Abstract
Eukaryotic cells express a family of eukaryotic translation initiation factor 2 alpha (eIF2α) kinases (eg, PKR, PERK-PEK, GCN2, HRI) that are individually activated in response to distinct types of environmental stress. Phosphorylation of eIF2α by one or more of these kinases reduces the concentration of eIF2–guanosine triphosphate (GTP)–transfer ribonucleic acid for methionine (tRNAMet), the ternary complex that loads tRNAMet onto the small ribosomal subunit to initiate protein translation. When ternary complex levels are reduced, the related RNA-binding proteins TIA-1 and TIAR promote the assembly of a noncanonical preinitiation complex that lacks eIF2-GTP-tRNAMet. The TIA proteins dynamically sort these translationally incompetent preinitiation complexes into discrete cytoplasmic domains known as stress granules (SGs). RNA-binding proteins that stabilize or destabilize messenger RNA (mRNA) are also recruited to SGs during stress. Thus, TIA-1 and TIAR act downstream of eIF2α phosphorylation to promote SG assembly and facilitate mRNA triage during stress. The role of the SG in the integration of translational efficiency, mRNA stability, and the stress response is discussed.
In living cells, key components of the translational apparatus (eg, messenger ribonucleic acid [mRNA] and its associated proteins, ribosomal subunits, translation initiation factors) move between the nucleus and the cytoplasm in a regulated manner. These components differentially associate with cellular structures (eg, cytoskeletal elements) and organelles (eg, endoplasmic reticulum) in ways that regulate their availability and, thus, their activity. Because of this, biochemical studies of protein translation performed using cell lysates cannot provide a complete description of the translational regulatory pathways that operate in live cells. A striking example of this tenet is the assembly and disassembly of mammalian stress granules (SGs). These cytoplasmic microdomains are sites at which untranslated mRNAs that accumulate in stressed cells are transiently routed. SGs form when translation is initiated under conditions in which the concentration of the active eukaryotic translation initiation factor 2 (eIF2)–guanosine triphosphate (GTP)–transfer RNA for methionine (tRNAMet) ternary complex is reduced. The assembly of translationally inactive initiation complexes lacking eIF2 allows the RNA-binding proteins TIA-1 or TIAR (or both) to redirect untranslated mRNAs from polyribosomes to SGs. By regulating the equilibrium between polysomes and SGs, TIA-1 and TIAR may influence the frequency with which individual transcripts are sorted for translation or triage in both stressed and unstressed cells.
TRANSLATIONAL INITIATION
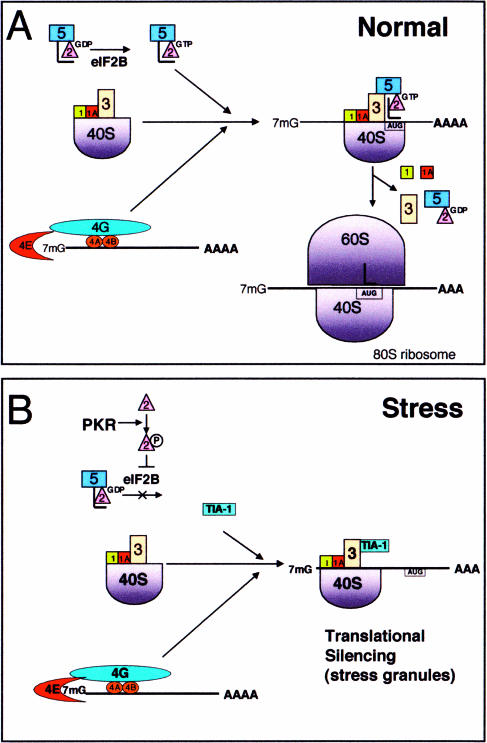
Eukaryotic translational initiation in vitro begins with the assembly of a 43S preinitiation complex that is formed when eIF1, eIF2, eIF3, and eIF5 combine with the 40S ribosomal subunit (Fig 1A) (Dever 1999; Pestova and Hellen 1999; Asano et al 2000; Hershey and Merrick 2000; Phan et al 2001). This 43S complex recruits a 7-methyl guanosine-capped mRNA and its associated initiation factors (eg, eIF4E, eIF4G, and poly(A)-binding protein) to produce the canonical 48S preinitiation complex. The 48S complex scans the 5′ untranslated region (UTR) of the mRNA transcript, coming to rest at an initiation codon (typically AUG) that is recognized by the anticodon of tRNAMet. Recognition of the initiation codon triggers hydrolysis of eIF2-associated GTP, a reaction catalyzed by eIF5. The early initiation factors (eg, eIF2–guanosine diphosphate [GDP], eIF3, eIF5, eIF1A) then dissociate from the 40S subunit, and the 60S subunit is recruited to form a functional 80S ribosome. As additional ribosomes assemble at the 5′ cap, the mRNA is converted into a polyribosome.
Fig. 1.
