PEP, phosphoenolpyruvate; PGP, 1,2-diacyl-sn-glycerol-3p; pntAB, proton-translocating transhydrogenase; PolyP, polyphosphate; PPP, pyrophosphate-energized proton pump; Ptd-L-Ser, phosphatidylserine; Pyr, pyruvate; 1,3-bPG, 1,3-bisphosphoglyceric acid; Ri15P2, ribulose 1,5P2; Ri5-P, ribose 5-phosphate; Ru5P, ribulose 5-phosphate; S7-P, sedoheptulose-7-phosphate; SBP, sedoheptulose 1,7-bisphosphate; X5P, xylulose 5-phosphate; 3HB-CoA, (R)-3-hydroxy-butanoyl-CoA; 2-PG, 2-phosphoglycerate; 3-PG, 3-phosphoglyceric acid.
One of the salient features of this evolutionary model is the abundance of laterally acquired genes involved in the distinctive carbon metabolism of the PAO phenotype, including: glycogen degradation (CAP2UW1_0254, CAP2UW1_0255, CAP2UW1_2663), glycolysis (CAP2UW1_2124–2127, CAP2UW1_2662, CAP2UW1_2666, CAP2UW1_2669, CAP2UW1_3196, CAP2UW1_0487, CAP2UW1_1890), PHB metabolism (phaC—CAP2UW1_0143, CAP2UW1_3185, CAP2UW1_3191), pyruvate ferredoxin oxidoreductase (PFOR—CAP2UW1_2510–2512) and acetate activation to acetyl-CoA (CAP2UW1_1515, CAP2UW1_2035). Another prominent laterally derived set of genes is P transport (PHO4—CAP2UW1_3785, CAP2UW1_3788) and regulation (phoR—CAP2UW1_1995; phoB- CAP2UW1_1996; phoR/phoB -CAP2UW1_1997; phoU—CAP2UW1_3786, CAP2UW1_3787, CAP2UW1_3789). Additional derived transporters arising from HGT included magnesium transport (corA—CAP2UW1_3581, CAP2UW1_2797) and ferrous iron transport (FeoA—CAP2UW1_0420; FeoB—CAP2UW1_0421, CAP2UW1_3321). Other notable HGT include genes involved in energy metabolism, such as NADP/NADPH transhydrogenase (CAP2UW1_4179—CAP2UW1_4180) and cytochrome-c oxidase (CAP2UW1_1790, CAP2UW1_1791). Finally, laterally derived genes were also identified to be involved in regulation and signaling, including two-component redox signaling (RegB—CAP2UW1_0008, RegA—CAP2UW1_0009) (Figure 7, see Supplementary Figure 4 for evolutionary model with locus tags). A prominent absence of laterally derived genes is seen in both the TCA cycle and in polyP metabolism.
Expression profiles of laterally derived genes
Recent metatranscriptomic investigations resulted in the identification of co-expressed gene clusters and of highly expressed genes in CAP2UW1 (Oyserman et al., 2015). Of the 135 putative HGT genes within the derived genome that have KEGG functional annotations, 31 genes were highly expressed. These included glycogen degradation (CAP2UW1_0255, CAP2UW1_2663), glycolysis (CAP2UW1_2124, CAP2UW1_2126–2127, CAP2UW1_2662, CAP2UW1_2666, CAP2UW1_3196, CAP2UW1_0487), PHB metabolism (CAP2UW1_3185, CAP2UW1_3191), pyruvate ferredoxin oxidoreductase (PFOR—CAP2UW1_2510–2512), ferrous iron transport (FeoA—CAP2UW1_0420) and NADP/NADPH transhydrogenase (PntAB—CAP2UW1_4179—CAP2UW1_4180) (Supplementary Spreadsheet 4, Sheet 1, Column J). Furthermore, the metatranscriptomic analysis demonstrated that of the 135 laterally derived genes identified in this study, 114 displayed co-expression patterns related to known environmental variables such as anaerobic acetate contact, including PhaC (CAP2UW1_3191) within the PHA synthesis modulon (Oyserman et al., 2015, also see Supplementary Spreadsheet 5, Sheet 2, Column Q).
