Abstract
We propose a method for detecting evolutionary forces underlying community assembly by quantifying the strength of community–environment relationships hierarchically along taxonomic ranks. This approach explores the potential role of phylogenetic conservatism on habitat preferences: wherein, phylogenetically related taxa are expected to exhibit similar environmental responses. Thus, when niches are conserved, broader taxonomic classification should not diminish the strength of community–environment relationships and may even yield stronger associations by summarizing occurrences and abundances of ecologically equivalent finely resolved taxa. In contrast, broader taxonomic classification should weaken community–environment relationships when niches are under great divergence (that is, by combining finer taxa with distinct environmental responses). Here, we quantified the strength of community–environment relationships using distance-based redundancy analysis, focusing on soil and seawater prokaryotic communities. We considered eight case studies (covering a variety of sampling scales and sequencing strategies) and found that the variation in community composition explained by environmental factors either increased or remained constant with broadening taxonomic resolution from species to order or even phylum level. These results support the niche conservatism hypothesis and indicate that broadening taxonomic resolution may strengthen niche-related signals by removing uncertainty in quantifying spatiotemporal distributions of finely resolved taxa, reinforcing the current notion of ecological coherence in deep prokaryotic branches.
Introduction
Clarifying the mechanisms by which community composition changes in response to environmental variability is important for understanding biodiversity, productivity and ecosystem stability (Hector and Bagchi, 2007; Ives and Carpenter, 2007). Traditional measures of community composition typically focus on species-level organization because species has long been considered the fundamental unit of biological classification (Magurran, 2004). For example, preparing a site-by-species matrix (observed species across sampling sites) is usually a first step when examining the dynamics of community composition across time and space (Magurran, 2004). Further, a common monitoring and assessment practice attempts to characterize community–environment relationships by combining the variation of species composition with environmental measurements (Margules and Pressey, 2000; Hansen et al., 2001). Note, however, that species-level community data are not always available because of constraints on time and the expertise required to properly identify all individuals to species level (Warwick, 1993; Bailey et al., 2001; Terlizzi et al., 2003). Accordingly, the concept of taxonomic surrogacy or sufficiency (that is, using broadly resolved taxonomic data as a substitute for species-level data) has been subjected to extensive examination in bioassessment studies (Warwick, 1993; Bailey et al., 2001; Terlizzi et al., 2003). Despite some controversial opinions about general applications for particular organisms and ecosystems, the use of broadly resolved data (that is, genus-, family- or even phylum-level classification) has been demonstrated to portray similar community-scale responses to environmental variability as species-level data in many empirical studies (Olsgard et al., 1998; Heino and Soininen, 2007; Heino, 2008; Bhusal et al., 2014; Heino, 2014).
Broad-level taxonomic data have been widely applied to various types of organisms in terrestrial, freshwater and marine environments as a means of optimizing the cost-effectiveness of detecting environmental impact on community organization (Terlizzi et al., 2003; Bennett et al., 2014). However, little attention has been devoted toward a mechanistic understanding of why broadly and finely resolved data can be similarly efficacious for the assessment of community–environment relationships (but see Bevilacqua et al., 2013). From the evolutionary perspective, the high efficacy of broadly resolved data may be explained by the conservation of habitat preferences among phylogenetically related taxa (Webb et al., 2002; Wiens and Graham, 2005; Wiens et al., 2010). In contrast, habitat differentiation between closely related taxa (Debussche and Thompson, 2003) and/or convergent evolution between distinctly related taxa (Cavender-Bares et al., 2004) is likely to be vital in those cases, which fail to detect ecologically meaningful patterns using broadly resolved data. Review of some earlier studies (Olsgard et al., 1998; Heino and Soininen, 2007; Heino, 2008; Bhusal et al., 2014; Heino, 2014) suggests that varying the taxonomic resolution of composition data exerts an influence on the strength (usually quantified as explained variance R2) of community–environment relationships, with the best taxonomic resolution differing for various targeted organisms and/or study regions. Thus, there are a variety of potential mechanisms by which taxonomic resolution may influence the strength of community–environment relationships. Here we aim to bridge the concepts of taxonomic surrogacy and phylogenetic conservatism with an emphasis on the impact of niche conservatism or niche divergence on observed macro-evolutionary patterns (Figure 1; Supplementary Figure S1).
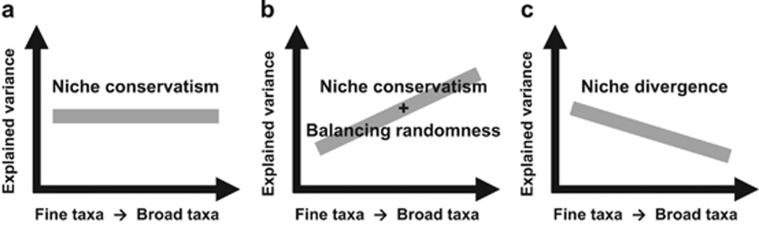
Figure 1.
A conceptual scheme for revealing the evolutionary effect regarding niche conservatism or niche divergence on community assembly. The strength of the community–environment relationship can be quantified by explained variance (R2) in multiple linear regression or redundancy analysis. (a) When niches are conserved over evolutionary time, the strength of community–environment relationships would remain constant with broadening taxonomic resolution, as phylogenetically related taxa exhibit similar responses to environmental filtering. (b) Moreover, under strong niche conservatism, the strength of community–environment relationships may increase with broadening taxonomic resolution, especially if broader taxonomic units that combine occurrences and abundances of ecologically equivalent taxa can balance the inherent spatiotemporal randomness of finer units. (c) In contrast, when niches experience quick divergence over evolutionary time, the strength of community–environment relationships would decrease with broadening taxonomic resolution, as each more finely resolved taxonomic unit carries unique responses to the environment. See Supplementary Figure S1 for a conceptual example.
