Abstract
Newly hatched Euprymna scolopes squid obtain their specific light-organ symbionts from an array of Vibrio (Allivibrio) fischeri strains present in their environment. Two genetically distinct populations of this squid species have been identified, one in Kaneohe Bay (KB), and another in Maunaloa Bay (MB), Oahu. We asked whether symbionts isolated from squid in each of these populations outcompete isolates from the other population in mixed-infection experiments. No relationship was found between a strain's host source (KB or MB) and its ability to competitively colonize KB or MB juveniles in a mixed inoculum. Instead, two colonization behaviors were identified among the 11 KB and MB strains tested: a ‘dominant' outcome, in which one strain outcompetes the other for colonization, and a ‘sharing' outcome, in which two strains co-colonize the squid. A genome-level comparison of these and other V. fischeri strains suggested that the core genomic structure of this species is both syntenous and highly conserved over time and geographical distance. We also identified ~250 Kb of sequence, encoding 194 dispersed orfs, that was specific to those strains that expressed the dominant colonization behavior. Taken together, the results indicate a link between the genome content of V. fischeri strains and their colonization behavior when initiating a light-organ symbiosis.
Introduction
Understanding the relationships between environmental adaptation, population dynamics and genomic diversity is a key goal of the field of microbial ecology (McInnes et al., 2004; Cordero and Polz, 2014). The study of microbial symbioses has provided a valuable platform from which this goal can be addressed. For instance, co-evolution or co-divergence of bacteria and their hosts has been extensively studied in vertically transmitted associations, and can involve events like gene sharing and genome reduction within the microbial partner(s) (Kikuchi et al., 2009; Moran and Bennett, 2014). Similarly, horizontally transmitted symbioses of plants and invertebrates have offered the opportunity to examine the evolution of species specificity, revealing evidence for strategies such as congruent evolution and partner choice (Chaston and Goodrich-Blair, 2010; Sudakaran et al., 2015). In the symbiosis between the squid Euprymna scolopes and the luminous bacterium Vibrio fischeri, as in other environmentally acquired, non-obligatory symbionts, one can experimentally expose an aposymbiotic host to genetically distinct V. fischeri strains, either singly or in combination, and monitor the establishment and dynamics of the resulting symbiont population. By manipulating this highly specific association, one can address fundamental questions concerning the role of strategies like host sanctioning in the evolution of persistent symbiotic associations (Koch et al., 2014).
V. fischeri is the specific light-organ symbiont of several families of fishes and squids, including the Hawaiian bobtail squid, E. scolopes. In exchange for nutrients, the bacterium provides counterillumination for the night-active squid by producing bioluminescence that obscures the shadow of the host from its predators (Jones and Nishiguchi, 2004). The squid hatches without V. fischeri cells, but rapidly acquires this specific bacterium from the surrounding environment (Ruby and Asato, 1993). The bacteria enter the nascent light organ through six surface pores, three on either side of the organ. They then traverse several different tissue environments before entering the six epithelium-lined crypts deep within the organ, where they establish a lifelong, albeit dynamic, symbiosis (Nyholm and McFall-Ngai, 2004). In nature, mature squids contain at least three to five strains (Wollenberg and Ruby, 2009), indicating the possibility of mixed-strain colonizations occurring in the crypts. Each morning for the 8–10-month lifetime of the host, the light organ expels 95% of its symbionts (Lee and Ruby, 1994b) and, within a few hours, the organ is repopulated by the growth of the remaining symbionts.
Distinct populations of squid occur within two bays on different sides of the Hawaiian Island of Oahu (Kimbell et al., 2002): Maunaloa Bay (MB) and Kaneohe Bay (KB). Using a DNA-fingerprint technique (VfRep-PCR) to characterize light-organ symbionts obtained from the two host populations, three genetically and phenotypically distinct groups of V. fischeri were described (Wollenberg and Ruby, 2009) that are found associated with hosts from both geographic locations. Group-A strains (Wollenberg and Ruby, 2012) exhibited an advantage when colonizing juvenile squid from the MB population; and they were less fit than other symbionts when incubated in natural seawater in the absence of the host, suggesting divergent ecological strategies.
In this study, we performed both functional and genomic analyses of 11 symbiotic V. fischeri strains that are representative of those described in Wollenberg and Ruby, 2012, and whose genomes we have recently sequenced. In the functional approach, we first asked whether symbionts of squid from one bay were more effective at colonizing juveniles from this same bay. We next examined the relative fitness of these symbionts by determining their behavior during an experimental co-colonization: specifically, when pairs of strains were co-inoculated, did one outcompete the other or did a co-colonization of the light organ result. Finally, we compared the genomes of squid light-organ symbionts, together with the fish light-organ symbiont, strain MJ11 (Mandel et al., 2009), to ask whether there were genes that were always found in the squid symbionts, but not in the fish symbiont.
Materials and methods
Media and strains
All V. fischeri strains were grown at 28 °C in either (i) seawater tryptone medium containing (per liter) 700 ml of a marine salts solution Instant Ocean (Spectrum Brands, Madison, WI, USA), 300 ml deionized water, 5 g Bacto-Tryptone, 3 g yeast extract and 3 ml of glycerol; or (ii) Luria-Bertani (LB) Salt media (Graf et al., 1994), containing (per liter) 20 g NaCl, 50 ml of 1 M Tris-HCl (pH 7.5), 10 g Bacto-Tryptone and 5 g yeast extract, with the addition of erythromycin (5 μg ml−1) for strains carrying a gene encoding a green or red fluorescent protein (RFP) in the Tn7 site (McCann et al., 2003). Derivatives of Escherichia coli DH5α were grown at 37 °C in LB medium (10 g Bacto-Tryptone, 5 g yeast extract and 10 g NaCl per liter). When necessary, LB medium was supplemented with kanamycin (50 μg ml−1), carbenicillin (100 μg ml−1) or chloramphenicol (25 μg ml−1).