Discussion
The transition from non-PAO to PAO, hypothesized to have occurred at the Accumulibacter LCA, was accompanied by significant molecular evolution in key carbon pathways, transporters, energy metabolism and regulatory elements. The changes in these pathways ranged from considerable, such as in glycolysis, to nearly no change at all such as in the TCA cycle (Figures 6a and b). Below we provide a detailed discussion of key laterally derived genes in the context of known aspects of PAO metabolism and the measured stoichiometry/kinetics of Accumulibacter Clade IIA identified in this study. In addition, we incorporate previous metatranscriptomic analyses (Oyserman et al., 2015) to postulate the relative importance of these derived genes in optimizing and linking key pathways in the Accumulibacter-type PAO phenotype. Finally, we discuss the broader implications of how these findings will change the search for additional PAO.
Acetate activation
The primary route for carbon acquisition in Accumulibacter is through the anaerobic uptake of volatile fatty acids, such as acetate, and the subsequent synthesis of the storage polymer PHA. After anaerobic acetate contact, acetate is transported into the cell via both passive and active transport (Saunders et al., 2007; Burow et al., 2008) and activated to acetyl-CoA (Figures 4 and 5). The activation of acetyl-CoA occurs either through acetyl-P or acetyl-AMP intermediates. The primary route for the activation of acetate is currently unknown, however higher relative expression of genes involved in acetyl-CoA synthetase suggest that the primary route is via acetyl-AMP (Oyserman et al., 2015). Although no laterally derived acetate transporters were identified, both routes for acetate activation contain laterally derived genes (CAP2UW1_1515 and CAP2UW1_2035) (Figure 7). Numerous copies of acetyl-CoA synthetase are found in the CAP2UW1 genome, including flexible (CAP2UW1_1069, CAP2UW1_2247, CAP2UW1_3266) and an ancestral gene (CAP2UW1_3755). Of these, the laterally derived gene had the lowest transcription rates while the ancestral copy (CAP2UW1_3755) was one of the most highly expressed genes in the CAP2UW1 genome (Oyserman et al., 2015). In contrast, no redundant copies for acylphosphatase are annotated in the CAP2UW1 genome aside from the laterally derived gene (CAP2UW1_1515) and this gene is also not highly expressed (Oyserman et al., 2015). This analysis suggests that despite containing laterally derived genes, the evolution of acetate activation at the Accumulibacter LCA may not have contributed substantially to transitioning from non-PAO to PAO.
PHB synthesis
Once acetate has been transported into the cell and activated to acetyl-CoA, it enters the PHB synthesis pathway. The synthesis of PHB (7 C-mmol (gVSS-h)−1) in Accumulibacter Clade IIA occurs at twice the rate of the degradation (3.4 C-mmol (gVSS-h)−1) and is also greater than the acetate uptake rate (4.8 C-mol (gVSS-h)−1) (Figure 5 and Supplementary Figure 5). The kinetic disparity between PHA synthesis, degradation and acetate uptake is due to the additional intracellular flux of carbon from anaerobic glycogen degradation via pyruvate, acetyl-CoA and finally to PHB. Together, these kinetic parameters suggest that a strong evolutionary pressure for rapid PHB synthesis exists. Of the three enzymes in the PHA synthesis pathway (PhaA, PhaB and PhaC), only PhaC contains laterally derived genes. Of the four copies of the PhaC gene in the CAP2UW1 genome, three of these are laterally derived (CAP2UW1_0143, CAP2UW1_3191 and CAP2UW1_3185) and two are among the most highly transcribed genes in CAP2UW1 (CAP2UW1_3191 and CAP2UW1_3185). In addition, CAP2UW1_3191 is co-expressed with a predicted PHA modulon controlled by the ancestral core regulatory protein phaR (CAP2UW1_3918) (Oyserman et al., 2015).