Translational initiation in the absence or presence of stress. (A) Normal: when the eukaryotic translation initiation factor 2 (eIF2)–guanosine triphosphate (GTP)–transfer ribonucleic acid for methionine (tRNAMet) ternary complex is available, a canonical 48S preinitiation complex is assembled at the 5′ end of capped transcripts and scanning begins. Upon recognition of the initiation codon by the anticodon of tRNAMet, eIF5 promotes GTP hydrolysis, and early initiation factors are displaced by the 60S ribosomal subunit. (B) In stressed cells the phosphorylation of eIF2α prevents GDP-GTP exchange by eIF2B, which lowers the effective concentration of eIF2-GTP-tRNAMet. Under these conditions, TIA-1 is included in a noncanonical preinitiation complex that is translationally silent. TIA-1 self-aggregation then promotes the accumulation of these complexes at discrete cytoplasmic foci known as stress granules
The process of translational initiation is under tight regulatory control. Excessive translational initiation can lead to cellular transformation, whereas inadequate translational initiation leads to cell death. This close association between translational initiation and cell survival is dramatically revealed by the functional effects of eIF2-GTP-tRNAMet, the ternary complex that loads the initiator tRNAMet onto the small ribosomal subunit during the assembly of the 43S preinitiation compex. eIF2 is composed of 3 subunits: α, β, and γ (Kimball 1999). The α subunit is the target of a family of serine or threonine kinases (ie, PKR, PERK-PEK, GCN2, HRI) that are activated by different forms of environmental stress. For example, PKR senses heat, ultraviolet (UV) irradiation, viral infection, and oxidative stress (Williams 2001), whereas PERK-PEK detects endoplasmic reticulum stress (eg, secretory pathway constipation caused by inhibitors of N-lined glycosylation such as tunicamycin) (Harding et al 2000). GCN2 senses amino acid starvation (Kimball 2001), and HRI monitors changes in the availability of heme during erythrocyte differentiation (Han et al 2001; Lu et al 2001). Each of these stress-activated kinases phosphorylates eIF2α on serine 51, a modification that increases the affinity of eIF2 for eIF2B, a GDP-GTP exchange factor that charges the eIF2-GTP-tRNAMet ternary complex (Kimball 2001). By functioning as a competitive inhibitor of eIF2B, phospho-eIF2α reduces the concentration of the active ternary complex, prevents the assembly of the 43S preinitiation complex, and halts protein translation. Consequently, overexpression of PKR results in phosphorylation of eIF2α and translational arrest. Overexpressed PKR is also a potent inducer of apoptotic cell death (Gil and Esteban 2000). Although the molecular mechanism whereby PKR induces cell death is not known, the translational arrest of constitutively synthesized survival factors may contribute to this process. Remarkably, a phosphomimetic mutant of eIF2α (S51D) also arrests protein translation and induces apoptotic cell death (Srivastava et al 1998). Conversely, kinase-dead PKR mutants and nonphosphorylatable eIF2α mutants (S51A) inhibit apoptosis and induce cellular transformation (Barber 2001). It follows that the PKR/PERK/GNC2/HRI-eIF2 pathway plays a critical dual role in regulating both translational initiation and cell survival (Barber 2001).
A remarkable feature of stress-induced translational arrest is its specificity. Whereas translation of constitutive transcripts encoding “housekeeping” proteins is inhibited, translation of stress-induced transcripts encoding heat shock proteins and selected transcription factors (eg, GCN4 and ATF4) is preserved and actually increased in stressed cells (Hinnebusch 1996, 1997; Harding et al 2000). Although the molecular features that distinguish between constitutive and stress-induced transcripts are not well understood, these 2 classes of transcripts are physically separated in the cytoplasm of heat-stressed tomato cells. Whereas mRNAs encoding housekeeping proteins are sequestered in phase-dense cytoplasmic particles known as heat shock granules (Nover et al 1983), mRNAs encoding inducible heat shock proteins (Hsps) are excluded from these granules (Nover et al 1989; our unpublished observations). By sequestering most of the cytoplasmic mRNAs from the translational machinery, the SG may promote the selective translation of stress-induced mRNA transcripts.
TIA-1 AND TIAR
Similar phase-dense heat shock granules are found in the cytoplasm of heat shocked mammalian cells (Collier and Schlesinger 1986; Arrigo et al 1988; Collier et al 1988). TIA-1 and TIAR, related RNA-binding proteins, were discovered to be robust markers of these cytoplasmic foci (Kedersha et al 1999). Although TIA-1 and TIAR are concentrated in the nuclei of cycling cells, heterokaryon analysis reveals that both proteins shuttle continuously between the nucleus and the cytoplasm (unpublished observations). In response to environmental stress, TIA-1 and TIAR accumulate in the cytoplasm, where they aggregate at the SGs. In situ hybridization using oligo-dT probes has shown that poly(A)+ RNA also accumulates at TIA-1/TIAR+ SGs (Kedersha et al 1999). Thus, mammalian SGs, like their plant counterparts, are discrete cytoplasmic foci at which mRNA accumulates in the stressed cells. Using TIA-1, TIAR, and poly(A)+ RNA as markers, we determined that a wide variety of stresses (eg, heat, UV irradiation, oxidative conditions, hyperosmolarity) induce the assembly of SGs. Hsps are components of SGs induced by heat but are not present in SGs induced by the other stress stimuli. Thus, the cytoplasmic heat shock granules first described in mammalian systems (Collier and Schlesinger 1986; Arrigo et al 1988; Collier et al 1988) constitute a specialized subset of mammalian SGs, all of which contain the TIA proteins and poly(A)+ mRNA but only some of which contain Hsps (Kedersha et al 1999).