All of the wild-type strains used in this study have been previously described (Ruby et al., 2005; Mandel et al., 2009; Wollenberg and Ruby, 2009) (Supplementary Table S1), and derivatives were constructed that chromosomally carried genes encoding either green fluorescent protein (GFP) or RFP to simplify their differentiation (Supplementary Table S2). To construct plasmids pCBNR6 and pCBNR7, we amplified the segments pLacZ-GFP and pLacZ-RFP, from pVSV102 and pVSV208, respectively (Dunn et al., 2006), and inserted them into pEVS107 (McCann et al., 2003). We amplified pVSV102 and pVSV208 by PCR using the following primers: (5′-GCATGCCTAGGTTTACACTTTATGCTTCCGGC-3′) and (5′-CGTACGGTACCTCAGTTGTACAGTTCATCCATG-3′) for pCBNR6; and (5′-CGTACGGTACCCTACAGGAACAGGTGGTGG-3′) for pCBNR7. The primers were synthetized by Integrated DNA Technologies (Coralville, IA, USA). Subsequent PCR digestion by KpnI-HF and AvrII, ligation and transformation were all performed according to standard methods. We then transferred the fragment containing the promoter, fluorescent protein and erythromycin-resistance gene (erm) from the plasmid to the Tn7 insertion site of wild-type strains by tetra-parental mating (McCann et al., 2003). An additional screening process was used to check for colony fluorescence using a Leica MZFLIII fluorescence dissecting scope (Leica Microsystems Inc., Buffalo Grove, IL, USA), equipped with a set of G, TXR and GFP2 filters.
Light-organ colonization assay
To determine the relative colonization effectiveness of different strains of light-organ isolates, inocula were prepared that contained either an individual strain or various pairs of competing strains (Supplementary Table S2). The inocula were prepared by growing each strain overnight in LB Salt medium, before diluting them 1:100 into seawater tryptone medium medium. The bacteria were then grown to mid-exponential phase (OD ~0.3) before diluting them in filter-sterilized Instant Ocean (FSIO). Newly hatched squids were then transferred into the inoculated FSIO containing either a single strain or, for competition assays, a pair of strains, targeting a 1:1 ratio. The pairs were differentially labeled by their expression of either GFP or RFP. In all cases, we aimed for a total inoculum of around 5000 colony-forming units (CFU) per ml. After 3 h of exposure to the inoculum, each squid was removed to a vial containing 4 ml of bacteria-free FSIO; at 24 h, the FSIO was changed, and at 48 h, the squids were frozen at −80 °C. Squids were then homogenized, dilutions spread on LB Salt agar, and the number of CFU per light organ was calculated. For co-colonized squid, the number of GFP and RFP CFUs were counted using a fluorescence dissecting scope.
To compare the total CFU present in different conditions, we applied a Kruskal–Wallis test using GraphPad Prism version 6.0. When squids were co-colonized with two strains, we determined the logarithm of the relative competitive index to the base 10 (Stabb and Ruby, 2003), and applied a one-way analysis of variance followed by a Sidak's multiple-comparisons test to compare between the different conditions. The relative competitive index was used to describe the relative colonization effectiveness of two competing strains, and was calculated as: (ratio of strain abundance in the light organ at 48 h)/(ratio of strain abundance in the initial inoculum). The sensitivity of the assay was 0.05% that is, it was possible to detect the presence of as few as 280 cells of either strain in the light organ.
Genome sequencing and analysis
We sequenced 11 previously characterized strains of E. scolopes symbionts that were isolated from KB or MB adult squid light organs. When the first five characters of the strains' names are the same, it means that they were isolated from the same side of a single light organ (for example, MB15A4 and MB15A5) (Wollenberg and Ruby, 2009). The 12 total V. fischeri squid symbionts in our study (the previously sequenced strain ES114 and the 11 sequenced here) were isolated across 17 years (that is, strain ES114 in 1988, ES213 in 1990 and the others in 2005), while the sequenced fish light-organ symbiont, MJ11, was isolated from the Japanese pinecone fish, Monocentris japonica in 1994 (Mandel et al., 2009). In all cases, genomic DNA was extracted using the protocol of the QIAgen genomic DNA buffer set with QIAgen genomic Tip 100G (QIAGEN Inc., Valencia, CA, USA).
Sequencing libraries were prepared using Nextera XT DNA Sample Preparation Kit (Illumina Inc., San Diego, CA, USA; #FC-131-1024) and Illumina Nextera XT DNA Sample Preparation Index Kit (Illumina Inc. #FC-131-1001). Sequencing libraries were normalized before sequencing with a modified protocol. All samples were sequenced on an Illumina MiSeq (PE 250+250) at MIT's BioMicroCenter. The resulting genomes were annotated with RAST (Aziz et al., 2008), and bioinformatically analyzed using both MAUVE (Darling et al., 2010) and OrthoMCI (Li et al., 2003). The accession numbers of the different strains are listed in Supplementary Table S1. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number XXXX00000000. The version described in this paper is XXXX01000000.
Results
Host preference
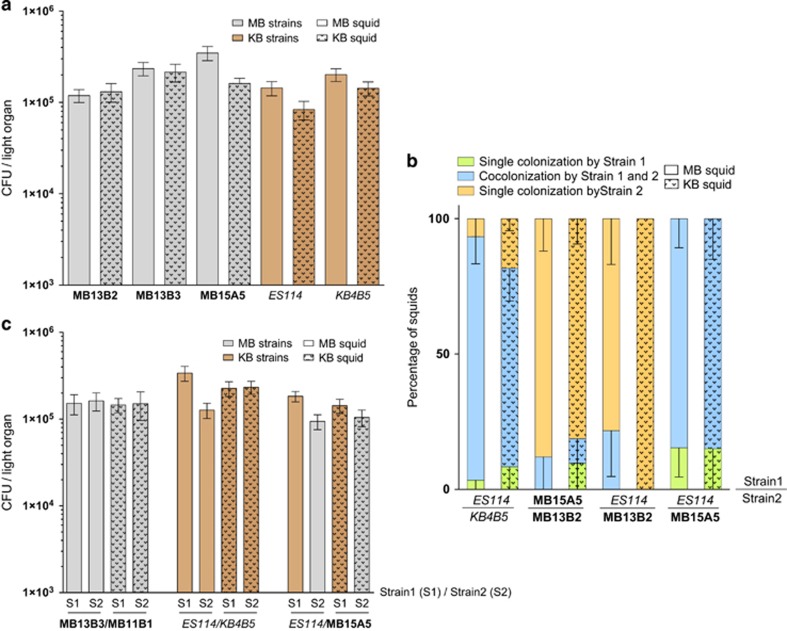
We first asked whether symbionts isolated from the light organs of squid in one geographic population—either Maunalua Bay (MB) or KB—were more effective at colonizing newly hatched juveniles from that same population. Specifically, juvenile squid of each population were exposed to a V. fischeri strain isolated from either MB or KB light organs, and the level of CFU achieved in the juvenile light organ was determined after 48 h (Figure 1a, Supplementary Figure S1). When the squids were exposed to these single strains of V. fischeri, there was no significant difference in the level of colonization by the strain between the two populations of squid, regardless of whether the strain originated from MB (MB13B2, MB13B3, MB15A5) or KB (ES114, KB1A98).