Thus, in contrast to the activation of the acetate to acetyl-CoA, the polymerization of 3-hydroxybutyryl-CoA to PHB is likely to occur primarily through laterally derived genes, suggesting that evolution of PHB metabolism in Accumulibacter was significant in transitioning from non-PAO to PAO. It is noteworthy that the laterally derived PhaC genes represent both class I and III PHA synthase (CAP2UW1_3191 and CAP2UW1_3185, respectively) (Yuan et al., 2001; Rehm, 2003) and that these genes were highly expressed and showed dissimilar expression profiles from each other (Oyserman et al., 2015). The dissimilar expression profiles of related but functionally divergent PhaC suggests these genes contribute differentially to the PAO metabolism of Accumulibacter, however more research is required to make such a conclusion. Regardless, dose effect (for example, numerous copies of PhaC) has been shown to increase PHA synthesis capabilities (Maehara et al., 1998).
Anaerobic reducing equivalents: glycolysis, glycogen degradation and PntAB
Anaerobic PHB synthesis requires both ATP and reducing equivalents. One strategy used by Accumulibacter to meet this demand is to use stored glycogen (Schuler and Jenkins, 1994). As noted earlier, a striking number of genes involved in glycogen degradation (starch/sucrose metabolism) and glycolysis are laterally derived genes (Figures 6 and 7). These include glycogen degradation via glucose phosphorylase (CAP2UW1_0255, CAP2UW1_2663), glucose-6-phosphate isomerase (CAP2UW1_2124), fructose-bisphosphate aldolase (CAP2UW1_2669, CAP2UW1_3196), phosphoglycerate kinase (CAP2UW1_0487), phosphopyruvate hydratase (CAP2UW1_2666) and pyruvate kinase (CAP2UW1_1890) (Figure 7).
Although glycolysis produces reducing equivalents in the form of NADH, NADPH is generally required for PHB synthesis (Peoples and Sinskey, 1989; Steinbüchel et al., 1993; Madison and Huisman, 1999; Kim et al., 2014). A recent investigation demonstrating hydrogen gas production during anaerobic acetate contact in Accumulibacter enriched bioreactors suggests the regeneration of NAD+ may represent a bottleneck in PAO metabolism that is alleviated through hydrogenase activity (Oyserman et al., 2015). Furthermore, metatranscriptomic evidence from this same study suggests that the demand for the conversion of NADH to NADPH is met by the NADPH/NADH transhydrogenase PntAB (CAP2UW1_4179, CAP2UW1_4180; Oyserman et al., 2015). Although the hydrogenases are ancestral (CAP2UW1_0999, CAP2UW1_2286), interestingly, both complexes of PntAB are laterally derived. Furthermore, these complexes are highly expressed, as well as many of the laterally derived genes involved in glycogen degradation and glycolysis (CAP2UW1_2124, CAP2UW1_2126, CAP2UW1_2127, CAP2UW1_2662, CAP2UW1_2663, CAP2UW1_2666, CAP2UW1_0255, CAP2UW1_3196, CAP2UW1_0487, CAP2UW1_1890, CAP2UW1_4179, CAP2UW1_4180; Oyserman et al., 2015). Together, this evidence suggests that considerable selective pressures to optimize the production of reducing equivalents in the form of NADPH via glycogen degradation, glycolysis and the activity of NADPH/NADH transhydrogenase existed at the LCA of Accumulibacter and is an important adaptation for the storing PHA anaerobically.