We propose that evaluating the strength of community–environment relationships hierarchically along taxonomic ranks might serve as a means to infer the importance of evolutionary forces underlying community assembly. It is theoretically possible that broadening taxonomic resolution does not weaken the strength of community–environment relationships if broader taxonomic units carry niche-related signals as strong as those associated with finer units (Figure 1a; Supplementary Figure S1a). This scenario might be expected when taxa belonging to the same lineage tend to exhibit similar responses to environmental factors (Warwick, 1993; Bailey et al., 2001). Moreover, when finely resolved taxa are hypothesized to be ecologically equivalent (that is, they have equivalent fitness and potentially occupy the same niche space) (Leibold and McPeek, 2006), broad-level taxonomic classification might be expected to enhance the strength of community–environment relationships (Figure 1b; Supplementary Figure S1b), as broader taxonomic grouping can balance the randomness (caused by neutral-stochastic processes) in spatiotemporal distributions associated with finer taxa, by summarizing occurrences and abundances of those ecologically equivalent units as a signal responder (Warwick, 1993; Bailey et al., 2001). In contrast, the strength of community–environment relationships might be expected to decrease with the combination of finer taxonomic groups that exhibit distinct environmental responses (Figure 1c; Supplementary Figure S1c). This scenario could happen if habitat preferences diverge quickly over evolutionary time; for instance, phylogenetically related species undergo adaptive diversification in habitat-use (Schluter, 2000; Gavrilets and Losos, 2009; Pfennig and Pfennig, 2009).
To test our approach, we used prokaryotic communities to evaluate the niche-based community assembly hypothesis (here the niche is characterized by environmental conditions), as prokaryotes are functionally diverse and believed to be sensitive to the changes in environmental conditions, owing to large population sizes with short generation times (Whitman et al., 1998; Torsvik et al., 2002). Here, we used modern amplicon sequencing or shotgun sequencing of metagenomes (Table 1) to determine prokaryotic community compositions. For prokaryotic identification, phylogenetic relatedness and taxonomic classification can be uniformly determined by a single genetic marker (that is, the 16S ribosomal RNA (rRNA) gene) (Woese, 1987), which permits a systematic comparison of prokaryotic compositions across various environments. For hierarchical grouping of taxonomic units (Yarza et al., 2014), two main types of hierarchical systems exist in parallel: (1) based on molecular criteria, defining operational taxonomic unit (OTU) ranks through sequence similarity cutoffs (for example, 97%, 94%, 91%); and (2) based on database annotation, following traditional taxonomic classification (that is, species, genus, etc). In this study, we used both types of hierarchical systems to generate composition profiles at fine to broad taxonomic resolutions and evaluated how the strength of community–environment relationships changed along taxonomic ranks. In addition, we calculated the phylogeny-based UniFrac metric (Lozupone and Knight, 2005) as a standard to contrast our multi-level taxonomy-based composition analyses, as this metric has recently gained popularity in microbial community analyses and believed to be more informative than taxonomy-based estimates (Lozupone and Knight, 2005; Swenson, 2014).
Table 1. Characteristics of sequence-based prokaryotic community data sets for analysis in this study.
| Case | Targeted community | Study location | Scale | Sequencing approach | 16S region (primers) | Read length | Subsampled read | Data point | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Soil prokaryotes (surface) | North and South America | Global | Amplicon-454 | V1–V2 (27F/338R) | 312 | 400 | 88 | Lauber et al. (2009) |
| 2 | Soil prokaryotes (surface) | Canadian, Alaskan, and European Arctic | Global | Amplicon-454 | V1–V2 (27F/338R) | 312 | 400 | 40 | Chu et al. (2010) |
| 3 | Soil prokaryotes (surface) | New York City's Central Park, USA | Local | Amplicon-Illumina | V4–V5 (515F/806R) | 292 | 37 000 | 580 | Ramirez et al. (2014) |
| 4 | Soil prokaryotes (surface) | Long Island's vineyards, New York, USA | Local | Amplicon-Illumina | V4–V5 (515F/806R) | 292 | 1500 | 94 | Zarraonaindia et al. (2015) |
| 5 | Seawater prokaryotes (surface) | Tara Oceans (Atlantic, Pacific, Indian Oceans...) | Global | Metagenome-Illumina | Varied (shotgun) | 164 | 35 000 | 59 | Sunagawa et al. (2015) |
| 6 | Seawater prokaryotes (Chl maximum) | Tara Oceans (Atlantic, Pacific, Indian Oceans...) | Global | Metagenome-Illumina | Varied (shotgun) | 164 | 35 000 | 39 | Sunagawa et al. (2015) |
| 7 | Seawater prokaryotes (surface) | Station L4 of the Western English Channel | Local | Amplicon-454 | V6 (967F/1064R) | 98 | 4200 | 61 | Gilbert et al. (2012) |
| 8 | Seawater prokaryotes (surface) | A short transect of the Southern East China Sea | Local | Amplicon-454 | V6 (967F/1064R) | 98 | 9800 | 27 | Yeh et al. (2015) |
Moreover, given the potential scale dependency (Levin, 1992; Cavender-Bares et al., 2006, 2009; Swenson et al., 2006, 2007) in community–environment relationships, we considered case studies covering sampling sites from either global or local scales (Table 1) and which may provide insight into the scale dependency of microbial community assembly. Previous studies on plant communities (focused on within-community phylogenetic structure) have identified concerns about spatiotemporal and taxonomic scales on the detected pattern. For example, the signal of phylogenetic clustering (that is, taxa within a given community contain lower mean phylogenetic distances than expected from random; as evidence for habitat filtering) has been found to be relatively evident when considering samples from a larger geographic region and/or a broader span of taxonomy (Cavender-Bares et al., 2006, 2009; Swenson et al., 2006, 2007). In this study, rather than detecting the within-community structure, we concentrated our analyses on the inter-community pattern (that is, how taxonomic compositions differ across sites in response to environmental factors) to assess whether changing taxonomic resolution affects the explained variance of community–environment relationships (Figure 1). This analytical framework might be directly relevant to the question of how functional traits are conserved through macro-evolutionary time (Lennon et al., 2012; Martiny et al., 2013; Tresedera and Lennon, 2015). Moreover, plant ecologists have suggested that traits related to large-scale distributions (that is, beta-niche; along temperature and precipitation gradients) are generally conserved over time, whereas traits linked to small-scale distributions (that is, alpha-niche; local coexisting patterns) tend to be evolutionarily labile (Cavender-Bares et al., 2004, 2009; Silvertown et al., 2006a, 2006b). With this notion, we expect that the evidence for niche conservatism may be more easily detected by global data sets, whereas the evidence for niche divergence may be better revealed with local data sets.
Here, we used eight case studies (Table 1) to demonstrate our approach and explore empirical patterns of prokaryotic community responses to environmental variables in soil and marine ecosystems. We argue that an approach such as ours, which assesses community–environment relationships while taking into consideration hierarchical taxonomic information, has the potential to advance our understanding regarding the evolutionary effects such as niche conservatism or niche divergence on community assembly, improving our ability to estimate community characteristics corresponding to environmental conditions.