Figure 1.
Colonization of light organs of squids from either Maunalua Bay (MB; open bars) or KB (dotted bars) populations, by symbiont strains isolated from either MB (gray) or KB (brown) adult light organs. Inoculations were made with either single strains (a) or with pairs of strains (b and c). In all experiments, squid were incubated for 3 h with 103–104 CFU of bacteria per ml of FSIO, transferred to uninoculated FSIO and assayed after another 45 h for CFU per light organ. Values are means plus or minus the standard error of the mean (s.e.m.). (a) None of the values for the pairs of bars (open and dotted) were significantly different from each other. (b) Animals inoculated with pairs of strains were found to be co-colonized either by both strains (blue) or by only Strain 1 (green) or only Strain 2 (orange), and the percentage of the three classes are indicated by a fractionated bar graph. (c) In addition, when organs were analyzed that had been co-colonized by either two MB strains (MB11B1 and MB13B2), two KB strains (ES114 and KB4B5) or an MB and a KB strain (ES114 and MB15A5), the relative numbers of the two co-occurring strains were approximately equal in hosts from both populations, regardless of which population the strain had been originally isolated from.
Because the multiple V. fischeri strains naturally present in the ambient seawater of newly hatched juveniles are expected to compete during colonization of the squid (Lee and Ruby, 1994a), we also performed co-inoculation assays to determine whether a strain isolated from one squid population had a competitive advantage in colonizing juveniles from that population. The squids were inoculated with pairs of bacteria from either the same (for example, ES114 and KBAB5; or MB15A5 and MB13B2) or different (for example, ES114 and MB13B2; or ES114 and MB15A5) locations. Both the percentage (Figure 1b) and the total number (Supplementary Figure S2) of squids colonized by one or both strains suggested that the degree of competitive dominance was independent of the geographical source of either the bacteria or the juveniles being colonized. Similarly, in these same competition experiments, the relative abundance of each strain in a co-colonized symbiont population was not significantly influenced by the origin of the squid (Figure 1c,Supplementary Figure S3), indicating that the origin of the two partners does not affect the competitive dominance of the V. fischeri strain in the symbiosis. Taken together, even this limited study provided no evidence of co-evolution (Wollenberg and Ruby, 2009) among symbionts and hosts that are found in the same location but, instead, suggested that although the host populations are genetically distinct (Kimbell et al., 2002), the pool of potential symbionts in the waters around the island of Oahu is effectively continuous.
Colonization behaviors of different strains
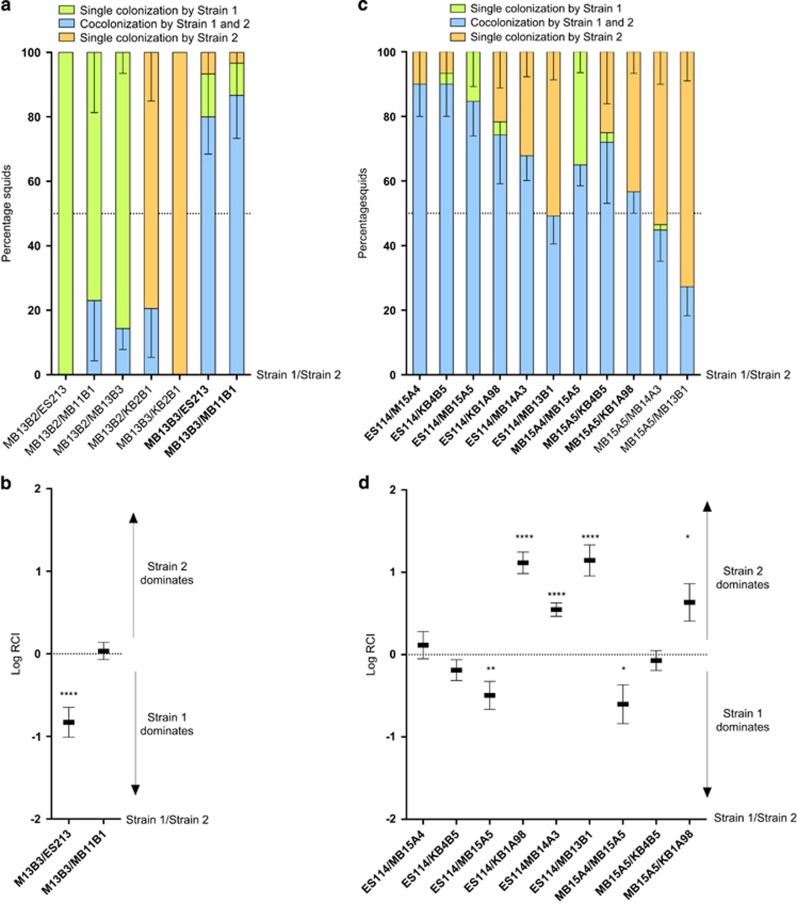
We next asked whether there were any patterns in the co-colonization outcomes for the different symbiont isolates. Interestingly, in a high percentage of cases in which one (92%) or both (71% Figure 2a) of the two competing strains were of the Group-A designation (Wollenberg and Ruby, 2012), one of the strains clearly dominated; that is, over 50% of the squids were colonized by only that strain. We defined this as a ‘dominance', or D-type, outcome (Supplementary Figure S4), and have renamed the Group-A strains as ‘D-type strains' based on this shared behavior. In contrast, when neither of the two co-inoculated strains were D-type, there was generally (82% of the cases) a co-colonization of the squid; that is, 50% or more of the squids were colonized by both strains (Figure 2c). We designated this as a ‘sharing', or S-type, outcome because the strains were sharing the light-organ habitat. Interestingly, when co-colonizations occurred in either the D-type or the S-type strain pairs, one was often more competitive, as indicated by their ratio in the organ's symbiotic population (Figures 2b and d). Taken as a whole, the results illustrate that a given V. fischeri strain generally exhibits one of two kinds of behavior when colonizing the light-organ niche: the first is dominance, that is, D-type strains effectively exclude other, less dominant strains from the light organ creating a hierarchy (Supplementary Figure S5). The second behavior is one of sharing, that is, S-type strains typically will share the niche with another S-type strain. To determine whether the dominance phenotype was expressed in culture as well as in the light organ, we measured relative growth rates and yields among strains in rich medium. No significant difference in growth parameters was observed between the two types of strains. Moreover, 24 h after co-inoculation of seawater (FSIO) with both a D-type and an S-type strain, both were present, and the D-type strains were not always more abundant (data not shown). Thus, the D- and S-type behaviors during squid colonization do not reflect any intrinsic growth differences in the bacteria.
