Abstract
Molecular chaperone complexes containing heat shock protein (Hsp) 70 and Hsp90 are regulated by cochaperones, including a subclass of regulators, such as Hsp70 interacting protein (Hip), C-terminus of Hsp70 interacting protein (CHIP), and Hsp70-Hsp90 organizing factor (Hop), that contain tetratricopeptide repeats (TPRs), where Hsp70 refers to Hsp70 and its nearly identical constitutive counterpart, Hsc70, together. These proteins interact with the Hsp70 to regulate adenosine triphosphatase (ATPase) and folding activities or to generate the chaperone complex. Here we provide evidence that small glutamine-rich protein/viral protein U–binding protein (SGT/UBP) is a cochaperone that negatively regulates Hsp70. By “Far-Western” and pull-down assays, SGT/UBP was shown to interact directly with Hsp70 and weakly with Hsp90. The interaction of SGT/UBP with both these protein chaperones was mapped to 3 TPRs in SGT/UBP (amino acids 95–195) that are flanked by charged residues. Moreover, SGT/UBP caused an approximately 30% reduction in both the intrinsic ATPase activity of Hsc70 and the ability of Hsc70 to refold denatured luciferase in vitro. This negative effect of SGT/UBP on Hsc70 is similar in magnitude to that observed for the cochaperone CHIP. A role for SGT/UBP in protein folding is also supported by evidence that a yeast strain containing a deletion in the yeast homolog to SGT/UBP (ΔSGT/UBP) displays a 50-fold reduction in recovery from heat shock compared with the wild type parent. Together, these results are consistent with a regulatory role for SGT/UBP in the chaperone complex.
INTRODUCTION
The multiprotein chaperone complex promotes the correct cotranslational folding and refolding of denatured proteins, intracellular transport, and targeted degradation of substrate polypeptides (reviewed in Frydman 2001). Heat shock protein 70 (Hsp70) and its nearly identical constitutive counterpart, Hsc70, are important components of the chaperone complex. Both proteins (together referred to here as Hs70) contain intrinsic adenosine triphosphatase (ATPase) domains located in the N-terminal segment and cycle between an ATP-bound form and an adenosine diphosphate (ADP)-bound form. ATP-bound Hs70 has relatively low affinity for substrate peptides, whereas the ADP-bound form has higher affinity and promotes more efficient protein folding. When a protein substrate occupies the substrate binding site of ADP-bound Hs70, a conformational change in the C-terminus takes place that results in tight association between Hs70 and the substrate (Ha and McKay 1995; Johnson and McKay 1999). The ATP-bound Hs70 does not undergo this conformational change, and this accounts for the difference between high- and low-affinity substrate binding of Hs70. Although Hs70 can independently refold denatured proteins in vitro, in the chaperone complex Hs70 transfers some protein substrates to Hsp90, which is another important constituent of the multiprotein chaperone complex. Hsp90 can actively promote the refolding of protein substrates.
Cochaperones regulate the activity of the chaperone complex by interacting with Hs70 and Hsp90. One important class of cochaperones contains contiguous copies of a motif called the tetratricopeptide repeat (TPR). A TPR motif is a 34–amino acid segment that forms 2 amphipathic alpha-helices punctuated by a turn, facilitating interaction with other proteins through their hydrophobic face (Hirano et al 1990). Moreover, tandem multiple TPRs can be arranged in a parallel array leading to a regular series of antiparallel alpha-helices that constitute an amphipathic groove (Das et al 1998). TPR-containing cochaperones that affect Hs70 interaction include Hs70 interacting protein (Hip; p48), which stabilizes the more active ADP-bound form of Hs70 (Hohfeld et al 1995), and C-terminus of Hs70 interacting protein (CHIP), which inhibits the formation of ADP-bound Hs70 (Ballinger et al 1999). TPR-containing cochaperones that affect Hsp90 include the peptidylprolyl isomerases CyP-40 and FKBP52, and the protein phosphatase PP5 (Ratajczak and Carrello 1996; Das et al 1998). CHIP and Hs70-Hsp90 organizing factor (Hop) can interact with both Hs70 and Hsp90 (Smith et al 1993; Chen and Smith 1998). Hop interacts simultaneously with Hs70 and Hsp90 as a tether in the chaperone complex and promotes the transfer of substrate proteins from Hs70 to Hsp90 (Chen and Smith 1998). When bound to Hs70 or Hsp90, CHIP is able to alter the chaperone complex such that specific protein substrates are targeted for degradation rather than folding (Connell et al 2001). Moreover, CHIP can function as an E3 ubiquitin ligase and contains a U-box motif that is important for the ability of CHIP to target substrate proteins for degradation (Jiang et al 2001).
Viral protein U (Vpu)–binding protein (UBP) was identified initially as a human protein that interacts with Vpu and with Group specific Antigen (Gag), the major structural protein of the viral capsid. These are 2 proteins of human immunodeficiency virus (HIV)-1 (Callahan et al 1998). Small glutamine-rich protein (SGT) is the rat homolog of SGT/UBP and was identified independently as a protein that interacts with the nonstructural protein NS-1 from autonomous parvovirus H-1 (Cziepluch et al 1998). NS-1 is required for viral deoxyribonucleic acid (DNA) replication and is found together with SGT in nuclear foci at the site of H-1 DNA synthesis (Cziepluch et al 2000). Recently SGT/UBP was also identified as part of a trimeric chaperone composed of Hsp70, a cysteine string protein located in synaptic vesicles, and SGT/UBP (Tobaben et al 2001). To gain further insight into the normal function of SGT/UBP, we searched for human cellular proteins that stably interact with human SGT/UBP. Hs70 is the principle protein that interacts with SGT/UBP. On the basis of biochemical and genetic analysis, SGT/UBP is a cochaperone that negatively affects the activity of Hs70. SGT/UBP also interacts with Hsp90. Thus, the protein chaperone complex may be involved in Vpu function and HIV particle exit and in parvovirus DNA replication.