Materials and methods
Sequence-based prokaryotic community data sets
We used eight data sets: (1) Lauber ‘America-Soils' study (Lauber et al., 2009) (referred to as case #1 hereafter), (2) Chu ‘Arctic-Soils' study (Chu et al., 2010) (referred to as case #2 hereafter), (3) Ramirez ‘NYpark-Soils' study (Ramirez et al., 2014) (referred to as case #3 hereafter), (4) Zarraonaindia ‘NYfarm-Soils' study (Zarraonaindia et al., 2015) (referred to as case #4 hereafter), (5) Sunagawa ‘TaraSur-Seawaters' study (Sunagawa et al., 2015) (referred to as case #5 hereafter), (6) Sunagawa ‘TaraChl-Seawaters' study (Sunagawa et al., 2015) (referred to as case #6 hereafter), (7) Gilbert ‘WEC-Seawaters' study (Gilbert et al., 2012) (referred to as case #7 hereafter) and (8) Yeh ‘SECS-Seawaters' study (Yeh et al., 2015) (referred to as case #8 hereafter) to test our theoretical framework regarding how the strength of community–environment relationships varies with changes in taxonomic resolution (Figure 1). We summarize the characteristics of these sequence-based prokaryotic community data sets in Table 1. Detailed information regarding data sources and data properties can be found in Supplementary Methods. The lists of community samples and environmental factors used in the present study are provided in Supplementary Tables S1–S8.
Processing of sequence data sets to community data sets
Sequence data sets were processed using QIIME version 1.9.0 (Caporaso et al., 2010), following the settings of Qiita (http://qiita.microbio.me). To satisfy our requirement for accurate taxonomic assignments along fine to broad taxonomic resolutions, we adopted the classification system of the GreenGenes database (providing a set of 16S rRNA gene references and linkage maps of reference sequences) (DeSantis et al., 2006a) for hierarchical grouping of sequence reads. A closed-reference OTU picking protocol against the GreenGenes otu97 core set (version 13_8) was carried out using SortMeRNA (Kopylova et al., 2012). To avoid inaccurate taxonomic assignments because of short reads, we only used OTUs with significant hits (similarity >0.97 and coverage >0.97) to the GreenGenes otu97 core set for community profiles. Then, we used CopyRighter (Angly et al., 2014) to adjust relative abundance profiles, which correct the bias of the 16S rRNA gene copies among different prokaryotic genomes (Acinas et al., 2004; Vetrovsky and Baldrian, 2013). This initial site-by-otu97 matrix was then aggregated into a series of community matrices with fine to broad taxonomic resolutions, according to GreenGenes OTU linkage maps (DeSantis et al., 2006b) and GreenGenes explicit taxonomic ranks (McDonald et al., 2012).
Two types of hierarchical community matrices were generated: (1) site-by-OTU matrices relying on sequence similarity; and (2) site-by-taxon matrices relying on database annotation. For site-by-OTU matrices: the initial site-by-otu97 matrix was transformed into the site-by-otu94 matrix according to the GreenGenes otu97-otu94 linkage map (DeSantis et al., 2006b), and a series of site-by-OTU matrices were analogously generated in line with the corresponding OTU linkage maps (a total of 10 ranks, from otu97 to otu70 with an interval of 3% sequence similarity). For site-by-taxon matrices: hierarchical taxonomic annotation of the initial site-by-otu97 matrix was retrieved according to the GreenGenes explicit taxonomic ranks (McDonald et al., 2012), and a series of site-by-taxon matrices were consequently generated (a total of six ranks, from species to phylum). Example files of site-by-OTU matrices, site-by-taxon matrices, and related R codes for community analyses along taxonomic ranks (see below) can be found in Supplementary Data.
To provide a reliable comparison in community structure across sampling sites, all the following community analyses were based on 100 subsampled data sets with an equal number of sequence reads per community (based on the minimum read number; Table 1).
Compositional variation measured at distinct taxonomic resolutions
Compositional variation across sampling sites was assessed with the Bray–Curtis dissimilarity index (Bray and Curtis, 1957), which was calculated for each of 10 site-by-OTU matrices and 6 site-by-taxon matrices, using the ‘vegdist' function of the ‘vegan' package (version 2.2-1) (Oksanen et al., 2015) in R (version 3.1.3) (R Development Core Team, 2015). In addition, we applied abundance-weighted UniFrac (unique fraction metric) (Lozupone and Knight, 2005) to quantify the phylogeny-based compositional variation, based on the site-by-otu97 matrix with the pruned GreenGenes phylogenetic tree (containing only taxa found in each data set), using the QIIME platform (Caporaso et al., 2010).
Evaluating the similarity of inter-community relationships
Before incorporating environmental measurements into community analyses, the similarity of the inter-community relationships (that is, the Bray–Curtis results) measured at different taxonomic resolutions was evaluated in order to estimate: (1) whether broad-level classification data can reveal inter-community relationships as fine-level classification data; and (2) whether two types of hierarchical systems (site-by-OTU and site-by-taxon) can generate comparable composition profiles from fine to broad levels. To evaluate the similarity of inter-community relationships at different taxonomic resolutions, the compositional variation quantified at each taxonomic resolution was displayed in an ordination diagram using multi-dimensional scaling (Legendre and Legendre, 2012); similarly, the phylogeny-based compositional variation (that is, the UniFrac distance matrix) was also displayed using multi-dimensional scaling ordination. Then, Procrustes analysis (Peres-Neto and Jackson, 2001) was conducted to detect the correlation of each pair of multi-dimensional scaling ordination plots, using the ‘protest' function of the ‘vegan' package (version 2.2-1) (Oksanen et al., 2015) in R (version 3.1.3) (R Development Core Team, 2015).
Evaluating the strength of community–environment relationships
To quantify the strength of community–environment relationships along taxonomic ranks, the explained variance (adjusted R2) (Peres-Neto et al., 2006) of distance-based redundancy analysis (Legendre and Anderson, 1999) was used to estimate the influence of environmental factors on community compositional variation (that is, the Bray–Curtis dissimilarity or UniFrac distance matrix), using the ‘capscale' function of the ‘vegan' package (version 2.2-1) (Oksanen et al., 2015) in R (version 3.1.3) (R Development Core Team, 2015). In order to equitably compare environmental influence on community structure at different taxonomic resolutions, we performed distance-based redundancy analysis with the whole set of environmental variables (that is, the whole set of soil or water factors; standardized to zero mean and unit variance) for each case. Moreover, the analyses based on each environmental variable were also applied to assess the importance of each factor on community compositional variation.