Figure 2.
State of colonization of juvenile squid after co-inoculation with a mixture of either two D-type strains (a and b) or two S-type strains (c and d); mean of at least two experiments per condition. Animals inoculated with pairs of strains were found to be co-colonized either by both strains (blue), or by only Strain 1 (green) or by only Strain 2 (orange), and the percentage of the three classes are indicated by a fractionated bar graph (a and c). When more than 50% of the squids were co-colonized (indicated with bold strain names), we determined the relative competitive index (RCI) of the pair of strains (b and d). The RCI was calculated as the ratio of the two strains in the light organ after 48 h, divided by their ratio in the inoculum. Those RCIs that are significantly different from zero are indicated (T-test: *P<0.1; **P<0.01; ****P<0.0001).
Comparative genomics
To gain insight into the kinds of genetic differences that may underlie the capacity of certain strains to outcompete others during the initiation of symbiosis, we compared the genomes of 13 V. fischeri isolates: the 12 squid symbionts tested above for colonization hierarchy, plus the fish light-organ symbiont MJ11 (Mandel et al., 2009). Two distinct subsets could be seen when comparing genome size: one averaging 4.25 Mb that comprised all of the S-type strains, and a second averaging 4.50 Mb that included the D-type strains, as well as MJ11 (Supplementary Table S1).
The core genome common to all of the strains contained 3170 orthologous proteins, which represent an average of 80.4% of the total proteins encoded within these V. fischeri. This relatively large percentage indicates a surprisingly small variability among the strains, considering the geographical and temporal breadth of their origins; that is, Hawaii to Japan, and 1988 to 2005 (Supplementary Table S1). Beyond this core, we found 194 proteins specific to the D-type strains, producing 173 distinct blast hits; 69 of these were involved in different bacterial functions as defined by blastp, and the remaining 104 represented hypothetical proteins (Supplementary Table S3). The genes encoding these proteins are dispersed along the genomes of the D-type strains; for example, they are localized on 41 of the 78 contigs of MB11B1. In this strain, there are 5 contigs that each harbor more than 10 genes specific to D-type strains, 6 contigs harboring between 5 and 10 such genes, and 30 contigs harboring the remaining 59 D-type specific genes. This organization argues against a single 250-Kb ‘dominance island' within the genomes of these strains, a conclusion that is supported by a closer examination of the distribution of these genes on the contigs. For instance, on contig #8, genes encoding 31 proteins specific to D-type strains are spread among two distinct regions. The first region contains 14 D-specific genes interspersed among genes having orthologous in S-type strains (Supplementary Figure S6). Thus, we find evidence for a number of D-specific loci, any one (or more) of which could be necessary for the dominance of the D-type strains. Both the dispersed nature of the genes and the high number of hypothetical proteins encoded complicate the identification of a candidate(s) for the dominance phenotype; nevertheless, future work may identify such a gene(s). In contrast to the pattern just described, no proteins were found to be specific to the S-type strains as a group, suggesting that the dominance of the D-type strains is unlikely to arise from their loss of a specific gene(s) promoting the S-type sharing phenotype.
Because the fish symbiont, MJ11, was reported to not encode at least one gene required for efficient colonization of the squid (Mandel et al., 2009), we asked whether we could identify other genes present in all the squid symbionts, but absent from this strain (Table 1) that might be additional candidates for specificity factors. Of these 23 genes, there are 8 that encode hypothetical proteins and 11 that are in two predicted operons of 5 and 6 genes; most of the 23 are predicted to encode surface or membrane proteins. As expected, we found two orfs, rscS (VFA0237) and qsrP (VFA1058), that were previously shown to have roles in initiating colonization (Callahan and Dunlap, 2000; Mandel et al., 2009). Interestingly, the transcription of 9 of the 23 genes (including qsrP) is altered by the presence of N-3-oxohexanoyl-L-homoserine lactone (3-oxo-C6-HSL), indicating that they are regulated by quorum signaling (Antunes et al., 2007). In addition, the transcription of 7 of the 23 squid-symbiont-specific genes is modulated during the diel cycle of the adult squid symbiosis (Wier et al., 2010). Further study of these genes may prove to reveal additional specificity factors.
Table 1. Genes present in all squid symbionts, but absent in the fish symbiont MJ11a.
| Gene | Annotationb |
|---|---|
| VF0154 | Hypothetical protein |
| VF1161c | Periplasmic protein of efflux system |
| VF1162d | Outer membrane protein TolC |
| VF1163 | Export ABC transporter permease |
| VF1164 | Export ABC transporter permease |
| VF1165 | Macrolide ABC transporter ATP-binding/membrane protein |
| VF1246 | Outer membrane protease |
| VF1334 | Oxalate/formate antiporter |
| VF1349 | Subtilisin-like serine protease |
| VF1471 | Hypothetical protein |
| VF1787 | Hypothetical protein |
| VF2634 | Hypothetical protein |
| VFA0023 | Enoyl-CoA hydratase |
| VFA0024 | LysR family transcriptional regulator |
| VFA0237e | Hybrid sensor kinase RscS |
| VFA0894 | Surface protein |
| VFA0980 | Hypothetical protein |
| VFA0981 | Hypothetical protein |
| VFA0982 | Hypothetical protein |
| VFA0983 | Hypothetical protein |
| VFA0984 | Transglycosylase |
| VFA0985 | Lipoprotein |
| VFA1058e | LuxR-regulated periplasmic protein |
Bar indicates the number of nucleotide substitutions per site.
Determined using OrthoMCl to be present in all squid symbionts, but not the non-colonizing fish symbiont, MJ11.
From NCBI.
Names italicized were previously identified as differentially expressed in response to the quorum signal 3-oxo-C6-HSL (Antunes et al., 2007).
Names in bold were previously identified as differentially expressed in the light organ on a day/night rhythm (Wier et al., 2010).
Previously shown to be key during initiation of colonization (Callahan and Dunlap, 2000; Mandel et al., 2009).