MATERIALS AND METHODS
Cell lines and antibodies
HeLa CCL-2 cells (American Type Tissue Culture Collection) were used for analysis of HIV-1 particle release. Cells were grown in monolayer cultures at 37°C in Dulbecco modified Eagle medium (DMEM; GIBCO, Gaithersberg, MD, USA) supplemented with 5% fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT, USA) and maintained using standard techniques. Polyclonal antibodies to HIV Gag and SGT/UBP have been described previously (Callahan et al 1998). The anti-Hsp70 and anti-Hsp90 monoclonal antibodies were purchased from Stressgen (Victoria, British Columbia, Canada). Antiglutathione s-transferase (GST) polyclonal antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Goat anti-rabbit and anti-mouse antibodies conjugated with alkaline phosphatase (AP) were purchased from Sigma Immunochemicals (St Louis, MO, USA).
DNA constructions and proteins
Several of the plasmids used, including pGST-SGT/UBP, have been described previously (Callahan et al 1998). Derivatives of pGST-SGT/UBP contained the following SGT/UBP segments: Δ1–93, Δ95–195, Δ288–313, N1/2 (amino acids [a.a.] 1–145), C1/2 (a.a. 145–313), and TPR2–4 (a.a. 288–313). For yeast SGT/UBP knockout studies, the entire y-SGT/UBP gene (GenBank Accession number U43491) was directly cloned into the BamHI site of pRS316. Yeast SGT/UBP including the flanking nontranslated regions was amplified with the following primers: upstream, 5′-CGCGGATCCAGAAGATTCCAGGTTCAAG-3′ and downstream, 5′-GCTGGATCCAGTTCTATACAGATTTACAT-3′, where the BamHI sites are underlined. The resulting construct was referred to as pRS316-y-SGT/UBP. To generate the y-SGT/UBP gene targeting construct, pRS316-y-SGT/UBP was digested with Bgl II at nt 751 of the SGT/UBP open reading frame (ORF), and a histidine biosynthetic marker gene flanked by BamHI sites was directly ligated to the Bgl II ends. This resulted in a targeting construct (pRS316-His-y-SGT/UBP) in which the His marker was flanked by more than 1 kb of SGT/UBP DNA on each end. To produce purified SGT/UBP, a vector expressing histidine-tagged SGT/UBP (pHis-SGT/UBP) was created by cloning the SGT/UBP ORF directly into the pQE30 vector via the BamHI and HindIII restriction sites.
Protein purification
GST-SGT/UBP, used for “Far-Western blots,” was expressed in Escherichia coli used standard techniques. Bacteria pelleted from a 1-L culture were treated with 10 mL of lysis buffer (100 mM NaCl, 10 mM Tris [pH 8.0], 0.1 mM ethylenediamine-tetraacetic acid (EDTA), and 0.5% Triton X-100). Lysozyme was added to a final concentration of 1.8 mg/mL and the mixture incubated for 30 minutes at 37°C. The lysate was then subjected to 3 freeze-thaw cycles. The protein was centrifuged at 10 000 × g in a Beckman J2-21 centrifuge, and the supernatant was recovered. The protein fraction was then applied to a glutathione-Sepharose column (Pharmacia, Uppsala, Sweden), washed, and eluted according to the manufacturer's specifications. For ATPase and luciferase refolding assays, purified His-SGT/UBP was used. Similarly, His-SGT/UBP expressed in bacteria was applied to a 1-mL nickel-nitroloacetyl acid (NTA) resin column in the appropriate buffer as prescribed by the manufacturer (Qiagen, Valencia, CA). The column was washed and eluted with 400 mM imidazole. All purified proteins were analyzed by Coomassie gel and quantified by the Bradford assay (Bio-Rad, Hercules, CA, USA). Pure Hsp70, Hsc70, and Hsp90 used in several experiments were purchased from Stressgen.
Western and Far-Western blotting
Western blots were performed using standard techniques. Briefly, 30 mg HeLa cytoplasmic extract or 20 ng of Hsc70, Hsp70, or Hsp90 was denatured in 1× sodium dodecyl sulfate (SDS) sample buffer. The samples were loaded on a 10% polyacrylamide gel. Separated protein was transferred to nitrocellulose membranes. The membranes were blocked in 5% dry milk protein in Tris-buffered saline (TBS; 20 mM Tris [pH 7.4], 100 mM NaCl) for 45 minutes. The gels with proteins separated on them were transferred to nitrocellulose and blocked against 5% dry milk protein in TBS for 45 minutes. Primary antibody, anti-Hsp70 or anti-Hsp90 monoclonal antibody, or anti-SGT/UBP rabbit polyclonal antibody was incubated with the blots at appropriate concentrations (1:1000 dilutions for each) in 5% milk protein in TBS for 2 hours at 4°C with gentle rocking. The membranes were washed with TBS for 10 minutes. Secondary antibody, either alkaline phosphatase–conjugated goat anti-mouse IgG or alkaline phosphatase–conjugated goat anti-rabbit IgG diluted to 1:5000 in 5% milk in TBS was incubated with the blots for 2 hours at 4°C with gentle rocking. The blots were washed in TBS and then rinsed in AP buffer (100 mM Tris [pH 9.5], 100 mM NaCl, 5 mM MgCl2). The blots were developed by incubation with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (Sigma) at concentrations of 0.33 and 0.17 mg/mL, respectively, in 5 mL of AP buffer.
For the Far-Western blots, after the blocking step, GST-SGT/UBP was incubated with the blot in 1% milk protein in TBS overnight at 4°C with gentle rocking. Subsequently, the Far-Western blot was developed by incubation with anti-GST mouse monoclonal antibody followed by incubation with alkaline phosphatase–conjugated goat anti-mouse IgG (Angeletti and Engler 1996, 1998).