Randomization tests
To further provide statistical assurance concerning application of our method, we conducted randomization tests to evaluate whether the detected trends of community–environment relationships along taxonomic ranks derived from the real data sets significantly differ from the predictions based on randomized data sets. For multi-level taxonomy-based Bray–Curtis, we shuffled linkages between fine-taxa and broad-taxa level by level (remaining topology), and generated randomized data sets based on 100 random linkage maps. In addition, for phylogeny-based UniFrac, we shuffled tip labels of the phylogenetic tree and re-calculated UniFrac based on 100 random phylogenetic trees, using the ‘phyloshuffle' function of the ‘phylotools' package (version 0.1.2) (Zhang et al., 2012) in R (version 3.1.3) (R Development Core Team, 2015). The explained variance of community–environment relationships for Bray–Curtis at each taxonomic level and phylogeny-based UniFrac generated from the randomized data were re-examined as described above as the null expectation.
Additional analyses regarding data quality
The eight case studies used here cover distinct sampling scales and sequencing strategies (Table 1), allowing us to explore empirical patterns of community–environment relationships. In addition to the analyses mentioned above, as a variety of sequencing techniques with different sequencing efforts exist for modern sequence-based community data sets, we further conducted sensitivity tests to evaluate the effects of varying read length and read depth on our detected patterns along taxonomic ranks. Basically, we found that the detected trends and strengths of the community–environment relationships along taxonomic ranks were robust, regardless of the sequencing length or depth. The detailed methods and results of these analyses are provided in Supplementary Methods and Supplementary Results.
Moreover, although describing taxonomic composition with relative abundance information should be more precise when assessing community dynamics (Jost, 2007), however, for some early ecological surveys, only incidence data (detection or non-detection of each species) are available. Thus, in addition to analyses based on abundance-weighted data, we also evaluated community–environment relationships based on transformed presence–absence matrices for the sake of comparison. Compared with abundance-weighted results, the compositional variation explained by environmental factors was reduced markedly when using presence–absence data, although the detected trends along taxonomic ranks generally remained and showed a distinction from the randomization patterns. The detailed results can be found in Supplementary Results.
Results
Inter-community relationships measured at distinct taxonomic resolutions
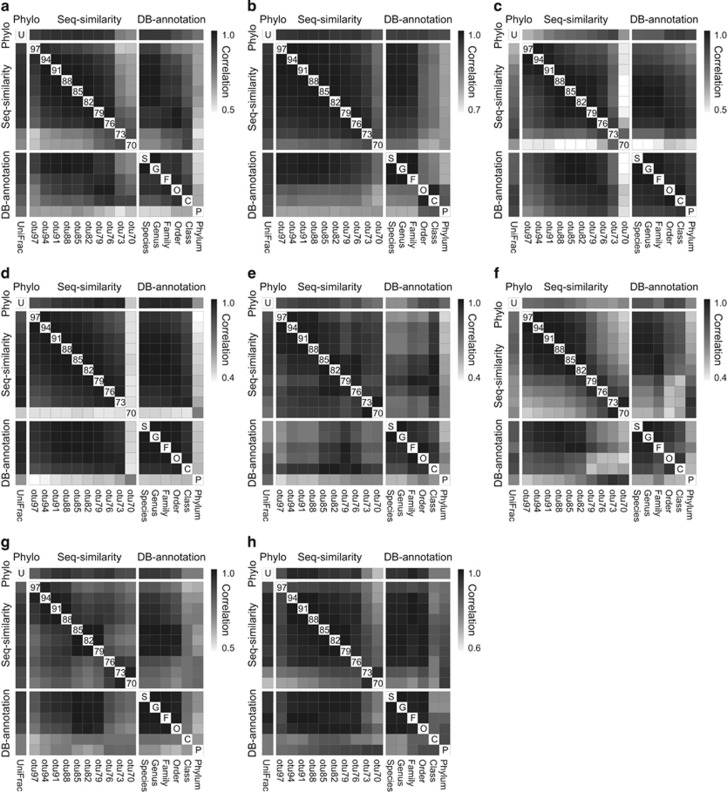
In all eight cases, community dissimilarities in abundance-weighted compositions measured at distinct taxonomic resolutions are significantly correlated to each other (Procrustes correlations ranging from 0.41 to 0.99; Figure 2). These strong Procrustes correlations indicate that the inter-community relationships revealed by broadly resolved taxonomic data are largely consistent with the patterns revealed by finely resolved taxonomic data (Supplementary Figures S2–S9). Specifically, measures at two adjacent taxonomic levels conveyed a higher similarity in their ordination diagrams than those at more separated levels (Figure 2; Supplementary Figures S2–S9). In addition, fine to broad levels of composition profiles extracted from two types of hierarchical matrices (site-by-OTU and site-by-taxon) were found to generally match (Figure 2). For example, otu97-otu94 levels corresponded to species-genus levels, with strong correlations among those pairs of measures (Procrustes correlations >0.9 in most cases; Figure 2); similarly, otu79-otu70 levels were comparable with the phylum level. Moreover, we found that the phylogeny-based composition profiles (UniFrac) were significantly correlated with composition profiles at all taxonomic resolutions, showing the greatest similarity with family- or order-level (otu88-otu79) profiles in most cases (Procrustes correlations >0.8; Figure 2).
Figure 2.
Pairwise correlations of abundance-weighted compositions measured at different taxonomic resolutions based on Procrustes analyses. The community analyses are conducted on eight distinct data sets: (a) case #1 America-Soils, (b) case #2 Arctic-Soils, (c) case #3 NYpark-Soils, (d) case #4 NYfarm-Soils, (e) case #5 TaraSur-Seawaters, (f) case #6 TaraChl-Seawaters, (g) case #7 WEC-Seawaters and (h) case #8 SECS-Seawaters. Measures of taxonomy-based compositions are calculated following two types of hierarchical classification (sequence similarity: otu97 to otu70 and database annotation: species to phylum), and compared with the phylogeny-based composition using UniFrac metric.