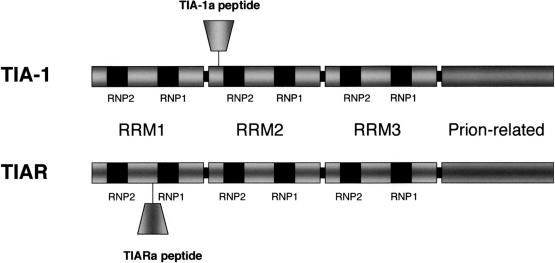
TIA-1 and TIAR are members of the RNA-recognition motif (RRM) family of RNA-binding proteins (Tian et al 1991; Kawakami et al 1992). Both proteins possess 3 RRMs at their amino termini (RRM1, 2, and 3) and a glutamine-rich domain at their carboxyl termini (Fig 2). Two major isoforms of both proteins are the products of alternative mRNA splicing (Kawakami et al 1994; Beck et al 1996). TIA-1a includes an 11–amino acid mini exon within RRM2, whereas TIA-1b lacks this exon. TIARa includes a 17–amino acid mini exon within RRM1, which is missing from TIARb. The functional consequences of the inclusion or exclusion of these TIA-1 or TIAR exons are not known; however, a similar situation exists in hnRNPD-AUF1, wherein the inclusion of a mini exon within the RRM decreases the affinity with which the protein binds to RNA (Wagner et al 1998). In vitro selection analysis has shown that the RNA-binding domains of TIA-1 and TIAR preferentially recognize uridine-rich RNAs (Dember et al 1996). The second RRM is sufficient to confer binding to mRNAs encoding uridine-rich elements (Dember et al 1996). Surprisingly, the first RRM does not bind to mRNA in vitro, whereas the third RRM binds to mRNA without apparent sequence specificity (Dember et al 1996). Thus, TIA-1 and TIAR are capable of both general and sequence-specific RNA binding.
Fig. 2.
TIA-1 and TIAR are related ribonucleic acid (RNA)–binding proteins that possess 3 RNA-recognition motifs and a prion-related domain. Alternative splicing creates 2 isofoms of both proteins
The carboxyl termini of TIA-1 and TIAR are structurally related to prion protein (Tian et al 1991). When a recombinant protein encoding the isolated prion-related domain of TIA-1 is overexpressed in COS cells, the truncated protein forms spontaneous cytoplasmic microaggregates that coaggregate and sequester endogenous TIA-1 and TIAR (Kedersha et al 1999). This demonstrates that the prion-related domain of TIA-1, like the native prion protein, appears to be capable of self-oligomerization in vivo. The TIA prion-related domain is required for the formation of SGs (see later). Thus, the basic structure of the TIA proteins provides the requisite features needed to assemble SGs, eg, the ability to bind RNA and the ability to self-assemble within the cytoplasm.
Targeted disruption of either TIA-1 or TIAR results in partial embryonic lethality, indicating that these related proteins play an important role in vertebrate development (Beck et al 1998; Piecyk et al 2000). The penetrance of embryonic lethality is strain dependent, suggesting that epistatic influences can modify the functional effects of these proteins. In mice lacking TIAR the rate of embryonic lethality is 100% in the BALB/c strain and 90% in the C57BL/6 strain. In mice lacking TIA-1 the rate of embryonic lethality is approximately 50% in both BALB/c and C57BL/6 strains. TIAR nullizygotes that survive to birth are sterile because of defective germ cell maturation (Beck et al 1998), whereas TIA-1 nullizygotes are fully fertile (Piecyk et al 2000). These different phenotypes indicate that TIA-1 and TIAR have distinct functions during embryogenesis. Breeding experiments designed to obtain double nullizygotes have been unsuccessful: embryos lacking both TIAR and TIA-1 are not produced, suggesting that one or the other of these proteins is essential for some aspect of early embryonic development.