Yeast SGT/UBP gene disruption
The diploid yeast strain DG401a, which was used in gene-knockout experiments was the gift of David Brow (UW-Madison). DG401a has the following genotype: mat a/α, ura3–52/ura3–52, his3–200/his3–200, lys2–801/lys2–801, trp1–901/trp1–901, ade2–101/ade2–101, met−/met−, gal4–542/gal4–542, gal80–538/gal80–538. SGT/UBP gene disruptions were carried out essentially as described by Rothstein (1991). The targeting construct was excised from pRS316-His-y-SGT/UBP with a BamHI digest. Approximately 200 ng of the linear fragment was recovered and directly transformed into 105 yeast cells using the standard LiAc method (Schiestl et al 1993). Transformants were plated on media lacking His, and colonies were isolated after 2–3 days. Correct integrants were identified by polymerase chain reaction (PCR) amplification using a primer in the chromosomal region upstream of SGT/UBP (5′-CTAATCACAACACTTAGC-3′) and a primer internal to the His biosynthetic gene (5′-ACTAGAGGAGGCCAAGAG-3′). Correct integrants gave rise to a 1.4-kb PCR product, whereas nonhomologous integrants gave rise to no product (data not shown). Positive integrants were subjected to tetrad analysis. To induce sporulation, the diploid strain was grown in nitrogen-deficient sporulation media and the asci were analyzed for viability by tetrad analysis. Haploid sporulates were then analyzed for SGT/UBP disruption by PCR.
Heat shock analysis of y-SGT/UBP
Haploid yeast, either wild type or those containing a SGT/UBP deletion, were grown in liquid culture in yeast peptone dextrose medium (YPD) to a density of 0.5 at OD600. The yeast were then subjected to heat treatment at 55°C for 1 hour (Nicolet and Craig 1991; Hampsey 1997). As a control, a duplicate aliquot of cells was left untreated. The yeast were then plated on YPD at dilutions appropriate for counting colonies. After 3 days colonies were counted, and the data were expressed as the percent total survivors after heat treatment normalized to wild type.
Hsc70 ATPase assays
ATPase assays were performed essentially as described by Liberek et al (1991). In a reaction volume of 25 μL, pure Hsc70 at a final concentration of 28 nM was incubated with 1 μCi (13 nM) [α-32P]ATP (Amersham, Piscataway, NJ) and various amounts of pure SGT/UBP or bovine serum albumin (BSA). The reactions were carried out at 30°C in ATPase buffer (30 mM 4-[-2 hydroxymethl]-1-piperazineethanesulfonic acid [HEPES] buffer [pH 7.5], 40 mM KCl, 50 mM NaCl, 5 mM MgCl2, and 2 mM dithiothreitol). For the dose-curve experiment, 0, 12, 58, 118, or 176 nM BSA or SGT/UBP was incubated with Hsc70 for 1 hour. A 5-μL aliquot of each reaction was directly loaded onto a polyethylamine-cellulose–thin layer sheet (Selecto Scientific, Suwanee, GA, USA). The products were separated from the substrate by chromatography using 1 M formic acid/1 M LiCl (1:1, vol/vol) as a vehicle. The location of ADP and ATP on the plates was visualized by use of a Phosphorimager (Molecular Dynamics, San Jose, CA, USA). The data were quantified and expressed as the percent total Hsc70 ATPase activity normalized to the control reactions lacking either BSA or SGT/UBP. In the time course experiment, Hsc70 (28 nM) was incubated with SGT/UBP at a final concentration of 28 nM. Five-microliter aliquots of each reaction were taken at time points of 0, 15, 30, 45, 60, and 75 minutes. The samples were analyzed as previously described. Data were quantified and expressed as the percent ATP hydrolysis.
Luciferase refolding assays
The assay for the refolding of heat-denatured luciferase was performed as described by Lu and Cyr (1998) and Ballinger et al (1999). Pure luciferase was diluted to 129 nM in refolding buffer (25 mM HEPES, [pH 7.4], 50 mM KCl, 5 mM MgCl2). Luciferase was denatured by incubation at 42°C for 20 minutes. Refolding reactions contained 28 nM Hsc70 and 1 mM ATP in the presence or absence of pure SGT/UBP at a final concentration of 28 nM. Two microliters of denatured luciferase was added to each reaction, and incubations were carried out for 0–100 minutes at 30°C. Samples were analyzed for luciferase activity using a monolight luminometer (Analytical Luminescence Laboratory, Ann Arbor, MI, USA). Data were compiled and graphed using Microsoft Excel.
GST-SGT/UBP/Hsc70 pull-down assay
Glutathione-Sepharose beads (4B; Pharmacia) were equilibrated in GST binding buffer (100 mM NaCl, 20 mM Tris [pH 7.9], 1 mM EDTA, 5% glycerol, 0.02% NP40 + 1 mM phenylmethane sulfonyl fluoride). Either GST or GST-SGT/UBP protein was incubated with a 100-μL bed volume of beads in a total volume of 300 μL of binding buffer at 4°C with gentle rocking for 2 hours. The beads were washed and then blocked with 5% BSA in binding buffer. The beads were washed in binding buffer and then incubated with 200 ng of Hsc70 in binding buffer for 2 hours at 4°C. The beads were again washed with binding buffer, and then 20 μL of beads were added to individual tubes. Each tube was washed 3 times with binding buffer containing 100, 200, 300, or 500 mM NaCl. The beads were resuspended in 1× Laemmli buffer. The samples were heated to 80°C for 3 minutes, and then the supernatants recovered by centrifugation were analyzed by Western blot for Hsc70.