Pyruvate metabolism
Anaerobic glycogen degradation provides both ATP and NADH, but also produces abundant pyruvate that must be converted to PHB via acetyl-CoA. In general, two complexes exist that may convert pyruvate to acetyl-CoA, pyruvate-ferredoxin oxidoreductase (PFOR) and pyruvate dehydrogenase (PDH). These multi-enzyme complexes differ in that PFOR uses ferredoxin and is often coupled with hydrogen production (Chabrière et al., 1999), while PDH uses NAD+ and is inhibited by high levels of NADH (Snoep et al., 1993). Both of these complexes in CAP2UW1 are highly expressed and form separate operons (PFOR, CAP2UW1_2510-CAP2UW1_2512; pyruvate dehydrogenase CAP2UW1_1838-CAP2UW1_1840). However, because PFOR is the primary route from pyruvate to acetyl-CoA under NADH rich conditions (Patel and Roche, 1990; Blamey and Adams, 1993; Townson et al., 1996), it likely fills this role in Accumulibacter PAO metabolism, contributing to the hydrogen gas production recently reported (Oyserman et al., 2015). Interestingly, the PFOR operon in Accumulibacter is composed of laterally derived genes (Figure 7). Thus, the kinetic, evolutionary and transcriptional data all suggest that the ability to efficiently shunt pyruvate to PHB via acetyl-CoA anaerobically is an essential adaptation for the Accumulibacter-type PAO phenotype, without which a build-up of pyruvate would likely inhibit glycogen degradation and stall the anaerobic metabolism of Accumulibacter.
Phosphorus and counter cation transport
PolyP is a source of ATP in anaerobic PAO metabolism (Comeau et al., 1986). Thus, one of the key metabolic processes in Accumulibacter is the degradation and synthesis of polyP. Transport of P into and out of the cell must accompany the degradation and synthesis of polyP, as well as the transport of counter cations that are used to balance the negative charge of phosphate. Indeed, the stoichiometric analysis in this investigation demonstrates that P transport of Accumulibacter is linked to the counter cations magnesium and potassium at a nearly 1:1 molar equivalent ratio (Figure 5 and Supplementary Figure 5). Despite the obvious linkage between polyP metabolism and the transport of P, Mg and K, the evolutionary histories of these genes differ significantly. The polyP metabolism of Accumulibacter is ancestral, whereas many of the transporters involved in P (Pit CAP2UW1_3785, CAP2UW1_3788; PstS, CAP2UW1_1747 PstB, CAP2UW1_1751–1752 PstC CAP2UW1_1749) and magnesium transport (corA CAP2UW1_3581, CAP2UW1_2797) are laterally derived genes. The kinetic/stoichiometric and evolutionary data presented here suggests that an increased capability to transport P and counter cations such as Mg was an important adaptation at the Accumulibacter LCA, supporting and expanding upon previous hypotheses that inorganic P transporters may be absolutely required for the Accumulibacter-PAO phenotype (Saunders et al., 2007; Kristiansen et al., 2013; Nobu et al., 2014).
Ferrous iron transport
Iron is an essential co-factor in many enzymes, and bacteria have evolved many diverse strategies for the transport and acquisition of iron from the environment (Andrews et al., 2003; Wandersman and Delepelaire, 2004). When reducing (that is, anaerobic) environmental conditions prevail, ferrous iron predominates over ferric iron. Under these conditions, ferrous iron transport using the Feo pathway is favored over alternative ferric transporter mechanisms, such as siderophores (Cartron et al., 2006). The Feo system was laterally acquired at the Accumulibacter LCA suggesting that anaerobic demand for iron-containing enzymes, such as by the highly expressed PFOR and hydrogenases, is an important adaptation for the Accumulibacter-type PAO phenotype.
Signaling and regulation
It has been demonstrated that Accumulibacter transcriptionally regulates genes correlating with carbon, P and oxygen availability (Oyserman et al., 2015). In order to accurately respond to such environmental cues, bacteria rely primarily upon two-component systems (Chang and Stewart, 1998). Furthermore, HGT of two-component systems is an important mechanisms for niche adaptation, reflecting the selective pressures of the environment (Alm et al., 2006). In Accumulibacter, both phosphate limitation (PhoR CAP2UW1_1995, PhoB CAP2UW1_1996, PhoR-PhoB CAP2UW1_1997) and redox signaling (RegB CAP2UW1_0008, RegA CAP2UW1_0009) two-component systems are laterally derived at the LCA. Although it is difficult to surmise what specific genes may be under control of these two-component systems without additional molecular evidence, metatranscriptomic analysis identified many co-expressed genes responding to aerobic (1844) and low P (438) conditions (Supplementary Spreadsheet 5; Oyserman et al., 2015), which may be good candidates for further study in this regard.