Composition–environment relationships measured at distinct taxonomic resolutions
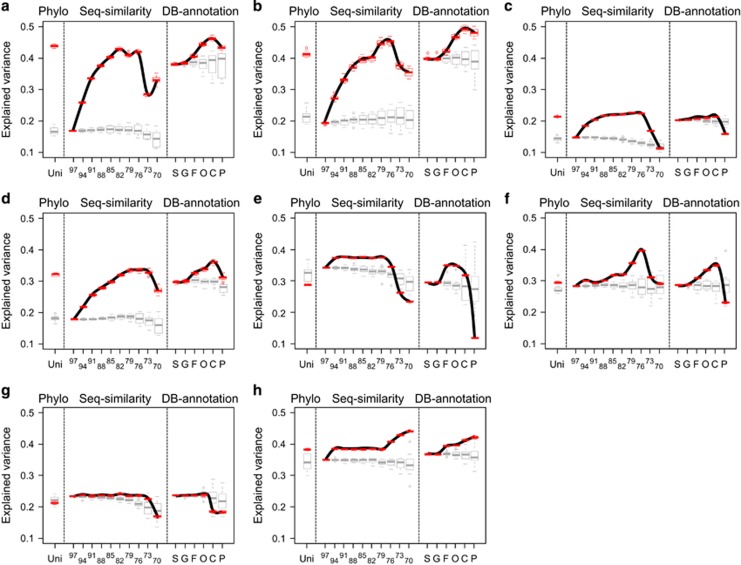
Compositional variation at all taxonomic resolutions can be significantly explained by environmental factors for all of the eight case studies (Figure 3), suggesting that habitat filtering is a strong force determining prokaryotic community structure. More importantly, the explained variation tends to increase or remain constant with broadening taxonomic resolution (the red boxplots connected by the black line; Figure 3), suggesting that taxa within the same lineages generally show similar responses to variability of environmental conditions. For cases #1 to #4, which targeted surface soil prokaryotes, the explained variation gradually increased up to the class level (otu82-otu76), and then decreased at the phylum level (Figures 3a–d). For cases #5 and #6, which represented seawater prokaryotic communities from two distinct epipelagic layers of the global oceans, the trends of explained variation achieved the maximum values at middle taxonomic levels, although the patterns differed in detail (Figures 3e and f). For case #7 of a seasonally sampled, temperate coastal, prokaryotic community dynamics, the explained variation remained constant from species to order levels, and then dropped at class and phylum levels (Figure 3g). Whereas the explained variation for the prokaryotic community in subtropical shelf waters (case #8) continually increased from species to the phylum level (Figure 3h). Considering the effect of scale dependency, the habitat filtering (implied by explained variation) seems to be more evident in the two global soil cases than the two local soil cases, whereas this effect is unclear for the seawater cases.
Figure 3.
The influence of environmental factors on variation of abundance-weighted compositions evaluated at fine to broad taxonomic resolutions. The community analyses are conducted on eight distinct data sets: (a) case #1 America-Soils, (b) case #2 Arctic-Soils, (c) case #3 NYpark-Soils, (d) case #4 NYfarm-Soils, (e) case #5 TaraSur-Seawaters, (f) case #6 TaraChl-Seawaters, (g) case #7 WEC-Seawaters and (h) case #8 SECS-Seawaters. Taxonomy-based compositional variation is calculated following two types of hierarchical classification (sequence similarity: otu97 to otu70 and database annotation: species to phylum), whereas phylogeny-based compositional variation is calculated using UniFrac metric for comparison. The influence of environmental factors on composition (adjusted R2, the explained variance) is quantified by distance-based redundancy analysis (db-RDA). The red boxplots (connected by the black line along taxonomic ranks) represent the explained variance of the db-RDA results based on 100 subsamples derived from the original data sets, whereas the gray boxplots represent the results based on the randomized data sets.
The detected patterns derived from original data sets are significantly different from the expectation based on randomized data sets (Figure 3). In the randomization null results (the gray boxplots; Figure 3), the explained variance remained constant or slightly decreased for community compositions at fine to broad taxonomic levels, with increasing uncertainty at broader levels (that is, higher variability as indicated by the boxplots). For soil cases, the amount of variation explained at the middle level (genus to class) was significantly higher in real data sets than what was expected from random. For seawater cases, the trends in explained variation along taxonomic ranks for real data sets were also significantly different from those predicted by randomization. Furthermore, considering the explained variation for the phylogeny-based composition (UniFrac), it showed roughly the same degree as the family- or order-level estimates in original data sets, whereas it became similar to the species-level estimates in the randomization results (Figure 3). These findings indicate the importance of considering evolutionary history among species when surveying and evaluating the deterministic ecological processes acting on community composition. Nevertheless, UniFrac, which weighs the phylogenetic information of the whole tree, may not necessarily be the optimal index to investigate environmental effects; that is, when to use UniFrac should depend on research purposes. For example, in the cases #5 and #7 (Figures 3e and g), the explained variation for real UniFrac results was lower than expected by chance, as a phylogeny-based metric might function as a summary indicator, providing an average estimate for composition measures across all taxonomic levels (Figures 2 and 3). Overall, these results show that analyzing multi-level taxonomic data instead of focusing on a single taxonomic level can provide additional information regarding the impact of niche conservatism or niche divergence on observed macro-evolutionary patterns.
Moreover, with respect to the results based on a single environmental variable (Supplementary Figures S10–S17), the strengthened or constant trends in explained variation of composition–environment relationships along taxonomic ranks are also evident in those cases, with pH accounting for the most compositional variation for soil samples (cases #1 to #4; Supplementary Figures S10–S13) and temperature or nitrogen source accounting for the most compositional variation for seawater samples (cases #5 to #8; Supplementary Figures S14–S17). Interestingly, for case #7, similar trends and strengths of composition–environment relationships can be detected when using day-length or artificial temporal indices as environmental variables (Supplementary Figure S16).
Discussion
Niche-related signals strengthen or remain along taxonomic ranks
We examined prokaryotic communities in soil and seawater samples to evaluate whether niche-related signals remain, strengthen or vanish with changes in taxonomic resolution (Figure 1). We detected strong correlations among compositions at fine and broad taxonomic resolutions for all eight case studies (Figure 2) and found an increasing or constant strength of the composition–environment relationships with broadening taxonomic resolution from species to order or even phylum level (Figure 3). These findings support the hypothesis of phylogenetic niche conservatism and further suggest that broader taxonomic classification may balance the distribution uncertainty associated with finer taxonomic units and strengthen niche-related signals. Overall, these results are consistent with the current notion of ecological coherence in deep prokaryotic branches (Philippot et al., 2009, 2010), which suggests that members of the same prokaryotic clades generally maintain similar ecological characteristics over evolutionary time (Martiny et al., 2015).