RESULTS
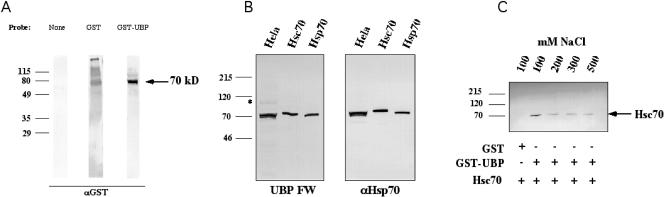
To elucidate the normal role of SGT/UBP in cells, we attempted to identify other cellular proteins that interact stably with SGT/UBP. To this end, HeLa cell lysates were subjected to Far-Western analysis. This entailed separation of the cell proteins on SDS gels and electrophoretic transfer to an Immobilon P membrane. The membrane was then incubated with GST-SGT/UBP, and stable association of the SGT/UBP with transferred protein was detected using anti-GST antibody as in conventional Western analysis. Data from this experiment indicated that a protein of about 70 kDa was the primary species that stably interacted with SGT/UBP (Fig 1A). Using a yeast 2-hybrid screen for proteins that interact with Hsp70, Liu et al (1999) found that Hs70 interacts with SGT/UBP. Moreover, SGT/UBP contains TPR motifs in an array similar to that found in cochaperones such as Hip, Hop, and CHIP (Irmer and Hohfeld 1997; Chen and Smith 1998; Ballinger et al 1999). Thus, we used purified Hsp70 and Hsc70 in Far-Western analysis in parallel with HeLa cell proteins to see whether the 70-kDa protein was Hs70. This indicated that SGT/UBP does associate stably with both Hsp70 and Hsc70 in this in vitro assay (Fig 1B). To examine the stability of this SGT/UBP-Hs70 interaction, we subjected the protein complexes to washes of increasing NaCl concentrations. This indicated that the protein was maintained even in the presence of relatively high salt concentrations (0.5 M NaCl) and is consistent with a robust interaction between SGT/UBP and Hs70 (Fig 1C).
Fig 1.
Small glutamine-rich protein/viral protein U–binding protein (SGT/UBP) binds directly to heat shock protein 70 (Hsp70) and its constitutive counterpart, Hsc70, with high affinity. (A) HeLa cell lysate (30 μg) was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis in 3 identical lanes blotted to nitrocellulose and probed with no probe (none), GST alone, or GST-SGT/UBP. The Far-Western blot was then developed with anti-GST antibody to reveal bands that interact with SGT/UBP. The arrow to the right indicates a 70-kDa band detected by the GST-SGT/UBP fusion. The numbers to the left of each blot represent molecular weights in kilodaltons. (B) Identical blots containing 30 μg of HeLa lysate, 50 ng pure Hsc70, and 50 ng pure Hsp70, as indicated, were subjected to SGT/UBP Far-Western (SGT/UBP FW) or anti-Hsp70 Western analysis (αHSP70). The asterisk indicates the position of Hsp90, also detected in HeLa lysate by SGT/UBP Far-Western interaction. (C) Samples from the GST-SGT/UBP pull-down assay were analyzed by Western blot for the presence of Hsc70. At the bottom of the gel, the contents of each reaction are indicated by the plus symbols. The NaCl concentration of the washing buffer (100, 200, 300, and 500 mM) is indicated at the top of each lane. The arrow to the right of the blot indicates the position of the Hsc70 band
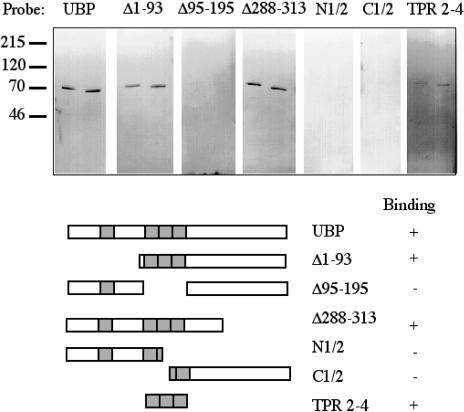
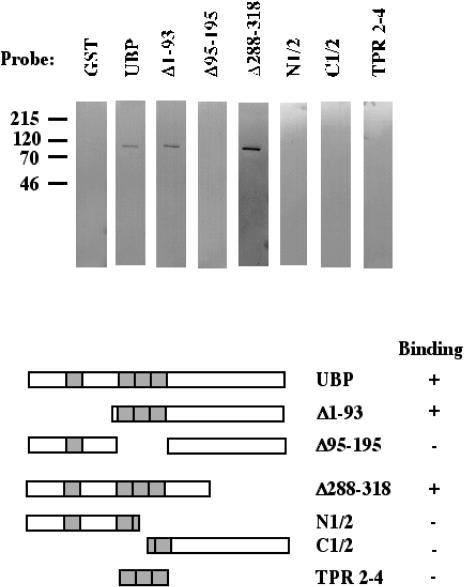
Several cochaperones that mediate the activity of Hs70 contain TPR motifs and interact with Hs70 by way of their TPR motifs. TPRs are generally considered to be motifs that mediate intermolecular interaction by way of a signature alpha-helix (Das et al 1998). Thus, we wanted to test the hypothesis that the TPRs in the N-terminal half of SGT/UBP are required for SGT/UBP-Hs70 interaction. Several deletion mutants of SGT/UBP were constructed that lack various segments of the protein, and these were tested for interaction with Hsc70 using Far-Western analysis (Fig 2) These data indicate that the TPRs of SGT/UBP are indeed necessary for interaction with Hs70. SGT/UBP mutants that maintained the 3 tandem TPRs (Δ1–93, Δ288–313, and TPR 2–4) were capable of stable interaction with Hsc70, whereas those that lacked the 3 TPRs (Δ95–195, N 1/2, and C 1/2) did not interact detectably with Hsc70. Further, the fact that N 1/2 and C 1/2, which contain a single intact TPR each, were unable to associate with Hs70 indicates that a single TPR is unlikely to be sufficient for association with Hsc70. TPR 2–4 is a mutant that expresses only the 3 tandem TPRs (a.a. 95–195). Because this mutant was able to interact stably with Hsc70, it appears that these 3 contiguous TPRs (a.a. 95–195) are sufficient for detectable SGT/UBP-Hs70 interaction. But in further experiments we found that this interaction was enhanced when a fragment contained TPR 2–4 as well as flanking charged residues (a.a. 81–209) such that the extent of binding was similar to that observed for SGT/UBP, Δ1–93, and Δ288–313 (data not shown).