MAMMALIAN SGs: COMPOSITION AND FUNCTION
Immunofluorescent microscopy using subunit-specific antiribosomal antibodies revealed that small, but not large, ribosomal subunits are components of mammalian SGs (Kedersha et al 2002). The absence of large ribosomal subunits eliminates the possibility that SGs are sites at which selected mRNAs are translated in stressed cells. At the same time, the presence of small ribosomal subunits suggests that the mRNPs comprising SGs might be structurally related to polysomes. Indeed, the 48S preinitiation complex that assembles on capped mRNAs includes the small, but not the large, ribosomal subunit (Fig 1A), suggesting that 48S preinitiation complexes might be the core constituents of SGs. Immunofluorescent microscopy using antibodies against individual components of the 48S preinitiation complex showed that this hypothesis is partially correct. Most components of the 48S preinitiation complex (eg, eIF3, eIF4E, eIF4G, and PABP) do colocalize with TIA-1 and poly(A)+ RNA at SGs. Translational regulatory factors that are not components of the 48S preinitiation complex (eg, PHAS-I, eIF2Bɛ) are not found at SGs. However, neither eIF2 nor eIF5, bona fide components of the 48S preinitiation complex, are components of SGs, suggesting that SGs are composed of eIF2-eIF5–deficient, noncanonical preinitiation complexes that are assembled during stress (Fig 1B).
One such model in which an eIF2-deficient 48S preinitiation complex is formed during stress has already been proposed in order to account for the preferential translation of stress-induced transcription factors, such as ATF4 in mammals (Harding et al 2000) and GCN4 in yeast (Hinnebusch 1996, 1997), which occurs in response to eIF2α phosphorylation. The transcripts encoding these proteins possess long 5′ UTRs that contain multiple small upstream open-reading frames (uORFs). These appear to prevent productive translation under normal conditions by causing scanning ribosomes to terminate before they reach the initiator codon of the stress-induced transcription factor. During stress conditions, the reduced availability of eIF2-GTP-tRNAMet may allow the assembly of eIF2-deficient 48S preinitiation complexes that scan past the uORFs without initiating translation. Delayed acquisition of eIF2-GTP-tRNAMet (ie, after the eIF2-deficient 48S complexes have scanned past these regulatory “decoy” uORFs) has been proposed to explain the preferred initiation at the major ORF. Both the induction of SG assembly by phospho-eIF2α and the composition of SGs suggest that they comprise eIF2-deficient 48S complexes; whether these are identical to those proposed to regulate the translation of ATF4 and GCN4 remains to be determined.
The absence of eIF5 from SGs may be explained by its specific interactions with eIF2. eIF5 links eIF2 to eIF3 during the assembly of the preinitiation complex (Asano et al 2000; Phan et al 2001). Although eIF2 and eIF3 do not interact directly with one another, eIF5 binds to both eIF2β and eIF3i, allowing the assembly of a multifactor initiation complex. Because eIF3 interacts independently with the 40S ribosomal subunit, it can join the preinitiation complex in the absence of other factors. If eIF2 were to provide the sole tether for eIF5, it would be absent from the eIF2-deficient preinitiation complexes. The assembly of a preinitiation complex lacking eIF2 and eIF5 would be expected to be translationally incompetent. Because eIF2-GTP-tRNAMet is required for the recognition of initiation codons, eIF2-eIF5–deficient complexes are unlikely to reside at the initiation codon. It remains to be determined whether these complexes scan past the 5′ cap.
SGs AND eIF2α
Phosphorylation of eIF2α is sufficient to induce translational arrest and SG assembly: the expression of a phosphomimetic mutant of eIF2α (S51D) inhibits translation and concurrently induces the assembly of SGs (Kedersha et al 1999). Phosphorylation of eIF2α is also necessary for the induction of translational arrest and SG assembly in cells subjected to selected environmental stresses (eg, arsenite) because the expression of a nonphosphorylatable eIF2α mutant (S51A) inhibits both arsenite-induced translational arrest and SG assembly (Kedersha et al 1999). However, metabolic inhibitors that deplete cellular adenosine triphosphate (eg, oligomycin, FCCP, 2-deoxyglucose) can induce the assembly of SGs without increasing the basal levels of phospho-eIF2α (Kedersha et al 2002). These treatments all induce energy starvation in the cell, apparently reducing the GTP levels necessary to generate eIF2-GTP-tRNAMet (Kedersha et al 2002). This suggests that the requisite signal for the assembly of SGs is a reduced concentration of eIF2-GTP-tRNAMet rather than eIF2α phosphorylation per se.
The effects of TIA-1 (and SGs) on protein expression were examined in vivo using a COS7 transfection system. Whereas overexpression of wild-type TIA-1 represses the expression of cotransfected reporter proteins (eg, luciferase, β-galactosidase) (Kedersha et al 2000), the overexpression of a dominant-negative truncation mutant (TIA-1ΔRRM) that prevents SG assembly promotes the expression of these same reporter proteins (Kedersha et al 2000). These effects on reporter gene expression are independent of the AU-rich TIA target sequences in the 3′ UTR of these genes (see later), suggesting that the sequence nonspecific RNA-binding properties of TIA are important in this context. Moreover, these effects were observed in the absence of stress, suggesting that the functional effects of TIA-1 are not restricted to stressed cells. Indeed, the appearance of microscopically visible SGs may be an exaggerated example of a normal translational control process.