Typically, community data sets at the finest taxonomic resolution are preferred over more broadly resolved classifications in environmental assessment studies (Terlizzi et al., 2003; Bevilacqua et al., 2013; Bennett et al., 2014), because data sets with detailed taxonomy are expected to provide more accurate information regarding community structure and niche partitioning patterns. However, our results indicate that sometimes the use of broadly resolved data might be sufficient, or even superior to fine-level data if the goal is to identify community responses to environmental variables. Our assessment of environmental effects on the variation of prokaryotic communities detected a strengthened or constant niche-related signal with broadening taxonomic resolution (Figure 3), suggesting that phylogenetically related taxa might have constrained ecological properties and do not distribute randomly across habitats (Andersson et al., 2010; Philippot et al., 2010; Martiny et al., 2015). These results are consistent with some general notions about the phylogenetic conservatism of functional traits in microorganisms (Martiny et al., 2013). For example, studies on soil bacteria and fungi have indicated that traits associated with moisture preferences are highly conserved, with a broad taxonomic level usually accounting for the greatest variation in a given trait (Lennon et al., 2012; Tresedera and Lennon, 2015). Given strong phylogenetic conservatism, broadly resolved data may be well-suited for the detection and interpretation of prokaryotic community dynamics, especially in studies covering broad sampling scales with clear environmental variability. In fact, the original studies of cases #1 and #2 have reported strong correlations between relative abundances of dominant phyla/classes and soil pH (Lauber et al., 2009; Chu et al., 2010). In addition to the spatial variation, the prokaryotic communities might be also sensitive to the seasonal variation, and this temporal dynamics might be also well represented by broadly resolved data. In fact, the original studies of cases #7 and #8 have reported clear seasonal community dynamics using order-level or class-level compositions (Gilbert et al., 2009; Yeh et al., 2015).
Prokaryotic communities in natural environments usually contain extremely high species-level diversity; however, the mechanisms maintaining such high diversity are still unclear (Torsvik et al., 2002; Lynch and Neufeld, 2015). We observed a sharp increase of explained variation when grouping otu97 into otu94 and broader taxa (for example, at least 50% increase in R2 from otu97 to otu94 for soil cases; Figure 3), suggesting that under conditions of strong habitat filtering, some sequence-derived finely resolved taxa might exhibit similar responses to environmental conditions and behave as the same ecologically cohesive units (Philippot et al., 2009, 2010). Previous studies have hypothesized ecological equivalence for finely resolved taxa such that a particular niche may be occupied by any of the suitable taxa drawn from a large pool of candidates (Lozupone et al., 2012; Prosser, 2012; DeLong, 2014). Our results follow this line of thinking, and show that broader taxonomic units, which group closely related taxa as responding unions may enhance our ability to identify environment-driven community dynamics. Conversely, breaking down broader taxa into finer units may result in more noise than information, as the distributions and abundances of those ecologically equivalent units exhibit a high degree of randomness caused by neutral-stochastic processes instead of niche-deterministic processes (Lozupone et al., 2012; Prosser, 2012; DeLong, 2014). Actually, previous studies have indicated that despite the considerable variability in species-level composition, prokaryotic communities in a given habitat usually have stable phylum-level composition as well as similar functional attributes (Fierer et al., 2007; Lauber et al., 2009; Burke et al., 2011; Fan et al., 2012; Lozupone et al., 2012); these findings further support the view that there is ecological coherence in deep prokaryotic branches (Philippot et al., 2009, 2010) and potentially many fine-level taxa are functionally redundant and fulfill the same ecological role (Lozupone et al., 2012; Prosser, 2012; DeLong, 2014).
Implications for further studies regarding taxonomic resolution
It should be recognized that no particular taxonomic level is well suited for all cases. Our findings for these prokaryotic case studies emphasize the profound value of broad-level taxonomic data when evaluating community–environment relationships, but do not imply that fine-level taxonomic data only capture or represent additional minor details. Rather, we highlight two considerations regarding community analyses at distinct taxonomic resolutions. First, as discussed by other researchers (Warwick, 1993; Bailey et al., 2001), the choice of taxonomic resolution for understanding variation in ecological systems should depend on the questions being asked. For example, in studies of benthic invertebrates, broad-level taxonomic data have been demonstrated to be reliable when the objective is to evaluate the impact of pollution events on biological assemblages (Warwick, 1993; Bailey et al., 2001). In contrast, fine-level taxonomic data are needed if we want to detect specific toxicological responses. Second, and perhaps more importantly, as certain processes are only apparent and drive ecosystem variation at a particular spatiotemporal scale (Levin, 1992), their effects may only be evident by examining communities at a particular taxonomic resolution. Following previous notions of scale dependency in plant communities (Levin, 1992; Cavender-Bares et al., 2006, 2009; Swenson et al., 2006, 2007) and the idea of beta- versus alpha-niche (Silvertown et al., 2006a, 2006b), we might postulate that large-scale processes such as climatic tolerances (beta-niche) may be better evidenced by broadly resolved community profiles (Figures 1a and b); whereas, small-scale processes such as biotic interactions and microhabitat differentiation (alpha-niche) may only be evident with fine-level classification data (Figure 1c). With respect to prokaryotes with small cell size and short generation time (as considered in this study), a typical sampling setup probably emphasizes the effect of β-niche (evidenced by broadly resolved taxonomic data), whereas the effect of α-niche may be underestimated because of measurement limitations for biotic properties and microhabitat conditions (Ranjard and Richaume, 2001; Armitage et al., 2012).
With respect to the scale-dependency issue (Levin, 1992; Cavender-Bares et al., 2006, 2009; Swenson et al., 2006, 2007), we acknowledge that the detected patterns can vary according to phylogenetic combinations or target groups of communities. For example, the best taxonomic resolution and key environmental factor might differ when considering whole prokaryotic communities versus focusing on a particular taxonomic group (Johnson et al., 2006; Martiny et al., 2009). The main objective of this study is to demonstrate the potential of our approach, which assesses community–environment relationships while taking into consideration hierarchical taxonomic information and the concept of phylogenetic niche conservatism; rather than stressing specific conclusions associated with any particular data set. Future studies incorporating both multi-level taxonomic data and multi-scale sampling setups with suitable environmental measurements should allow for: (1) a better resolution of the potential mechanisms involved in prokaryotic community assembly; and (2) an understanding of how these mechanisms shift with spatiotemporal scale and variable target clades.