MAUVE—gene distribution
Another bioinformatic approach for identifying possible colonization factors is to ask whether all squid symbionts encode particular genes that have been reported to support symbiosis initiation. For instance, using the genome of strain ES114 as the guide, we determined the distribution among the other strains of genes encoding certain sensing functions (for example, pili, methyl-accepting chemotaxis proteins (MCPs) and quorum signaling) (Table 2, Supplementary Figure S7). Our analysis, which used the alignment program MAUVE (Darling et al., 2010), was based on the assumption that, if a gene is not present in the genomes of all 12 of the strains that were isolated from squid light organs, then it is unlikely to be critical for light-organ symbiosis. For instance, type-4 pili (T4P) are a class of surface structures often associated with pathogenic or beneficial microbial colonization of host tissue (Craig et al., 2004; Dulla et al., 2012), and 10 distinct T4P gene clusters have been identified in V. fischeri strain ES114 (Ruby et al., 2005). Some of these putative pilus proteins (that is, MshA, PilT and PilA2) have been linked previously to the colonization process (Stabb and Ruby, 2003; Ariyakumar and Nishiguchi, 2009; Chavez-Dozal et al., 2012). As expected, these pili are encoded in the genomes of all the light organ-derived strains (Table 2). On the other hand, the genes encoding three other putative T4P (PilA1, Tcp and VirB2) are not present in all the strains, suggesting they are not required for colonization. That the Tcp structural genes (VFA0865-875) are not present in all strains was unexpected because the homologous pilus is a critical tissue-colonization factor in human pathogenesis by V. cholerae (Childers and Klose, 2007). However, when a V. fischeri strain with a mutation in the major pilin subunit, tcpA, was competed against wild type, no defect in light-organ colonization was detected (data not shown), an outcome that supports our present bioinformatic approach. In contrast, orthologs of tcpP and tcpH (VF0473-0474), two Tcp cluster genes that encode membrane-associated regulators of virulence in V. cholerae (Beck et al., 2004), are present in all V. fischeri squid symbionts, suggesting that these regulators may have had a role modulating other bacterial processes in ancestral Vibrio species, and were only recently recruited into modulating V. cholerae virulence. Further, the widespread distribution of the gene encoding TcpP homologs in a number of other Vibrio species is consistent with this hypothesis, while the limited distribution of that encoding TcpH suggests a more phylogenetically defined role for this regulatory protein.
Table 2. Distribution of type-4 pilus protein-encoding orthologs in V. fischeri strainsa.
| PILUS LOCIb | MshA | PilT | FlplA | PilA1 | PilA | PilA2 | Flp1 |
Tcp cluster |
VirB2 |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VF0355-VF0371 | VF0431-VF0432 | VF0510-VF0523 | VF0571-VF0572 | VF2184-VF2191 | VFA0148 | VFA0217-VFA0232 | VF1138 | VFA0473-VFA0474 | VFA0865-VFA0875 | VFB38-VFB44 | VFB54-VFB55 | |
| STRAINS | ||||||||||||
| ES114 | + | + | + | + | + | + | + | + | + | + | ||
| MB13B3c | + | + | + | + | + | + | + | + | + | |||
| MB11B1 | + | + | + | + | + | + | + | + | + | |||
| KB2B1 | + | + | + | + | + | + | + | + | + | |||
| MB13B2 | + | + | + | + | + | + | + | + | + | + | ||
| ES213 | + | + | + | + | + | + | + | + | + | + | + | |
| MB14A3 | + | + | + | + | + | + | + | + | ||||
| KB4B5 | + | + | + | + | + | + | + | + | + | |||
| MB15A5 | + | + | + | + | + | + | + | + | ||||
| KB1A98 | + | + | + | + | + | + | + | + | ||||
| MB13B1 | + | + | + | + | + | + | + | + | ||||
| MB15A4 | + | + | + | + | + | + | + | + | ||||
| MJ11 | + | + | + | + | + | + | + | + | ||||
Putative type-4 pilus proteins encoded by V. fischeri strain ES114 that have orthologs in other genome-sequenced squid symbiont strains.
+, ortholog present; blank, ortholog absent.
Bold indicates the five D-type strains.
Another set of important factors in bacterial colonization are the MCPs (Jones and Armitage, 2015). V. fischeri strain ES114 has 43 genes encoding known and putative MCPs (Supplementary Figure S7). Among squid symbionts, nine of these MCP genes are specific to strain ES114 and, therefore, are unlikely to be critical for colonization; however, it is possible that their functions are compensated by non-homologous members of the other strains' MCP repertoires, thereby complicating any inference of their importance in light-organ colonization. Although some chemoattractants of V. fischeri have been identified, the specific ligands of the MCPs present in V. fischeri are known for only three (DeLoney-Marino et al., 2003; Brennan et al., 2013; Nikolakakis et al., 2015), none of which is required for light-organ colonization.
Finally, because quorum signaling is involved in many cellular functions including light-organ colonization (Miller and Bassler, 2001; Verma and Miyashiro, 2013), we identified shared genes involved in this cell-cell communication mechanism. Of the 30-orf quorum-signaling regulon of strain ES114 (Antunes et al., 2007), 5 orfs, that is, VF1725, VF2034-2036 and VFA0090, are unlikely to be required for symbiosis as they are not conserved in the genomes of all the sequenced V. fischeri. In contrast, nine others (VF1161-VF1165, VF1246, VF1349, VFA0894 and VFA1058) are conserved in all squid symbionts but not in the fish symbiont (Table 1) and, thus, are promising yet untested candidates as squid-specific colonization factors.