In addition to the evolution of novel regulatory mechanisms in Accumulibacter, it is also possible for genes to integrate into existing regulatory networks; albeit this process often occurs slowly, with both recent and ancient laterally acquired genes generally showing lower degrees of co-expression than non-laterally transferred counterparts (Lercher and Pál, 2008). Currently, one of the most well-examined aspects of the Accumulibacter regulatory network is a putative PHA regulon likely controlled by the ancestral core regulatory protein (CAP2UW1_3918) (Oyserman et al., 2015). A key gene proposed to be in this regulon, a type III PhaC, is laterally derived providing evidence that laterally derived core genes integrated into existing ancestral regulatory networks. Thus, evolution of the regulatory networks through novel P and redox signaling, as well as through the integration of novel genes into existing regulatory networks such as the PHA regulon, likely contributed to the evolution of the PAO phenotype in Accumulibacter.
Uncertainty in reconstructions and future work
The analysis on Accumulibacter evolution was conducted within the constraints of our current knowledge into the phenotypic and genotypic diversity within the Rhodocyclaceae. We included all closely related, publically available, completed genomes (aside from the Accumulibacter genomes) at the time of the start of this analysis. Our understanding of the evolutionary and genomic capabilities of many lineages is continuously being re-written as the available data on a lineage increases. For example, recent investigations have expanded upon the definition of the Cyanobacteria phylum is based on new genomic information (Soo et al., 2014). One of the key uncertainties in our analysis is a lack of closely related non-Accumulibacter Rhodocyclaceae genomes that have been reconstructed from EBPR systems (for example, from Dechloromonas spp.). In addition, it remains difficult to distinguish ancient HGT events, especially if they are obfuscated by multiple gains and losses. Future discoveries may expand the diversity of Rhodocyclaceae involved in EBPR, either blurring or clarifying the delineation between PAO and non-PAO.
Conclusion
Here we report the first evolutionary study on the PAO phenotype through ancestral genome reconstructions, identification of HGT and chemical characterization. Through this analysis, we identified important metabolic transformations that occurred in the Accumulibacter LCA, where the transition from non-PAO to PAO is hypothesized to have occurred. Prominent lateral acquisitions include numerous genes involved in glycogen degradation, glycolysis, pyruvate metabolism and PHB pathways, as well as regulatory and sensory mechanisms involved in redox and P metabolism. In contrast, the TCA cycle and polyP metabolism are composed almost entirely of ancestral genes present before the Accumulibacter LCA. The molecular evolution that occurred in these pathways was likely necessary to overcome key stoichiometric and kinetic bottlenecks identified in PAO metabolism; specifically anaerobic carbon flux from glycogen to PHA via PFOR, P and counter cation transporters to maintain polyP synthesis, and anaerobic NADPH production from NADH via PntAB. Convergent evolution often occurs when non-related organisms under similar selective pressures independently evolve similar adaptations. Based on this assumption, the molecular evolution that occurred at the Accumulibacter LCA is likely representative of the general adaptations necessary for the Accumulibacter-type PAO phenotype to emerge. This analysis demonstrates the significance of differentiating the core genome of a lineage into ancestral and derived states when investigating a complex and phylogenetically cohesive phenotype.