Fig 2.
Mapping of the small glutamine-rich protein (SGT)/viral protein U–binding protein (UBP)/Hs70 interaction, where Hs70 represents heat shock protein (Hsp) 70 and its constitutive counterpart, Hsc70. Seven identical sets of blots containing Hsc70 and Hsp70 were subjected to SGT/UBP Far-Western analysis. The blots were probed with GST fusions as indicated: full-length SGT/UBP, Δ1–93, Δ95–195, Δ288–313, N1/2 (amino acids [a.a.] 1–145), C1/2 (a.a. 145–313), and tetratricopeptide repeats 2–4 (a.a. 95–195). The numbers to the left of the blots indicate the molecular weights in kilodaltons. The diagram below the blots indicates regions of SGT/UBP contained in each fusion construct. The column to the right summarizes the results; a plus symbol indicates binding and a minus symbol indicates no binding
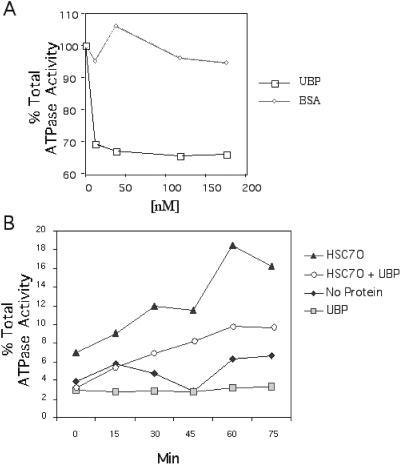
Hs70 contains an intrinsic ATPase activity that is important in the activity and function of the multiprotein chaperone complex. The ATP-bound form of Hs70 has relatively low affinity for protein substrates, whereas the ADP-bound form of Hs70 has relatively high affinity for protein substrates. Regulatory cochaperones that associate with Hs70, such as Hsp40, BAG-1, Hip, and CHIP, often exert their effect by affecting positively or negatively the ATPase activity of Hs70 (Irmer and Hohfeld 1997; Bimston et al 1998; Ballinger et al 1999). To see whether SGT/UBP might similarly affect the ATPase activity of Hsc70, we carried out in vitro ATPase assays in the presence of SGT/UBP. This analysis indicated that SGT/UBP affected negatively the Hsc70-mediated hydrolysis of ATP (Fig 3). The magnitude of this effect was similar to that of the cochaperone CHIP, a cochaperone that negatively affects the ATPase activity of Hs70. These data are consistent with the idea that SGT/UBP is also a cochaperone that affects the activity of Hs70.
Fig 3.
Small glutamine-rich protein/viral protein U–binding protein (SGT/UBP) inhibits the adenosine triphosphatase (ATPase) activity dependent on the constitutive counterpart, Hsc70, of heat shock protein (Hsp) 70. (A) Hsc70 (28 nM) was incubated with 0, 12, 58, 118, or 176 nM SGT/UBP or bovine serum albumin (BSA) and 1 μCi of [α-32P]ATP (13 nM) for 1 hour at 30°C. Adenosine diphosphate was separated from ATP by thin-layer chromatography and quantified using a Phosphorimager. Data are represented as the percent total ATPase activity, with the Hsc70 ATPase activity in the absence of BSA or SGT/UBP set to 100%. The Hsc70 ATPase activity in the presence of BSA (diamonds) or SGT/UBP (squares) is graphed as shown. (B) Hsc70 (28 nM) was incubated with 1 μCi of [α-32P]ATP with or without SGT/UBP at a final concentration of 28 nM. Hydrolysis of ATP was monitored in the presence of Hsc70 (triangle), Hsc70 and SGT/UBP (circle), SGT/UBP alone (squares), or with no protein (diamond). The data are represented as the percent ATP hydrolysis and are plotted as a function of time from 0 minutes to 75 minutes
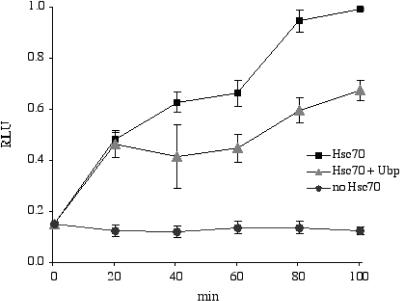
Although Hs70 functions as part of a multiprotein complex, the protein is able to promote independently the refolding of denatured protein in vitro in the presence of ATP. To see whether SGT/UBP can affect this refolding activity of Hs70, we carried out an assay to detect the refolding of denatured luciferase in the presence and absence of SGT/UBP (Fig 4). As expected, Hsc70 was able to catalyze the refolding of heat-denatured luciferase to functional form. SGT/UBP inhibited the Hs70-dependent refolding of luciferase by about 30%. This is consistent with the observed negative effect of SGT/UBP on the ATPase activity of Hsc70, similar in effect and magnitude to CHIP's affect on Hsc70-mediated protein refolding and again indicative of a likely role for SGT/UBP as a cochaperone.
Fig 4.