MAUVE—genetic synteny
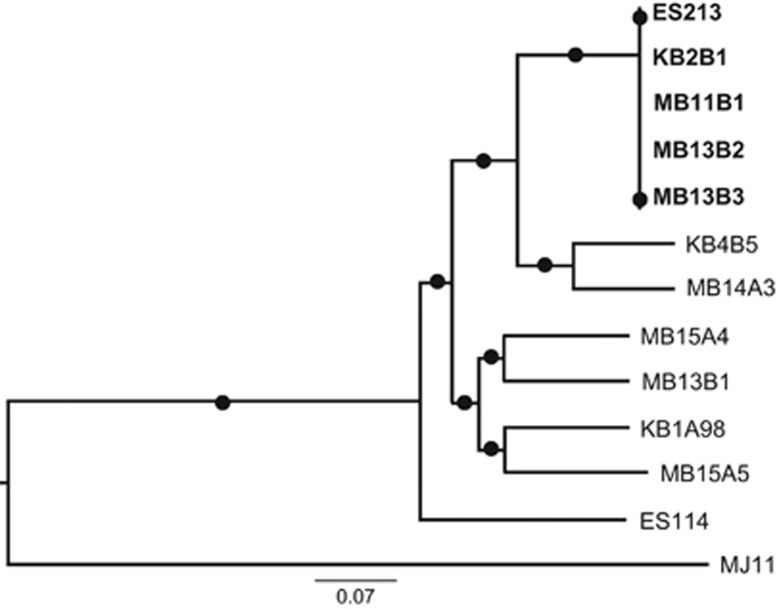
As a further indicator of genome diversification, we used MAUVE to determine the degree to which synteny was preserved within the symbiotic V. fischeri strains (Supplementary Figure S8). We concluded that the spatial organization of the genome is quite well conserved among the strains; that is, there are apparently no large-scale rearrangements in this species. When viewed on a maximum-likelihood phylogenetic tree based on the 3170 core genes, D-type and S-type strains clustered separately (Figure 3). Although this phylogenetic analysis does not resolve any separation among the D-type strains, taking into account the relationships across the entire genome (Supplementary Figure S9) permitted a greater discrimination of their relationships. As expected, the D-type strains continued to group together on an early branch, and the most efficiently colonizing strains (that is, KB2B1 and MB13B2) grouped closely. The location of the less competitive D-type strain, ES213, (Supplementary Figure S5) suggests that it may have lost an important genetic determinant for the phenotype of strong dominance.
Figure 3.
Maximum-likelihood phylogenetic tree, based on 3170 core-genome genes of V. fischeri symbionts, obtained with PhyML (Guindon et al., 2010). D-type strains (upper right) are indicated in bold. (a) Shimodaira-Hasegawa (SH) test was performed to evaluate likelihood of the bifurcations. Circles appear at nodes that were supported by an SH-test result>0.9. The scale bar indicates the number of nucleotide substitutions per site.
In contrast, the phylogenetic tree (Figure 3) suggested a closer common ancestor between the D-type strains and two of the S-type strains, KB4B5 and MB14A3; however, those two strains are not more competitive colonizers than other S-type strains (Supplementary Figure S5). Thus, the relationship between phylogeny and colonization hierarchy is more complex than a single divergence step between D-type and S-type strains.
Discussion
Studies linking the presence of genome content to colonization effectiveness have led to a greater understanding of not only individual microbe–host interactions, but also the driving role of population genetics in the evolution of symbiosis. For instance, the characterization of pathogenicity islands (Nieto et al., 2016) and symbiosis plasmids (Christie and Gordon, 2014) has provided insight into the mechanisms underlying the rapid evolution of bacterial associations with new hosts. Similarly, discovery of the roles that extreme genome diversification and behavioral specialization have in the evolution of bacterial populations (Shapiro et al., 2012; Yawata et al., 2014) has led to a realization that these events are a powerful force in microbial ecology, and especially host interactions.
The study here aimed to determine whether there is a link between the population biology, symbiotic characteristics and genomic content of 12 different light-organ-derived strains of V. fischeri. We found that (i) although there is a strong species specificity between the Hawaiian bobtail squid and V. fischeri, there is no evidence of any co-evolution within two genetically isolated host populations and their associated symbionts; instead, (ii) symbionts found in both populations can be divided into two groups defined by their behavior during the colonization process, and (iii) a genome-level comparison of these strains suggested that, while these behaviors correlate with a 250-Kb difference in genome size, comprising almost 200 dispersed orfs, the core genomic structure of V. fischeri strains is syntenous and highly conserved over time and location.
Despite the genetic separation of host populations from the northern and southern sides of the island of Oahu (Kimbell et al., 2002), there is no evidence of co-diversification among V. fischeri symbiont strains and the squid hosts from these two regions. This conclusion is analogous to that drawn at the species level for light-organ symbionts in the genus Photobacterium and their fish hosts (Dunlap et al., 2007; Urbanczyk et al., 2011), wherein more than one symbiont species is harbored. Thus, symbiont specificity can express itself at a number of phylogenetic levels among different bioluminescent symbioses; that is, while some host species will harbor more than one bacterial species (Fidopiastis et al., 1998), other hosts (for example, E. scolopes) are strongly species-specific, yet recognize many different strains as potential symbionts.
Nevertheless, we found that, regardless of which population of squid hosts they are isolated from, different V. fischeri strains will exhibit one of two strategies during competition for colonization of E. scolopes. Some strains, mainly those designated ‘D-type', outcompete others to become the dominant bacterium in the light organ. Although these D-type strains are more fit at initiating symbiosis, they may survive less well in seawater (Wollenberg and Ruby, 2012), a result that suggests symbiotic- and planktonic-niche specialization, and predicts a possible relationship between the frequency of these types of strains and the local density of host populations. One can speculate that such a dominant behavior may place the symbiosis at a greater risk because, in the face of environmental disturbance, a clonal population might be less robust (West et al., 2007). In contrast, the behavior exhibited by S-type strains, which more readily co-colonize the juvenile light organ, results in the mixed-strain population structure most often observed in field-caught adult E. scolopes (Wollenberg and Ruby, 2009). In such a strategy of sharing the tissue habitat, the mixed population may be less vulnerable to perturbations (Hodgson et al., 2002; Lee et al., 2014). Despite these two different behaviors, co-occurrence between D-type and S-type strains can be observed in the light organs of field-caught squid (Wollenberg and Ruby, 2009). Thus, it is possible for an S-type strain to escape the process that selects against it during initiation, perhaps by either (i) becoming established in a crypt before the recruitment of a D-type strain, thereby avoiding competition during the initiation phase, or (ii) existing as a clonal colonization within an individual crypt, thus escaping inter-strain competition. Future studies can resolve between these possibilities by tracking the trajectory of a mixed colonization as the animal matures (Koch et al., 2014), as well as by imaging the fate of mixed populations in each of the organ's six crypts (Wollenberg and Ruby, 2009). Whether the selection occurs at the intra-bacterial level, and/or the host is participating by a selective sanctioning (Nyholm et al., 2000; Koch et al., 2014), remains an intriguing and unresolved question. In either case, because each of the strains used in this study can, when inoculated by itself, initiate a light-organ symbiosis, it is likely that the bacteria have an important role in strain-level selection.