Acknowledgments
We thank Shaomei He, Sarah Stevens, Joshua Hamilton and Pamela Camejo for friendly review. KDM acknowledges funding from the US National Science Foundation (CBET-0967646 and MCB-1518130) and the UW-Madison Graduate School. The work described here would not have been possible without the ongoing support of scientists and programs at the US Department of Energy Joint Genome Institute.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alm E, Huang K, Arkin A. (2006). The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol 2: 1329–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodríguez-Quiñones F. (2003). Bacterial iron homeostasis. FEMS Microbiol Rev 27: 215–237. [DOI] [PubMed] [Google Scholar]
- Blamey JM, Adams MWW. (1993). Purification and characterization of pyruvate ferrodoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol 1161: 19–27. [DOI] [PubMed] [Google Scholar]
- Burow LC, Mabbett AN, McEwan AG, Bond PL, Blackall LL. (2008). Bioenergetic models for acetate and phosphate transport in bacteria important in enhanced biological phosphorus removal. Environ Microbiol 10: 87–98. [DOI] [PubMed] [Google Scholar]
- Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. (2006). Feo—transport of ferrous iron into bacteria. BioMetals 19: 143–157. [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chabrière E, Charon MH, Volbeda A, Pieulle L, Hatchikian EC, Fontecilla-Camps JC. (1999). Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat Struct Biol 6: 182–190. [DOI] [PubMed] [Google Scholar]
- Chan AP, Sutton G, DePew J, Krishnakumar R, Choi Y, Huang X-Z et al. (2015). A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol 16: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Stewart RC. (1998). The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol 117: 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau Y, Hall KJ, Hancock REW, Oldham WK. (1986). Biochemical model for enhanced biological phosphorus removal. Water Res 20: 1511–1521. [Google Scholar]
- Comeau Y, Hall KJ, Oldham WK. (1988). Determination of Poly-3-hydroxybutyrate and Poly-3-hydroxyvalerate in activated sludge by gas-liquid chromatography. Appl Environ Microbiol 54: 2325–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell JH. (1980). Diversity and the coevolution of competitors, or the ghost of competition past. Oikos 35: 131–138. [Google Scholar]
- Crocetti GR, Hugenholtz P, Bond PL, Schuler A, Keller J, Jenkins D et al. (2000). Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl Environ Microbiol 66: 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csurös M. (2010). Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26: 1910–1912. [DOI] [PubMed] [Google Scholar]
- Flowers JJ, He S, Malfatti S, del Rio TG, Tringe SG, Hugenholtz P et al. (2013). Comparative genomics of two ‘Candidatus Accumulibacter' clades performing biological phosphorus removal. ISME J 7: 2301–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JJ, He S, Yilmaz S, Noguera DR, McMahon KD. (2009). Denitrification capabilities of two biological phosphorus removal sludges dominated by different ‘Candidatus Accumulibacter' clades. Environ Microbiol Rep 1: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Martín H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC et al. (2006). Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol 24: 1263–1269. [DOI] [PubMed] [Google Scholar]
- Ghylin TW, Garcia SL, Moya F, Oyserman BO, Schwientek P, Forest KT et al. (2014). Comparative single-cell genomics reveals potential ecological niches for the freshwater acI Actinobacteria lineage. ISME J 8: 2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo JFJG. (1979). Regulation of phosphate accumulation in the uni- cellular cyanobacterium Synechococcus. J Bacteriol 140: 508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J, Carniel E. (2001). Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep 2: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Golding GB. (2006). The fate of laterally transferred genes: life in the fast lane to adaptation or death. Genome Res 16: 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Gall DL, McMahon KD. (2007). ‘Candidatus Accumulibacter' population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl Environ Microbiol 73: 5865–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Gu AZ, McMahon KD. (2008). Progress toward understanding the distribution of Accumulibacter among full-scale enhanced biological phosphorus removal systems. Microb Ecol 55: 229–236. [DOI] [PubMed] [Google Scholar]