Small glutamine-rich protein/viral protein U–binding protein (SGT/UBP) inhibits refolding of denatured firefly luciferase dependent on the constitutive counterpart, Hsc70, of heat shock protein (Hsp) 70. Heat-denatured luciferase was incubated with Hsc70 (square), Hsc70 and SGT/UBP, or no Hsc70 (circle). Luciferase activity was quantified using a luminometer. Data are plotted as the relative light units as a function of time from 0 minutes to 100 minutes
Hsp90 is another key protein that is found in the chaperone complex and works in conjunction with other members of the complex to ensure correct substrate structure. Some TPR-containing cochaperones, such as Hip and Hop, are able to interact with Hs70 and Hsp90 by way of their TPRs; Hop interacts with both Hs70 and Hsp90 by way of different TPR-containing binding domains (Johnson et al 1998). CHIP interacts with Hsc70 and can also interact with Hsp90 to redirect Hsp90 to function in the degradation of the glucocorticoid receptor (Connell et al 2001). Additional Hsp90-associated proteins, such as the peptidylprolyl isomerases, Cyp40 and FKBP52, and the protein phosphatase PP5, also interact with Hsp90 by way of TPR motifs (Ratajczak and Carrello 1996; Das et al 1998). In our Far-Western analysis, we noticed that SGT/UBP was able to associate with a 90-kDa protein from HeLa cells (indicated with an asterisk in Fig 1B), but the signal appeared to be markedly weaker compared with that for Hs70. To investigate this further, we analyzed purified Hsp90 using Far-Western analysis with SGT/UBP as a probe. On the basis of this experiment, it appeared that SGT/UBP could associate with Hsp90 in vitro (Fig 5). We then evaluated the ability of the various SGT/UBP deletion mutants to interact stably with Hsp90. This analysis revealed that the 3 tandem TPRs of SGT/UBP were necessary for interaction between SGT/UBP and Hsp90. But in contrast to SGT/UBP-Hs70, the interaction of Hsp90 with the 3 TPRs was too weak to detect, suggesting that the specificity provided by the flanking charged amino acids was required for this interaction. Far-Western analysis is only semiquantitative. But the overall binding of SGT/UBP to Hsp90 appears weaker than that observed with Hs70. Moreover, in comparison with the known chaperones, SGT/UBP appears to resemble CHIP more closely than other cochaperones such as Hip or Hop; both SGT/UBP and CHIP interact with both Hs70 and Hsp90, and these interactions are mediated by the 3 tandem TPRs.
Fig 5.
Mapping of the small glutamine-rich protein (SGT)/viral protein U–binding protein (UBP)/heat shock protein 90 (Hsp90) interaction. Identical blots containing 20 ng Hsp90 developed by SGT/UBP Far-Western analysis. The blots were probed with GST or GST fusions having full-length SGT/UBP, Δ1–93, Δ95–195, Δ288–313, N1/2 (amino acids [a.a.] 1–145), C1/2 (a.a. 145–313), and tetratricopeptide repeats 2–4 (a.a. 95–195). The numbers to the left of the blots indicate the molecular weights in kilodaltons. The regions of SGT/UBP contained in each fusion construct are shown below the blots. The right columns summarize the results; a plus symbol indicates binding and a minus symbol indicates no binding
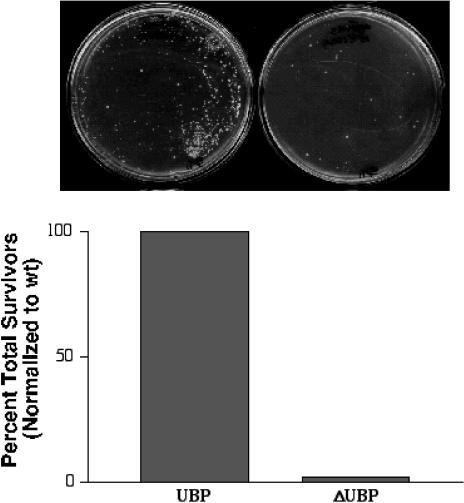
SGT/UBP appears to be a highly conserved gene, and diverse organisms including Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae contain apparent homologs to human SGT/UBP (Callahan et al 1998; Cziepluch et al 1998). To determine whether SGT/UBP may function as a cochaperone in yeast, we used homologous recombination to generate a knockout strain containing a deletion in the yeast SGT/UBP gene (y-SGT/UBP). This y-SGT/UBP mutant was indistinguishable from the parental strain when grown on a rich medium and at 37°C. Hsp70 and Hsp90 are members of a large class of “heat shock” proteins that were originally identified by increased expression after heat treatment. The increased expression of many of these heat shock proteins results in a corresponding increase in chaperone activity and concomitant refolding of heat-denatured proteins. Not surprisingly, many yeast strains that are deficient in appropriate cochaperone activity, and their regulation, are also poor in recovery from various heat shock treatments. We tested the ability of the y-SGT/UBP null mutant to recover from heat shock. The thermotolerance of the mutant was indistinguishable from that of the wild type when the cells were incubated at 18°C, 32°C, and 42°C (data not shown). But the y-SGT/UBP deletion mutant exhibited a marked reduction in viability after a high-temperature (55°C) heat shock (Fig 6). The viability of the mutant was reduced by at least 50-fold relative to the wild type by this treatment. Such a phenotype is similar to that observed for some yeast strains containing lesions in genes encoding protein chaperone functions, such as the gene HSP104 (Sanchez and Lindquist 1990). Thus, the phenotype of the y-SGT/UBP knockout mutant is consistent with a role for SGT/UBP as a functional component of the protein chaperone complex in yeast.
Fig 6.