The conservation of genomic synteny among V. fischeri strains is a characteristic also observed in V. parahaemolyticus (Chen et al., 2011), suggesting that the genomes of these two species are more stable than those of certain other Vibrio species. Although no genome-wide analysis has been performed yet, Vibrio strains in the beijerinckii clade have been reported to have surprisingly little sequence divergence, even among geographically dispersed isolates, collected 80 years apart (Figge et al., 2011). In contrast to these examples of conservation, populations of V. cholerae and V. vulnificus maintain much more genomic diversity due, in part, to the genes captured within a super-integron locus (Chen et al., 2003; Chun et al., 2009; Boucher et al., 2011). Similarly, V. harveyi uses extensive gene duplication and fixation to diversify its genome (Gu et al., 2009). However, the most striking example of extensive evolution with a Vibrio species is the remarkably high genomic diversity reported for V. splendidus strains; even environmentally co-occurring isolates have up to 1-Mb difference in gene content from one to another (Thompson et al., 2005; Le Roux et al., 2009). Perhaps the relatively high degree of conservation characteristic of V. fischeri strains collected in the Pacific Ocean over decades and across thousands of kilometers is due, in part, to a reduced level of environmental and niche challenges compared with some of its congeners.
The average total number of proteins (~4000) encoded by all of the 13 strains of V. fischeri is comparable with that of other Vibrio spp. (Gu et al., 2009). However, around 200 genes, found in 41 loci comprising two or more adjacent orfs, are present only in the genomes of D-type strains. The encoded proteins are attractive candidates through which the underlying cause(s) of the dominant behavior of this group of strains can be discovered and should be the focus of another work. Because there is no protein specifically encoded by the S-type strain group, and the D-type strains appear to be a newly diverged group (Figure 3), we predict that the D-type strains have evolved their dominance recently. One possible explanation for the appearance of their domination during initiation is that an ancestral V. fischeri acquired a gene (or block of genes) that subsequently became fixed in the genome. A well-studied example of such an event is the acquisition of the Tcp pilus genes by V. cholerae through their carriage by a lysogenic phage (Faruque and Mekalanos, 2012). In V. cholerae, this pilus is encoded at a single locus; however, in V. fischeri ES114, one of the few bacterial strains outside of V. cholerae to carry Tcp structural genes, the homologous orfs are widely dispersed across its two chromosomes (Ruby et al., 2005). It is possible that, like V. cholerae, an ancestor of ES114 also acquired the Tcp pilus genes as a block, but they have subsequently spread along the genome. Thus, the ancestor of the D-type strains may have also acquired one or more blocks comprising their 200 shared genes, which also have subsequently scattered within the descendants' genomes. This acquisition and dispersion appears to have occurred relatively recently as the D-type strains do not resolve even on a phylogenetic tree derived from the entire core genome (Figure 3). An alternate hypothesis that a common ancestor harbored this block, which was lost in all descendants except the D-type strains, is less likely because it requires at least four independent events of lost.
In the future, it will be informative to extend the investigation of both the symbiotic behavior and the genome content of the 12 E. scolopes-derived strains to those of other V. fischeri isolates that either are not light-organ symbionts (Wollenberg et al., 2012) or are specific to other squid or fish species (Nishiguchi et al., 1998; Urbanczyk et al., 2011). Because, over time, the host can sanction specific members of the symbiont population (Koch et al., 2014); it is critical to include the role of the host in the dynamics of the symbiont population (McFall-Ngai et al., 2010; Palmer et al., 2010). In any case, tracking the population dynamics of artificially constructed combinations of symbionts during the approximately 9-month lifetime of the squid may reveal whether the D- or S-type strain behaviors described here increase stability in the symbiosis or, in contrast, lead to undesirable outcomes for the association.
Acknowledgments
We thank the Ruby and McFall-Ngai lab members for their helpful discussions, and Phil Arevalo for his bioinformatics contribution. We also thank Samantha Mahoney and Emily Forster for their contribution with the strain colonization experiments. We are indebted to Michael Wollenberg and Toby Kiers for their advice on the manuscript. Support was provided by NIH grants from NIGMS (GM099507), NIAID (AI050661) and ORIP (RR012294/OD011024).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Antunes LC, Schaefer AL, Ferreira RB, Qin N, Stevens AM, Ruby EG et al. (2007). Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J Bacteriol 189: 8387–8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyakumar DS, Nishiguchi MK. (2009). Characterization of two host-specific genes, mannose-sensitive hemagglutinin (mshA and uridyl phosphate dehydrogenase (UDPDH) that are involved in the Vibrio fischeri-Euprymna tasmanica mutualism. FEMS Microbiol Lett 299: 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, Dejongh M, Disz T, Edwards RAF et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck NA, Krukonis ES, Dirita VJ. (2004). TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP. J Bacteriol 186: 8309–8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher Y, Cordero OX, Takemura A, Hunt DE, Schliep K et al. (2011). Local mobile gene pools rapidly cross species boundaries to create endemicity within global Vibrio cholerae populations. mBio 2: pii: e00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CA, DeLoney-Marino CR, Mandel MJ. (2013). Chemoreceptor VfcA mediates amino acid chemotaxis in Vibrio fischeri. Appl Environ Microbiol 79: 1889–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan SM, Dunlap PV. (2000). LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J Bacteriol 182: 2811–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston J, Goodrich-Blair H. (2010). Common trends in mutualism revealed by model associations between invertebrates and bacteria. FEMS Microbiol Rev 34: 41–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Dozal A, Hogan D, Gorman C, Quintanal-Villalonga A, Nishiguchi MK. (2012). Multiple Vibrio fischeri genes are involved in biofilm formation and host colonization. FEMS Microbiol Ecol 81: 562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Wu KM, Chang YC, Chang CH, Tsai HC, Liao TL et al. (2003). Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res 13: 2577–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Stine OC, Badger JH, GIL AI, Nair GB, Nishibuchi M et al. (2011). Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence. BMC Genomics 12: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers BM, Klose KE. (2007). Regulation of virulence in Vibrio cholerae: the ToxR regulon. Future Microbiol 2: 335–344. [DOI] [PubMed] [Google Scholar]
- Christie PJ, Gordon JE. (2014). The Agrobacterium Ti plasmids. Microbiol Spectr 2: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ et al. (2009). Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA 106: 15442–15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero OX, Polz MF. (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol 12: 263–273. [DOI] [PubMed] [Google Scholar]