- Jendrossek D. (2009). Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191: 3195–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. (2014). Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res 42: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S et al. (2007). Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3: e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chang JH, Kim E-J, Kim K-J. (2014). Crystal structure of (R)-3-hydroxybutyryl-CoA dehydrogenase PhaB from Ralstonia eutropha. Biochem Biophys Res Commun 443: 783–788. [DOI] [PubMed] [Google Scholar]
- Kong Y, Nielsen JL, Nielsen PH. (2005). Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Society 71: 4076–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-riché D. (1999). Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem 68: 89–125. [DOI] [PubMed] [Google Scholar]
- Kristiansen R, Nguyen HTT, Saunders AM, Nielsen JL, Wimmer R, Le VQ et al. (2013). A metabolic model for members of the genus Tetrasphaera involved in enhanced biological phosphorus removal. ISME J 7: 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Nylander JA, Bergman B. (2011). Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol Biol 11: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latysheva N, Junker VL, Palmer WJ, Codd Ga, Barker D. (2012). The evolution of nitrogen fixation in cyanobacteria. Bioinformatics 28: 603–606. [DOI] [PubMed] [Google Scholar]
- Lefébure T, Stanhope MJ. (2007). Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol 8: R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Pál C. (2008). Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol Biol Evol 25: 559–567. [DOI] [PubMed] [Google Scholar]
- Madison LL, Huisman GW. (1999). Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev 63: 21–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara A, Ikai K, Ueda S, Yamane T. (1998). Gene dosage effects on polyhydroxyalkanoates synthesis from n-alcohols in Paracoccus denitrificans. Biotechnol Bioeng 60: 61–69. [PubMed] [Google Scholar]
- Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40: D115–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maszenan AM, Seviour RJ, Patel BK, Schumann P, Burghardt J, Tokiwa Y et al. (2000). Three isolates of novel polyphosphate-accumulating gram-positive cocci, obtained from activated sludge, belong to a new genus, Tetrasphaera gen. nov., and description of two new species, Tetrasphaera japonica sp. nov. and Tetrasphaera australiensis sp. no. Int J Syst Evol Microbiol 50: 593–603. [DOI] [PubMed] [Google Scholar]
- McMahon KD, Read EK. (2013). Microbial contributions to phosphorus cycling in eutrophic lakes and wastewater. Annu Rev Microbiol 67: 199–219. [DOI] [PubMed] [Google Scholar]
- Mielczarek AT, Nguyen HTT, Nielsen JL, Nielsen PH. (2013). Population dynamics of bacteria involved in enhanced biological phosphorus removal in Danish wastewater treatment plants. Water Res 47: 1529–1544. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Latimer K. (2008). Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24: 319–324. [DOI] [PubMed] [Google Scholar]
- Nobu MK, Tamaki H, Kubota K, Liu WT. (2014). Metagenomic characterization of ‘CandidatusDefluviicoccus tetraformis strain TFO71', a tetrad-forming organism, predominant in an anaerobic-aerobic membrane bioreactor with deteriorated biological phosphorus removal. Environ Microbiol 16: 2739–2751. [DOI] [PubMed] [Google Scholar]
- Nowell RW, Green S, Laue BE, Sharp PM. (2014). The extent of genome flux and its role in the differentiation of bacterial lineages. Genome Biol Evol 6: 1514–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–304. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lerat E, Daubin V. (2005). Examining bacterial species under the specter of gene transfer and exchange. Proc Natl Acad Sci USA 102(Suppl): 6595–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyserman BO, Noguera DR, Glavina Del Rio T, Tringe SG, Mcmahon KD. (2015). Metatranscriptomic insights on gene expression and regulatory controls in Candidatus Accumulibacter phosphatis. ISME J 10: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer EA, Allen JP, Hauser AR. (2014). Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics 15: 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Lercher MJ. (2005). Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet 37: 1372–1375. [DOI] [PubMed] [Google Scholar]
- Patel MS, Roche TE. (1990). Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J 4: 3224–3233. [DOI] [PubMed] [Google Scholar]
- Peoples OP, Sinskey AJ. (1989). Poly-P-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16 identification and characterization of the PHB polymerase gene (phbC). J Biol Chem 264: 15298–15303. [PubMed] [Google Scholar]