Deletion mutants of small glutamine-rich protein/viral protein U–binding protein (SGT/UBP) in Saccharomyces cerevisiae are defective for recovery from severe heat shock. Wild type (wt) or SGT/UBP deletion strains were subject to heat shock at 55°C for 1 hour and then plated at appropriate dilutions to rich media. The plates at the top of the bar graph show representative surviving colonies from wild type or SGT/UBP-deletion strains. The number of surviving colonies was counted, and the data were expressed as the percent total survivor, with wild type SGT/UBP set to 100%
DISCUSSION
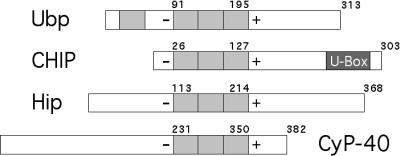
On the basis of multiple complementary lines of biochemical and genetic evidence, it appears that SGT/UBP is a cochaperone that regulates the activity of Hs70. Among the known cochaperones that regulate the activity of the chaperone complex, SGT/UBP appears to be resemble CHIP most in general in vitro activity and in spatial array of TPRs (Fig 7). Both proteins have a slight negative effect on the in vitro ATPase and refolding activity of Hs70, and the TPRs of both proteins are necessary for interaction with Hs70. Furthermore, our experiments show that the magnitude of SGT/UBP inhibition of Hsc70 ATPase activity exactly mirrors the SGT/UBP-dependent inhibition of Hsc70 refolding activity (Figs 3 and 4). SGT/UBP interacts with a TPR receptor site at the C-terminal regulatory domain of Hs70 (Liu et al 1999), and CHIP would also be expected to associate with the same binding site. The functional relationship between SGT/UBP and CHIP is unclear. CHIP plays a pivotal role in conversion of the chaperone complex from a protein-folding apparatus to a protein-degradation machine. This activity of CHIP is specific for particular substrate proteins; the CHIP-Hsp70 interaction targets selectively the cystic fibrosis transmembrane conductance regulator (CFTR) membrane protein for degradation in the endoplasmic reticulum (ER) (Meacham et al 2001). It is possible that SGT/UBP and CHIP are functionally redundant. But SGT/UBP lacks an obvious U-box, which is a motif found in both CHIP and the E4 family of ubiquitin ligases, and is required for the protein degradation function of CHIP. Thus, SGT/UBP could be an antagonist to CHIP and competitively inhibit CHIP-directed proteolysis mediated through the same binding site on Hsp70.
Fig 7.
Comparison of several tetratricopeptide repeat–containing cochaperones
SGT/UBP is able to interact with both Hsp70 and Hsp90 by way of the TPRs of SGT/UBP. But it is important to note that in our experiments the interaction between SGT/UBP and Hsp90 appeared to be significantly weaker than the interaction between SGT/UBP and Hsp90. Some TPR-containing cochaperones, such as Hop and CHIP, are also able to interact with both proteins. It appears that the binding of SGT/UBP to Hsp70 and Hsp90 resembles more the interaction of CHIP with Hsp70 and Hsp90 than that of Hop with Hsp70 and Hsp90. Hop is a bridge protein that interacts with both Hs70 and Hsp90 and is thought to promote the transfer of substrate proteins from Hsp70 to Hsp90 during the process of refolding (Johnson et al 1998). Hop associates with Hs70 and Hsp90 by way of separate, discrete sets of TPRs. In contrast, CHIP interacts separately with both Hsp70 and Hsp90 by way of a single set of TPRs. CHIP interacts functionally with Hsp90 to convert the chaperone complex such that it promotes the ubiquitination and proteosome-mediated degradation of the glucocorticoid receptor (Meacham et al 2001). On the basis of our in vitro binding data, SGT/UBP also interacts with both Hsp70 and Hsp90. As with CHIP, the 3 contiguous TPRs of SGT/UBP are necessary for binding to either Hsp70 or Hsp90, and concurrent binding to both Hs70 and SGT/UBP would not be feasible. Although it remains to be seen whether SGT/UBP affects Hsp90 activity, it again seems plausible that SGT/UBP and CHIP have related or antagonistic functions mediated through binding to a common TPR-receptor site on Hsp90.
The 3 tandem TPRs of SGT/UBP were sufficient for interaction with Hs70 in vitro. But in the case of other TPR-containing cochaperones, such as Hip and Hop, charged residues that flank the TPRs contribute to the binding of these proteins to Hs70 (Frydman 2001). Moreover, there are charged amino acids adjacent to the 3 TPRs of SGT/UBP, and it is possible that these residues, in conjunction with the TPRs, would facilitate a more specific and stronger interaction between SGT/UBP and Hs70.
SGT/UBP connects Vpu and Gag with the cellular chaperone complex. Although the exact mechanistic implications of this link remain ambiguous, it is possible that differential folding of Gag is elicited by Vpu and that Vpu affects the conformation of Gag by altering the cochaperone activity of the SGT/UBP/Hs70 complex. Another possibility is that Vpu targets the SGT/UBP/Hs70/Gag chaperone complex to the cell membrane, where particle assembly and release occurs. Stable membrane association of Gag is required for particle assembly and virus budding. Association of Gag with the membrane occurs by way of the N-terminus of Gag, which is composed of hydrophobic amino acids, that is modified by the addition of myristic acid to a penultimate Gly residue, and adjacent positively charged amino acids (Peitzsch and McLaughlin 1993; Matthews et al 1994; Paillart and Gottlinger 1999). On the basis of the 3-dimensional structure of HIV-1 matrix peptide (MA), it appears that the basic residues would all be displayed on the same face of the protein and would facilitate association with the head groups of acidic phospholipids on the cytoplasmic side of the plasma membrane. In addition, genetic data are consistent with interaction between separate domains of the HIV-1 MA leading to a conformational change in the peptide that results in correct display of the hydrophobic N-terminus.
Parvovirus H-1 and HIV-1 both encode proteins that interact with SGT/UBP. That these 2 diverse viruses both interact with SGT/UBP likely illustrates that both viruses have co-opted the chaperone complex, by way of SGT/UBP, for separate functions in the replication cycle. Parvovirus H-1 protein NS-1 is a nuclear protein required for viral DNA replication (Cziepluch et al 1998). But it appears that H-1 DNA replication occurs on nuclear structures designated “parvovirus-associated replication bodies.” SGT/UBP appears to be present in these structures (Cziepluch et al 2000). Interestingly, there is a precedent for a role of Hs70 and the protein chaperone complex in viral DNA replication. Initiation of phage lambda DNA replication requires the activity of Dna K (Hsp70) and the chaperone complex to facilitate the DNA unwinding activity of the essential DNA helicase, Dna B (Hoffmann et al 1992). Initiation of papillomavirus DNA replication by the viral helicase E1 involves the formation of a complex derived through direct interaction between E1 and Hs70 and Hsp40 (Liu et al 1998). SV40 T-Ag, which is required for viral DNA synthesis, interacts directly with Hs70 by way of a Dna J (Hsp40) domain (Li et al 2001). Therefore, it is plausible that parvovirus H-1 DNA synthesis requires the activity of the chaperone complex and that the complex is brought to the site of DNA synthesis by way of interaction of NS-1 and SGT/UBP.