- Craig L, Pique ME, Tainer JA. (2004). Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2: 363–378. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoney-Marino CR, Wolfe AJ, Visick KL. (2003). Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl Environ Microbiol 69: 7527–7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulla GF, Go RA, Stahl DA, Davidson SK. (2012). Verminephrobacter eiseniae type IV pili and flagella are required to colonize earthworm nephridia. ISME J 6: 1166–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap PV, Ast JC, Kimura S, Fukui A, Yoshino T, Endo H. (2007). Phylogenetic analysis of host–symbiont specificity and codivergence in bioluminescent symbioses. Cladistics 23: 507–532. [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. (2006). New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72: 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Mekalanos JJ. (2012). Phage-bacterial interactions in the evolution of toxigenic Vibrio cholerae. Virulence 3: 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidopiastis PM, Von Boletzky S, Ruby EG. (1998). A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J Bacteriol 180: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge MJ, Robertson LA, Ast JC, Dunlap PV. (2011). Historical microbiology: revival and phylogenetic analysis of the luminous bacterial cultures of M. W. Beijerinck. FEMS Microbiol Ecol 78: 463–472. [DOI] [PubMed] [Google Scholar]
- Graf J, Dunlap PV, Ruby EG. (1994). Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol 176: 6986–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Neary J, Cai H, Moshfeghian A, Rodriguez SA, Lilburn TG et al. (2009). Genomic and systems evolution in Vibrionaceae species. BMC Genomics 10(Suppl 1): S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hodgson DJ, Rainey PB, Buckling A. (2002). Mechanisms linking diversity, productivity and invasibility in experimental bacterial communities. Proc Biol Sci 269: 2277–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Nishiguchi MK. (2004). Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar Biol 144: 1151–1155. [Google Scholar]
- Jones CW, Armitage JP. (2015). Positioning of bacterial chemoreceptors. Trends Microbiol 23: 247–256. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Nikoh N, Meng XY, Kamagata Y, Fukatsu T. (2009). Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbell J, McFall-Ngai M, Roderick G. (2002). Two genetically distinct populations of Euprymna scolopes in the shallow waters of Oahu, Hawaii. Pac Sci 56: 347–355. [Google Scholar]
- Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG. (2014). Features governing symbiont persistence in the squid-vibrio association. Mol Ecol 23: 1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux F, Zouine M, Chakroun N, Binesse J, Saulnier D, Bouchier C et al. (2009). Genome sequence of Vibrio splendidus: an abundant planctonic marine species with a large genotypic diversity. Environ Microbiol 11: 1959–1970. [DOI] [PubMed] [Google Scholar]
- Lee KH, Ruby EG. (1994. a). Competition between Vibrio fischeri strains during initiation and maintenance of a light organ symbiosis. J Bacteriol 176: 1985–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Ruby EG. (1994. b). Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in Nature. Appl Environ Microbiol 60: 1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, Rice SA. (2014). Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J 8: 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJJr, Roos DS. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. (2009). A single regulatory gene is sufficient to alter bacterial host range. Nature 458: 215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J, Stabb EV, Millikan DS, Ruby EG. (2003). Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol 69: 5928–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Nyholm SV, Castillo MG. (2010). The role of the immune system in the initiation and persistence of the Euprymna scolopes—Vibrio fischeri symbiosis. Semin Immunol 22: 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes A, Thies J, Abbott L, Howieson J. (2004). Structure and diversity among rhizobial strains, populations and communities–a review. Soil Biol Biochem 36: 1295–1308. [Google Scholar]
- Miller MB, Bassler BL. (2001). Quorum sensing in bacteria. Annu Rev Microbiol 55: 165–199. [DOI] [PubMed] [Google Scholar]
- Moran NA, Bennett GM. (2014). The tiniest tiny genomes. Annu Rev Microbiol 68: 195–215. [DOI] [PubMed] [Google Scholar]
- Nieto PA, Pardo-Roa C, Salazar-Echegarai FJ, Tobar HE, Coronado I, Riedel CA et al. (2016). New insights about excisable pathogenicity islands in Salmonella and their contribution to virulence. Microbes Infect e-pub ahead of print 3 March 2016; doi:10.1016/j.micinf.2016.02.001. [DOI] [PubMed]
- Nikolakakis K, Monfils K, Moriano-Gutierrez S, Brennan CA, Ruby EG. (2015). Characterization of the Vibrio fischeri fatty acid chemoreceptors, VfcB and VfcB2. Appl Environ Microbiol 82: 696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi MK, Ruby EG, McFall-Ngai MJ. (1998). Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid-vibrio symbioses. Appl Environ Microbiol 64: 3209–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. (2004). The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol 2: 632–642. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. (2000). Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci USA 97: 10231–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer TM, Doak DF, Stanton ML, Bronstein JL, Kiers ET, Young TP et al. (2010). Synergy of multiple partners, including freeloaders, increases host fitness in a multispecies mutualism. Proc Natl Acad Sci USA 107: 17234–17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Asato LM. (1993). Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol 159: 160–167. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Urbanowski M, Campbell j, Dunn A, Faini M, Gunsalus R et al. (2005). Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci USA 102: 3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabo G et al. (2012). Population genomics of early events in the ecological differentiation of bacteria. Science 336: 48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, Ruby EG. (2003). Contribution of pilA to competitive colonization of the squid Euprymna scolopes by Vibrio fischeri. Appl Environ Microbiol 69: 820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakaran S, Retz F, Kikuchi Y, Kost C, Kaltenpoth M. (2015). Evolutionary transition in symbiotic syndromes enabled diversification of phytophagous insects on an imbalanced diet. ISME J 9: 2587–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J et al. (2005). Genotypic diversity within a natural coastal bacterioplankton population. Science 307: 1311–1313. [DOI] [PubMed] [Google Scholar]
- Urbanczyk H, Ast JC, Dunlap PV. (2011). Phylogeny, genomics, and symbiosis of Photobacterium. FEMS Microbiol Rev 35: 324–342. [DOI] [PubMed] [Google Scholar]
- Verma SC, Miyashiro T. (2013). Quorum sensing in the squid-vibrio symbiosis. Int J Mol Sci 14: 16386–16401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. (2007). The social lives of microbes. Annu Rev Ecol Evol Syst 38: 53–77. [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, Koroleva I et al. (2010). Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA 107: 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg MS, Preheim SP, Polz MF, Ruby EG. (2012). Polyphyly of non-bioluminescent Vibrio fischeri sharing a lux-locus deletion. Environ Microbiol 14: 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg MS, Ruby EG. (2009). Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol 75: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg MS, Ruby EG. (2012). Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J 6: 352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawata Y, Cordero OX, Menolascina F, Hehemann JH, Polz MF, Stocker R. (2014). Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci USA 111: 5622–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.