SGs AND POLYSOMES: A DYNAMIC EQUILIBRIUM
The paradoxically antagonistic effects of different pharmacological inhibitors of protein translation on SG assembly reveal the dynamic equilibrium between polysomes and SGs. Drugs that stabilize polysomes by freezing ribosomes on translating mRNAs (eg, cycloheximide and emetine) inhibit the assembly of SGs and actively dissolve them in the continued presence of both stress and eIF2α phosphorylation (Kedersha et al 2000). In contrast, drugs that destablize polysomes by releasing ribosomes from mRNA transcripts (eg, puromycin) promote the assembly of SGs (Kedersha et al 2000). The dynamic nature of the equilibrium between polysomes and SGs led us to propose that SGs may be the sites of mRNA triage at which untranslated mRNAs accumulate during stress prior to degradation, reinitiation, or repackaging as mRNPs (Kedersha et al 2000). When the accumulation of nonpolysomal mRNA exceeds the capacity for triage, untranslated mRNA transiently accumulates at the SG.
To examine this dynamic behavior at the molecular level, GFP–TIA-1 and GFP-PABP constructs were made. GFP–TIA-1 behaves like endogenous TIA-1 (ie, moves to SGs in response to various forms of stress), allowing us to monitor the assembly and disassembly of SGs in living cells (Kedersha et al 2000) (http://www.jcb.org/cgi/content/full/151/6/1257/DC1). In the absence of stress, GFP–TIA-1 is concentrated in the nucleus as expected. In response to arsenite-induced stress, GFP–TIA-1 (within 3–6 minutes) accumulates rapidly in the cytoplasm, where it is evenly and diffusely distributed. After approximately 10 minutes, the cytoplasmic GFP–TIA-1 aggregates into discrete foci that coalesce and slowly enlarge over the next 20 minutes. When arsenite is washed out of the cells, the SGs slowly disassemble with similar kinetics. However, this slow and steady accumulation of GFP–TIA-1 at SGs is misleading. Fluorescent recovery after photobleaching analysis reveals that GFP–TIA-1 shuttles in and out of SGs very rapidly, so that 50% of SG-associated GFP–TIA-1 is replaced every 2 seconds. Although the rate at which mRNA shuttles in and out of SGs has not yet been directly determined, GFP-poly(A)–binding protein was used as a surrogate marker for its associated mRNA. Interestingly, GFP-PABP shuttles in and out of SGs at a rate that is 10 times slower than that of GFP–TIA-1 (ie, 50% of SG-associated GFP-PABP is replaced every 20 seconds). Given these kinetics, and considering that the dominant negative mutant of TIA-1 (eg, TIA-1ΔRRM) prevents SG assembly altogether, it appears that TIA-1 actively escorts untranslated mRNA to SGs. These data reveal that SGs are highly dynamic structures despite their apparent stability in real-time microscopy. In this respect, SGs resemble several nuclear RNA-containing structures (eg, speckles, coiled bodies, Gemini of coiled bodies, and nucleoli) that are sites of active RNA metabolism. At the microscopic level, these metabolic domains appear to be stable structures that maintain a distinct morphology. Despite their stable appearance, many of the molecular components of these nuclear substructures are in constant flux. Metabolic disruption of the flux alters the morphology of the structure, just as a river comprised of water will vary during drought and flood. SGs appear to be analogous cytoplasmic “structural” domains, whose existence is the result of a sudden stress-induced flood of abortive initiation complexes that result from polysomes disassembled during stress.
GENERAL VS SPECIFIC TRANSLATIONAL SILENCING: TUMOR NECROSIS FACTOR α
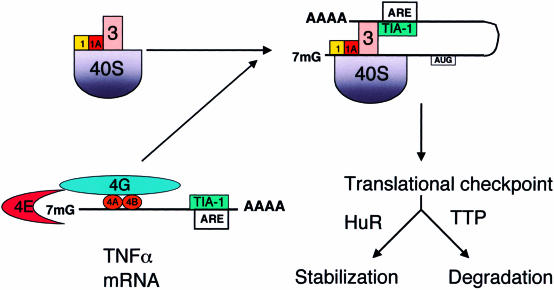
TIA-1 and TIAR were identified as components of a regulatory complex that targets an adenine-uridine–rich element (ARE) found in the 3′ UTR of tumor necrosis factor α (TNFα) transcripts, suggesting that the regulatory effects of these proteins are selective for specific transcripts (Gueydan et al 1999; Piecyk et al 2000). In fact, lipopolysaccharide (LPS)–activated macrophages derived from mice lacking either TIA-1 or TIAR were found to overexpress TNFα compared with the wild-type controls. In macrophages lacking TIA-1, the polysome profile of TNFα transcripts is shifted such that the percentage of TNFα transcripts associated with polysomes is increased compared with that of wild-type macrophages (Piecyk et al 2000). This suggests that TIA-1 represses the translation of TNFα by promoting the assembly of a nonpolysomal mRNP complex. We propose that this complex is structurally equivalent to the eIF2-eIF5–deficient preinitiation complex that is assembled during stress (Fig 1B). Because RRM2 of TIA-1 binds with high affinity to an AU-rich element in the 3′ UTR of TNFα transcripts, the probability that TIA-1 will also interact with an assembling preinitiation complex (possibly via the nonspecific binding ability of RRM3) will be markedly greater than with other transcripts (Fig 3). The tethering of TIA-1 to TNFα transcripts would increase the percentage of these transcripts found in nonpolysomal mRNP complexes, dampen the expression of TNFα, and increase the sensitivity of these transcripts to regulatory control. A similar mechanism is employed by the hnRNP proteins K and E1, proteins that are tethered to the 3′ UTR of 15-lipoxygenase transcripts, which induce the assembly of a translationally incompetent 48S preinitiation complex (Ostareck et al 2001). The hnRNP K-E1 proteins act by preventing the joining of the 60S ribosomal subunit (Ostareck et al 2001) while allowing the small ribosomal subunit to scan to the initiation codon, at which point the scanning is halted, and the transcript is silenced. As eIF2-GTP-tRNAiMet is required for the initiator codon recognition, it is likely that TIA-1–induced eIF2-eIF5–deficient complexes promote translational silencing via a different mechanism.