- Polz MF, Alm EJ, Hanage WP. (2013). Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet 29: 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. (2007). NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NN, Liu S, Kornberg A. (1998). Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol 180: 2186–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenhall M, Škunca N, Lassalle F, Dessimoz C. (2015). Inferring horizontal gene transfer. PLOS Comput Biol 11: e1004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm BHA. (2003). Polyester synthases: natural catalysts for plastics. Biochemistry 376: 15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Mabbett AN, McEwan AG, Blackall LL. (2007). Proton motive force generation from stored polymers for the uptake of acetate under anaerobic conditions. FEMS Microbiol Lett 274: 245–251. [DOI] [PubMed] [Google Scholar]
- Schluter D, Price T, Mooers AØ, Ludwig D. (1997). Likelihood of ancestor states in adaptive radiation. Evolution (N Y) 51: 1699–1711. [DOI] [PubMed] [Google Scholar]
- Schuler AJ, Jenkins D. (1994). Enhanced biological phosphorus removal from wastewater by biomass with different phosphorus contents, Part I: experimental results and comparison with metabolic models. Water Environ Res 75: 485–498. [DOI] [PubMed] [Google Scholar]
- Schuler AJ, Jenkins D. (2003). Enhanced biological phosphorus removal from wastewater by biomass with different phosphorus contents, Part III: Anaerobic sources of reducing equivalents. Water Environ Res 75: 512–522. [DOI] [PubMed] [Google Scholar]
- Seviour RJ, Mino T, Onuki M. (2003). The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol Rev 27: 99–127. [DOI] [PubMed] [Google Scholar]
- Skennerton CT, Barr JJ, Slater FR, Bond PL, Tyson GW. (2015). Expanding our view of genomic diversity in Candidatus Accumulibacter clades. Environ Microbiol 17: 1574–1585. [DOI] [PubMed] [Google Scholar]
- Snoep JL, de Graef MR, Westphal AH, de Kok A, Teixeira de Mattos MJ, Neijssel OM. (1993). Differences in sensitivity to NADH of purified pyruvate dehydrogenase complexes of Enterococcus faecalis, Lactococcus lactis, Azotobacter vinelandii and Escherichia coli: implications for their activity in vivo. FEMS Microbiol Lett 114: 279–283. [DOI] [PubMed] [Google Scholar]
- Soo RM, Skennerton CT, Sekiguchi Y, Imelfort M, Paech SJ, Dennis PG et al. (2014). An expanded genomic representation of the phylum Cyanobacteria. Genome Biol Evol 6: 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A, Hustede E, Liebergesell M, Pieper U, Timm A, Valentin H. (1993). Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids in bacteria. FEMS Microbiol Rev 10: 347–350. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P et al. (2009). Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5: e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson SM, Upcroft A, Upcroft P. (1996). Characterisation and purification of pyruvate:ferredoxin oxidoreductase from Giardia duodenalis. Mol Biochem Parasitol 79: 183–193. [DOI] [PubMed] [Google Scholar]
- van Dongen SM. (2000). Graph clustering by flow simulation. PhD Thesis, University of Utrecht, The Netherlands. [Google Scholar]
- Wandersman C, Delepelaire P. (2004). Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58: 611–647. [DOI] [PubMed] [Google Scholar]
- Welles L, Tian WD, Saad S, Abbas B, Lopez-Vazquez CM, Hooijmans CM et al. (2015). Accumulibacter clades type I and II performing kinetically different glycogen-accumulating organisms metabolisms for anaerobic substrate uptake. Water Res 83: 354–366. [DOI] [PubMed] [Google Scholar]
- Wilkinson JF. (1963). Carbon and energy storage in bacteria. J Gen Microbiol 32: 171–176. [DOI] [PubMed] [Google Scholar]
- Yuan W, Jia Y, Tian J, Snell KD, Müh U, Sinskey AJ et al. (2001). Class I and III polyhydroxyalkanoate synthases from Ralstonia eutropha and Allochromatium vinosum: characterization and substrate specificity studies. Arch Biochem Biophys 394: 87–98. [DOI] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, Viklund J, Zhao W, Ast J, Sczyrba A, Woyke T et al. (2013). Single-cell genomics reveal low recombination frequencies in freshwater bacteria of the SAR11 clade. Genome Biol 14: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Blasiak LC, Karolin JO, Powell RJ, Geddes CD, Hill RT. (2015). Phosphorus sequestration in the form of polyphosphate by microbial symbionts in marine sponges. Proc Natl Acad Sci USA 112: 4381–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles JL, Peccia J, Kim M, Hung C, Noguera DR. (2002). Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Society 68: 2763–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.