Acknowledgments
We thank Diccon Fiore, Katrin Talbot, Joshua Fischer, and Carmen Frias for technical assistance. This work was supported by grant RO1 GM63478 from the NIH. One of us (P.C.A) was supported by NIH grant T32 CA09075.
REFERENCES
- Angeletti PC, Engler JA. Tyrosine kinase-dependent release of an adenovirus preterminal protein complex from the nuclear matrix. J Virol. 1996;70:3060–3067. doi: 10.1128/jvi.70.5.3060-3067.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti PC, Engler JA. Adenovirus preterminal protein binds to the CAD enzyme at active sites of viral DNA replication on the nuclear matrix. J Virol. 1998;72:2896–2904. doi: 10.1128/jvi.72.4.2896-2904.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimston D, Song J, Winchester D, Takayama S, Reed JC, Morimoto RI. BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J. 1998;17:6871–6878. doi: 10.1093/emboj/17.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan MA, Handley MA, Lee YH, Talbot KJ, Harper JW, Panganiban AT. Functional interaction of human immunodeficiency virus type 1 Vpu and Gag with a novel member of the tetratricopeptide repeat protein family [published erratum appears in J Virol 1998 72: 8461] J Virol. 1998;72:5189–5197. doi: 10.1128/jvi.72.6.5189-5197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- Cziepluch C, Kordes E, Poirey R, Grewenig A, Rommelaere J, Jauniaux JC. Identification of a novel cellular TPR-containing protein, SGT, that interacts with the nonstructural protein NS1 of parvovirus H-1. J Virol. 1998;72:4149–4156. doi: 10.1128/jvi.72.5.4149-4156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cziepluch C, Lampel S, Grewenig A, Grund C, Lichter P, Rommelaere J. H-1 parvovirus-associated replication bodies: a distinct virus-induced nuclear structure. J Virol. 2000;74:4807–4815. doi: 10.1128/jvi.74.10.4807-4815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Ha JH, McKay DB. Kinetics of nucleotide-induced changes in the tryptophan fluorescence of the molecular chaperone Hsc70 and its subfragments suggest the ATP-induced conformational change follows initial ATP binding. Biochemistry. 1995;34:11635–11644. doi: 10.1021/bi00036a040. [DOI] [PubMed] [Google Scholar]
- Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+, Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- Hoffmann HJ, Lyman SK, Lu C, Petit MA, Echols H. Activity of the Hsp70 chaperone complex—DnaK, DnaJ, and GrpE—in initiating phage lambda DNA replication by sequestering and releasing lambda P protein. Proc Natl Acad Sci U S A. 1992;89:12108–12111. doi: 10.1073/pnas.89.24.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohfeld J, Minami Y, Hartl FU. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- Irmer H, Hohfeld J. Characterization of functional domains of the eukaryotic co-chaperone Hip. J Biol Chem. 1997;272:2230–2235. doi: 10.1074/jbc.272.4.2230. [DOI] [PubMed] [Google Scholar]
- Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;13:13. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- Johnson ER, McKay DB. Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein. Biochemistry. 1999;38:10823–10830. doi: 10.1021/bi990816g. [DOI] [PubMed] [Google Scholar]
- Li H, Soderbarg K, Houshmand H, You ZY, Magnusson G. Effect on polyomavirus T-antigen function of mutations in a conserved leucine-rich segment of the DnaJ domain. J Virol. 2001;75:2253–2261. doi: 10.1128/JVI.75.5.2253-2261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Skowyra D, Zylicz M, Johnson C, Georgopoulos C. The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J Biol Chem. 1991;266:14491–14496. [PubMed] [Google Scholar]
- Liu FH, Wu SJ, Hu SM, Hsiao CD, Wang C. Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J Biol Chem. 1999;274:34425–34432. doi: 10.1074/jbc.274.48.34425. [DOI] [PubMed] [Google Scholar]
- Liu JS, Kuo SR, Makhov AM, Cyr DM, Griffith JD, Broker TR, Chow LT. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J Biol Chem. 1998;273:30704–30712. doi: 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- Matthews S, Barlow P, and Boyd J. et al. 1994 Structural similarity between the p17 matrix protein of HIV-1 and interferon-gamma. Nature. 370:666–668. [DOI] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Nicolet CM, Craig EA. Inducing and assaying heat-shock response in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:710–717. doi: 10.1016/0076-6879(91)94052-e. [DOI] [PubMed] [Google Scholar]
- Paillart JC, Gottlinger HG. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of gag membrane targeting. J Virol. 1999;73:2604–2612. doi: 10.1128/jvi.73.4.2604-2612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitzsch RM, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- Ratajczak T, Carrello A. Cyclophilin 40 (CyP-40), mapping of its hsp90 binding domain and evidence that FKBP52 competes with CyP-40 for hsp90 binding. J Biol Chem. 1996;271:2961–2965. doi: 10.1074/jbc.271.6.2961. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Manivasakam P, Woods RA, Gietz RD. Introducing plasmid DNA into yeast by transformation. Methods Companion Methods Enzymol. 1993;5:79–85. [Google Scholar]
- Smith DF, Sullivan WP, Marion TN, Zaitsu K, Madden B, McCormick DJ, Toft DO. Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol Cell Biol. 1993;13:869–876. doi: 10.1128/mcb.13.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobaben S, Thakur P, Fernandez-Chacon R, Sudhof TC, Rettig J, Stahl B. A trimeric protein complex functions as a synaptic chaperone machine. Neuron. 2001;31:987–999. doi: 10.1016/s0896-6273(01)00427-5. [DOI] [PubMed] [Google Scholar]