Fig. 3.
Regulation of tumor necrosis factor α (TNFα) transcripts by TIA-1. An adenine-uridine–rich element (ARE) tethers TIA-1 to the 3′ untranslated region of TNFα transcripts. This increases the likelihood that TIA-1 will assemble at a noncanonical, translationally silent preinitiation complex, which delivers these transcripts to stress granules (SGs). The SG is proposed to function as a translational checkpoint that monitors mRNP composition and determines whether individual transcripts are stabilized or degraded. The ARE-binding proteins HuR and TTP are proposed to act downstream of the assembly of SGs to influence the functional fate of individual transcripts
The extent to which TIA-1 represses the production of TNFα is influenced by epistatic factors. LPS-activated macrophages derived fom BALB/c mice lacking TIA-1 secrete 2–3 times more TNFα than wild-type BALB/c controls do. However, LPS-activated macrophages derived from C57BL/6 mice lacking TIA-1 secrete 5–9 times more TNFα than wild-type controls do (Saito et al 2001). Although the genetic factors that modify the ability of TIA-1 to repress the expression of TNFα have not been identified, it is likely that the expression or activity (or both) of ARE-binding proteins, such as TTP, AUF1, and HuR, will influence the functional effects of TIA-1. The potential importance of interactions between different ARE-binding proteins is underscored by the observation that TIA-1 regulates the production of TNFα in macrophages, but not in lymphocytes, despite the fact that both cell types express similar amounts of TIA-1 protein (Saito et al 2001).
TIA-1 AND TIAR ALSO REGULATE SPECIFIC mRNA SPLICING
Their visible roles in the cytoplasm notwithstanding, TIA-1 and TIAR normally predominate in the nucleus, where they have been shown to act as selective regulators of alternative mRNA splicing (Del Gatto-Konczak et al 2000; Forch et al 2000; Le Guiner et al 2001). The binding of either TIA-1 or TIAR to uridine-rich elements found in intronic sequences located downstream of weak 5′ splice sites promotes the recruitment of U1snRNP and the inclusion of “cryptic” alternative exons that are otherwise excised from the heteronuclear RNA. Using in vitro assays, TIA-1 has been shown to promote the inclusion of alternatively spliced exons in heteronuclear RNAs encoding fibroblast growth factor receptor 2, male-specific lethal 2, and Fas. Although it is not yet clear whether TIA-1 has similar effects on these specific target genes in vivo, both TIA-1 and TIAR can regulate the splicing of their own heteronuclear RNAs in vivo (Le Guiner et al 2001). In this study, the overexpression of TIA-1 or TIAR results in the inclusion of cryptic exons in TIA-1 or TIAR transcripts that are normally excised. These exons introduce premature stop codons or frameshifts that are likely to inactivate the function of the expressed protein. By this novel mechanism, TIA-1 and TIAR may participate in a feedback regulatory loop that uses the splicing machinery to dampen their own protein expression. This may well occur in vivo, because cells and tissues from mice lacking TIA-1 express increased amounts of TIAR compared with wild-type cells. Conversely, cells lacking TIAR express increased amounts of TIA-1 compared with wild-type cells (unpublished observations).