Conclusion
A comprehensive understanding of biodiversity requires investigation at multiple scales, including scales of space, time and organization (Levin, 1992; Swenson et al., 2006; Cavender-Bares et al., 2009). Community studies have devoted considerable attention to issues of spatial and temporal scales, whereas similar examination and discussion of the organizational scale is limited. Here we used eight prokaryotic data sets to show that systematically investigating community structure at fine to broad taxonomic resolutions is practical and meaningful. Specifically, our detected patterns (Figure 3) reinforce the current notion of ecological coherence in deep prokaryotic branches (Philippot et al., 2009, 2010), which suggest that broad-level taxonomic classification might be useful for making generalizations about the biogeographic distribution of prokaryotic taxa, at least in the context of some environmental drivers. Through these cases, we demonstrate that tracking the strength of community–environment relationships along taxonomic ranks might serve as a means to uncover macro-evolutionary patterns, adding a new and important perspective to the evidence regarding niche conservatism or niche divergence in community assembly (Figure 1). We believe this approach could be applied to any community–environment data, including micro- and macro-organisms, to provide evolutionary insights into observed community structure.
Acknowledgments
We acknowledge helpful comments from Jay T Lennon, David W Armitage, Hon-Tsen Yu, and one anonymous reviewer. We thank Yu-Da Lyu for assistance with data processing. The pyrosequencing service for samples in case #8 was provided by the Genome Research Center, National Yang-Ming University, Taiwan. This work was supported by the National Center for Theoretical Sciences, Foundation for the Advancement of Outstanding Scholarship, and the Ministry of Science and Technology, Taiwan.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. (2004). Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol 186: 2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AF, Riemann L, Bertilsson S. (2010). Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISMEJ 4: 171–181. [DOI] [PubMed] [Google Scholar]
- Angly FE, Dennis PG, Skarshewski A, Vanwonterghem I, Hugenholtz P, Tyson GW. (2014). CopyRighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage DW, Gallagher KL, Youngblut ND, Buckley DH, Zinder SH. (2012). Milimeter-scale patterns of phylogenetic and trait diversity in a salt marsh microbial mat. Front Microbiol 3: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey RC, Norris RH, Reynoldson TB. (2001). Taxonomic resolution of benthic macroinvertebrate communities in bioassessments. J N Am Benthol Soc 20: 280–286. [Google Scholar]
- Bennett JR, Sisson DR, Smol JP, Cumming BF, Possingham HP, Buckley YM. (2014). Optimizing taxonomic resolution and sampling effort to design cost-effective ecological models for environmental assessment. J Appl Ecol 51: 1722–1732. [Google Scholar]
- Bevilacqua S, Claudet J, Terlizzi A. (2013). Best practicable aggregation of species: a step forward for species surrogacy in environmental assessment and monitoring. Ecol Evol 3: 3780–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhusal DR, Kallimanis AS, Tsiafouli MA, Sgardelis SP. (2014). Higher taxa vs. functional guilds vs. trophic groups as indicators of soil nematode diversity and community structure. Ecol Indic 41: 25–29. [Google Scholar]
- Bray JR, Curtis JT. (1957). An ordination of upland forest communities of southern Wisconsin. Ecol Monogr 27: 326–349. [Google Scholar]
- Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. (2011). Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA 108: 14288–14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavender-Bares J, Ackerly DD, Baum DA, Bazzaz FA. (2004). Phylogenetic overdispersion in Floridian oak communities. Am Nat 163: 823–843. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Keen A, Miles B. (2006). Phylogenetic structure of Floridian plant communities depends on taxonomic and spatial scale. Ecology 87: S109–S122. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Kozak KH, Fine PV, Kembel SW. (2009). The merging of community ecology and phylogenetic biology. Ecol Lett 12: 693–715. [DOI] [PubMed] [Google Scholar]
- Chu HY, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P. (2010). Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol 12: 2998–3006. [DOI] [PubMed] [Google Scholar]
- Debussche M, Thompson JD. (2003). Habitat differentiation between two closely related Mediterranean plant species, the endemic Cyclamen balearicum and the widespread C-repandum. Acta Oecol 24: 35–45. [Google Scholar]
- DeLong EF. (2014). Alien invasions and gut ‘island biogeography'. Cell 159: 233–235. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006. a). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM et al. (2006. b). NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34: W394–W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS et al. (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc Natl Acad Sci USA 109: E1878–E1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. (2007). Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Losos JB. (2009). Adaptive radiation: contrasting theory with data. Science 323: 732–737. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T et al. (2009). The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11: 3132–3139. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrueck L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AJ, Neilson RR, Dale VH, Flather CH, Iverson LR, Currie DJ et al. (2001). Global change in forests: responses of species, communities, and biomes. Bioscience 51: 765–779. [Google Scholar]
- Hector A, Bagchi R. (2007). Biodiversity and ecosystem multifunctionality. Nature 448: 188–190. [DOI] [PubMed] [Google Scholar]
- Heino J, Soininen J. (2007). Are higher taxa adequate surrogates for species-level assemblage patterns and species richness in stream organisms? Biol Conserv 137: 78–89. [Google Scholar]
- Heino J. (2008). Influence of taxonomic resolution and data transformation on biotic matrix concordance and assemblage-environment relationships in stream macroinvertebrates. Boreal Environ Res 13: 359–369. [Google Scholar]
- Heino J. (2014). Taxonomic surrogacy, numerical resolution and responses of stream macroinvertebrate communities to ecological gradients: are the inferences transferable among regions? Ecol Indic 36: 186–194. [Google Scholar]
- Ives AR, Carpenter SR. (2007). Stability and diversity of ecosystems. Science 317: 58–62. [DOI] [PubMed] [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311: 1737–1740. [DOI] [PubMed] [Google Scholar]
- Jost L. (2007). Partitioning diversity into independent alpha and beta components. Ecology 88: 2427–2439. [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noe L, Touzet H. (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28: 3211–3217. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Anderson MJ. (1999). Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol Monogr 69: 1–24. [Google Scholar]
- Legendre P, Legendre L. (2012) Numerical Ecology. Elsevier: Amsterdam, The Netherlands. [Google Scholar]
- Leibold MA, McPeek MA. (2006). Coexistence of the niche and neutral perspectives in community ecology. Ecology 87: 1399–1410. [DOI] [PubMed] [Google Scholar]
- Lennon JT, Aanderud ZT, Lehmkuhl BK, Schoolmaster DR Jr. (2012). Mapping the niche space of soil microorganisms using taxonomy and traits. Ecology 93: 1867–1879. [DOI] [PubMed] [Google Scholar]
- Levin SA. (1992). The problem of pattern and scale in ecology. Ecology 73: 1943–1967. [Google Scholar]
- Lozupone C, Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MDJ, Neufeld JD. (2015). Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13: 217–229. [DOI] [PubMed] [Google Scholar]
- Magurran AE. (2004) Measuring Biological Diversity. Blackwell Science: Oxford. [Google Scholar]
- Margules CR, Pressey RL. (2000). Systematic conservation planning. Nature 405: 243–253. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Tai APK, Veneziano D, Primeau F, Chisholm SW. (2009). Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol 11: 823–832. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Treseder K, Pusch G. (2013). Phylogenetic conservatism of functional traits in microorganisms. ISME J 7: 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JB, Jones SE, Lennon JT, Martiny AC. (2015). Microbiomes in light of traits: a phylogenetic perspective. Science 350: aac9323. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2015.) vegan: Community Ecology Package. R package version 2.2-1. http://CRAN.R-project.org/package=vegan.
- Olsgard F, Somerfield PJ, Carr MR. (1998). Relationships between taxonomic resolution, macrobenthic community patterns and disturbance. Mar Ecol Prog Ser 172: 25–36. [Google Scholar]
- Peres-Neto PR, Jackson DA. (2001). How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 129: 169–178. [DOI] [PubMed] [Google Scholar]
- Peres-Neto PR, Legendre P, Dray S, Borcard D. (2006). Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87: 2614–2625. [DOI] [PubMed] [Google Scholar]
- Pfennig KS, Pfennig DW. (2009). Character displacement: ecological and reproductive responses to a common evolutionary problem. Q Rev Biol 84: 253–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Bru D, Saby NPA, Cuhel J, Arrouays D, Simek M et al. (2009). Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ Microbiol 11: 3096–3104. [DOI] [PubMed] [Google Scholar]
- Philippot L, Andersson SGE, Battin TJ, Prosser JI, Schimel JP, Whitman WB et al. (2010). The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol 8: 523–529. [DOI] [PubMed] [Google Scholar]
- Prosser JI. (2012). Ecosystem processes and interactions in a morass of diversity. Fems Microbiol Ecol 81: 507–519. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2015) R: A Language and Environment for Statistical Computing. The R Foundation for Statistical Computing Platform: Vienna, Austria. [Google Scholar]
- Ramirez KS, Leff JW, Barberan A, Bates ST, Betley J, Crowther TW et al. (2014). Biogeographic patterns in below-ground diversity in New York City's Central Park are similar to those observed globally. Proc R Soc B-Biol Sci 281: rspb20141988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjard L, Richaume AS. (2001). Quantitative and qualitative microscale distribution of bacteria in soil. Res Microbiol 152: 707–716. [DOI] [PubMed] [Google Scholar]
- Schluter D. (2000). Ecological character displacement in adaptive radiation. Am Nat 156: S4–S16. [Google Scholar]
- Silvertown J, Dodd M, Gowing D, Lawson C, McConway K. (2006. a). Phylogeny and the hierarchical organization of plant diversity. Ecology 87: S39–S49. [DOI] [PubMed] [Google Scholar]
- Silvertown J, McConway K, Gowing D, Dodd M, Fay MF, Joseph JA et al. (2006. b). Absence of phylogenetic signal in the niche structure of meadow plant communities. Proc R Soc B-Biol Sci 273: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G et al. (2015). Structure and function of the global ocean microbiome. Science 348: 1261359. [DOI] [PubMed] [Google Scholar]
- Swenson NG, Enquist BJ, Pither J, Thompson J, Zimmerman JK. (2006). The problem and promise of scale dependency in community phylogenetics. Ecology 87: 2418–2424. [DOI] [PubMed] [Google Scholar]
- Swenson NG, Enquist BJ, Thompson J, Zimmerman JK. (2007). The influence of spatial and size scale on phylogenetic relatedness in tropical forest communities. Ecology 88: 1770–1780. [DOI] [PubMed] [Google Scholar]
- Swenson NG. (2014) Functional and Phylogenetic Ecology in R. Springer Science+Business Media: New York, NY, USA. [Google Scholar]
- Terlizzi A, Bevilacqua S, Fraschetti S, Boero F. (2003). Taxonomic sufficiency and the increasing insufficiency of taxonomic expertise. Mar Pollut Bull 46: 556–561. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Ovreas L, Thingstad TF. (2002). Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296: 1064–1066. [DOI] [PubMed] [Google Scholar]
- Tresedera KK, Lennon JT. (2015). Fungal traits that drive ecosystem dynamics. Microbiol Mol Biol Rev 79: 243–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetrovsky T, Baldrian P. (2013). The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. Plos One 8: e57923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick RM. (1993). Environmental impact studies on marine communities—pragmatical considerations. Aust J Ecol 18: 63–80. [Google Scholar]
- Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. (2002). Phylogenies and community ecology. Annu Rev Ecol Systemat 33: 475–505. [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. (1998). Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens JJ, Graham CH. (2005). Niche conservatism: integrating evolution, ecology, and conservation biology. Annu Rev Ecol Evol S 36: 519–539. [Google Scholar]
- Wiens JJ, Ackerly DD, Allen AP, Anacker BL, Buckley LB, Cornell HV et al. (2010). Niche conservatism as an emerging principle in ecology and conservation biology. Ecol Lett 13: 1310–1324. [DOI] [PubMed] [Google Scholar]
- Woese CR. (1987). Bacterial evolution. Microbiol Rev 51: 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P, Yilmaz P, Pruesse E, Glockner FO, Ludwig W, Schleifer KH et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12: 635–645. [DOI] [PubMed] [Google Scholar]
- Yeh YC, Peres-Neto PR, Huang SW, Lai YC, Tu CY, Shiah FK et al. (2015). Determinism of bacterial metacommunity dynamics in the southern East China Sea varies depending on hydrography. Ecography 38: 198–212. [Google Scholar]
- Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S et al. (2015). The soil microbiome influences grapevine-associated microbiota. Mbio 6: e02527–02514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Pei N, Mi X. (2012.) phylotools: Phylogenetic tools for Eco-phylogenetics. R package version 0.1.2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.