In their dual ability to regulate both mRNA splicing and translation, TIA-1 and TIAR resemble several other multifunctional RNA-binding proteins, including PTB, CUB-BP–related proteins, La, hnRNPK, and hnRNP A1 (Wilkinson and Shyu 2001; Ladomery 1997). The process of mRNA splicing has been shown to “mark” selected mRNA transcripts for both quality control in the nucleus and translational control in the cytoplasm (Le Hir et al 2000a, 2000b). It is therefore possible that heteronuclear mRNAs encoding introns that are recognized by TIA-1 or TIAR retain these proteins at the exon-exon junction after the removal of the intron upon splicing. Upon arrival in the cytoplasm, transcripts that are marked by TIA-1 or TIAR could be selectively regulated at the level of mRNA stability or translatability. This hypothetical explanation would allow for the nuclear loading of TIA-1 and TIAR onto selected transcripts, which are subject to translational silencing once in the cytoplasm. It also suggests a mechanism whereby stress-induced transcripts would be exempt from recruitment to SGs: mRNAs transcribed during stress would escape being marked by TIA proteins, whose normal shuttling appears disrupted when they are routed instead into cytoplasmic SGs.
CONCLUSIONS AND FUTURE QUESTIONS
To reprogram translation from a normal state to a stress state, the cell must rapidly reallocate its limited translational capacity to allow the preferred expression of stress response genes. Polysomes must be disassembled, their mRNAs sorted and processed, and the translation of newly synthesized stress response transcripts must be enhanced. Although it is immediately obvious that the cell needs to express new genes during stress, it is less obvious (but no less important) that in order to do so efficiently, it must first clear the old mRNAs away, hence the process of routing existing transcripts to the SG for triage. Regulation of the polysome-mRNP equilibrium is an active process, which also occurs normally in the absence of stress. The seemingly sudden appearance of the SG during stress is the result of the sudden influx of transcripts that are stalled because of lack of the ternary complex, which exceed the cell's processing capacity. The triage process is also linked to mRNA stability and is, therefore, likely to require RNA-binding proteins that regulate mRNA stability, such as HuR, hnRNPD/AUF1, and TTP. We and others have identified HuR as an SG component (Gallouzi et al 2000; Kedersha et al 2002) and have recently obtained data linking TTP to SGs (unpublished observations).
The efficiency of the triage process is likely to influence the eventual outcome of prolonged stress-induced translational arrest. If the damage is repaired before a critical time threshold, SGs are disassembled, and the translation of proteins essential for survival can resume and allow the cell to live (Kedersha et al 1999). If, on the other hand, stress-induced damage is not repaired, the SG persists, and the proteins essential for survival are not formed because of the continued preferential translation of Hsps, ATF4, etc (Kedersha et al 1999). When a critical threshold is reached, the “death by default” point is reached, and apoptosis ensues, a consequence of survival factor withdrawal. Cells that can most rapidly and efficiently sort and reprogram their translational repertoire would be best equipped for survival. We have been unable to obtain cell lines that stably overexpress the TIA-1ΔRRM dominant–negative mutant that blocks SG assembly, although cell lines that stably overexpress TIA-1 are readily obtained. This suggests that the normal function of the SG is essential in cells.
The available data suggest that eIF2-GTP-tRNAMet and TIA-1 and TIAR function as antagonists that promote the assembly of translationally competent and translationally incompetent preinitiation complexes, respectively. It remains to be determined whether these antagonistic translation factors compete for binding to a common site on the preinitiation complex. Regardless of the molecular mechanism, the functional antagonism between these factors could determine the number of times a given mRNA transcript is initiated before being subject to a checkpoint at which mRNP structure and composition is monitored. If the ratio of TIA-1 or TIAR to eIF2-GTP-tRNAMet were 1:10, one might predict that, on average, 10 productive initiation events would occur before a TIA-1– or TIAR-containing, translationally incompetent preinitiation complex is assembled. As translating ribosomes “run off” this mRNA, the eIF2-eIF5–deficient complex would be routed to an SG for mRNA triage. Depending on the availability and activity of other mRNA processing proteins, such as HuR and TTP, this mechanism could regulate the number of times a given transcript is translated before it is degraded.
In vivo, mRNA transport, quality control, and stability are coupled processes that are mediated by multifunctional shuttling proteins, such as TIA-1, TIAR, and HuR, which perform different but related functions in the nucleus and the cytoplasm. The identification of SGs as specialized cytoplasmic domains at which mRNA triage occurs suggests that similar dynamic structures dedicated to nuclear mRNA quality control may also exist in the nucleus (Spector 2001). Doubtless, more insights between function and structure will be forthcoming in the study of mRNA metabolism, where biochemistry and cell biology converge.
REFERENCES
- Arrigo AP, Suhan JP, Welch WJ. Dynamic changes in the structure and intracellular locale of the mammalian low-molecular-weight heat shock protein. Mol Cell Biol. 1988;8:5059–5071. doi: 10.1128/mcb.8.12.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. Host defense, viruses and apoptosis. Cell Death Differ. 2001;8:113–126. doi: 10.1038/sj.cdd.4400823. [DOI] [PubMed] [Google Scholar]
- Beck ARP, Medley QG, O'Brien S, Anderson P, Streuli M. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res. 1996;24:3829–3835. doi: 10.1093/nar/24.19.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck ARP, Miller IJ, Anderson P, Streuli M. RNA-binding protein TIAR is essential for primordial germ cell development. Proc Natl Acad Sci U S A. 1998;95:2331–2336. doi: 10.1073/pnas.95.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier NC, Heuser J, Levy MA, Schlesinger MJ. Ultrastructural and biochemical analysis of the stress granule in chicken embryo fibroblasts. J Cell Biol. 1988;106:1131–1139. doi: 10.1083/jcb.106.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier NC, Schlesinger MJ. The dynamic state of heat shock proteins in chicken embryo fibroblasts. J Cell Biol. 1986;103:1495–1507. doi: 10.1083/jcb.103.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Bourgeois CF, Le Guiner C, Kister L, Gesnel MC, Stevenin J, Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol Cell Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- Dever TE. Translation initiation: adept at adapting. Trends Biochem Sci. 1999;24:398–403. doi: 10.1016/s0968-0004(99)01457-7. [DOI] [PubMed] [Google Scholar]
- Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci U S A. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Esteban M. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis. 2000;5:107–114. doi: 10.1023/a:1009664109241. [DOI] [PubMed] [Google Scholar]
- Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J Biol Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- Han AP, Yu C, and Lu L. et al. 2001 Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20:6909–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC 2000 The pathway and mechanism of initiation of protein sythesis. In: Translational Control of Gene Expression, ed Sonenberg N, Hershey, JWB, Mathews MB. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 33–88. [Google Scholar]
- Hinnebusch AG 1996 Translational control of GCN4: Gene-specific regulation by phosphorylation of eIF2. In: Translational Control, ed Hershey JWB, Matthews MB, Sonenberg N. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 199–244. [Google Scholar]
- Hinnebusch AG. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- Kawakami A, Tian Q, Duan X, Streuli M, Schlossman SF, Anderson P. Identification and functional characterization of a TIA-1-related nucleolysin. Proc Natl Acad Sci U S A. 1992;89:8681–8685. doi: 10.1073/pnas.89.18.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami A, Tian Q, Streuli M, Poe M, Edelhoff S, Disteche CM, Anderson P. Intron-exon organization and chromosomal localization of the human TIA-1 gene. J Immunol. 1994;152:4937–4945. [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller I, Stahl J, and Anderson P 2002 Evidence that ternary complex (eIF2-GTP-tRNAMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell 13: 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho M, Li W, Yacono P, Chen S, Gilks N, Golan D, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR. Eukaryotic initiation factor eIF2. Int J Biochem Cell Biol. 1999;31:25–29. doi: 10.1016/s1357-2725(98)00128-9. [DOI] [PubMed] [Google Scholar]
- Kimball SR. Regulation of translation initiation by amino acids in eukaryotic cells. Prog Mol Subcell Biol. 2001;26:155–184. doi: 10.1007/978-3-642-56688-2_6. [DOI] [PubMed] [Google Scholar]
- Ladomery M. Multifunctional proteins suggest connections between transcriptional and post-transcriptional processes. Bioessays. 1997;19:903–909. doi: 10.1002/bies.950191010. [DOI] [PubMed] [Google Scholar]
- LeGuiner C, Lejeune F, Galiana D, Kister L, Breathnach R, Stevenin J, and Del Gatto-Konczak F 2001 TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J Biol Chem, 276: 40638–40646. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000a;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000b;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Lu L, Han AP, Chen JJ. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol Cell Biol. 2001;21:7971–7980. doi: 10.1128/MCB.21.23.7971-7980.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol. 1983;3:1648–1655. doi: 10.1128/mcb.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf K, Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D, Ostareck-Lederer A, Shatsky I, Hentze M. Lipoxygenase mRNA silencing in erythroid differentiation: the 3′ UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. Ribosome recruitment and scanning: what's new? Trends Biochem Sci. 1999;24:85–87. doi: 10.1016/s0968-0004(99)01356-0. [DOI] [PubMed] [Google Scholar]
- Phan L, Schoenfeld LW, Valasek L, Nielsen KH, Hinnebusch AG. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met. EMBO J. 2001;20:2954–2965. doi: 10.1093/emboj/20.11.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M, Wax S, and Beck A. et al. 2000 TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Chen S, Piecyk M, Anderson P. TIA-1 regulates the production of TNF alpha in macrophages, but not in lymphocytes. Arthritis Rheum. 2001;44:2879–2887. doi: 10.1002/1529-0131(200112)44:12<2879::aid-art476>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Spector D. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Srivastava SP, Kumar KU, Kaufman RJ. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J Biol Chem. 1998;273:2416–2423. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- Tian Q, Streuli M, Saito H, Schlossman SF, Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–639. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- Wilkinson MF, Shyu AB. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays. 2001;23:775–787. doi: 10.1002/bies.1113. [DOI] [PubMed] [Google Scholar]
- Williams BR. Signal integration via PKR. Sci: Signal Transduction Knowledge Environ. 2001;2001:RE2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]