Abstract
Mortalin/mthsp70/PBP74/Grp75 (called mortalin hereafter), a member of the Hsp70 family of chaperones, was shown to have different subcellular localizations in normal and immortal cells. It has been assigned to multiple subcellular sites and implicated in multiple functions ranging from stress response, intracellular trafficking, antigen processing, control of cell proliferation, differentiation, and tumorigenesis. The present article compiles and reviews information on the multiple sites and functions of mortalin in different organisms. The relevance of its differential distributions and functions in normal and immortal cell phenotypes is discussed.
IDENTIFICATION, CLONING, AND MULTIPLE SUBCELLULAR SITES
Mortalin/mthsp70/PBP74/Grp75 (mortalin) was first identified as a member of the Hsp70 family of proteins present in the cytoplasmic fractions of normal fibroblasts from CD1-ICR mouse (Wadhwa et al 1993a). Immortal fibroblasts from CD1-ICR, NIH Swiss, and Balb/c mice lacked this protein in their cytoplasmic fractions. An antibody raised against the full protein isolated from normal fibroblasts was highly specific to mortalin (did not cross-react with any other heat shock proteins, Wadhwa et al 1993a). Using this antibody for immunocloning, only 1 kind of mortalin complementary deoxyribonucleic acid (cDNA; named mot-1) was isolated. Immunocytochemical analysis with this antibody revealed a cytoplasmic staining of the protein in normal cells; immortal cells showed the immunofluorescence in the perinuclear region (Wadhwa et al 1993b). Immunocloning of cDNA from immortal cells led to the cloning of mortalin cDNA (named mot-2). Its sequence comparison with the cDNA isolated from normal mouse cells (mot-1) revealed a difference of 2 amino acids in the carboxy-terminus (Wadhwa et al 1993c). Genetic identities of 2 kinds of mortalin cDNAs (mot-1 and mot-2) in mouse were obtained from mouse family studies. mot-1 and mot-2 showed segregation in 2 mouse generations (Kaul et al 2000a), which illustrates that mot-1 and mot-2 are allelic in mouse, and were assigned to chromosome 18 (Kaul et al 1995; Ohashi et al 1995). Human normal and transformed cells also seem to have differential staining of mortalin. Whereas normal cells have pancytoplasmic staining, transformed cells showed 4 types of nonpancytoplasmic staining patterns that distinguished complementation groups of human transformed cells (Pereira-Smith and Smith 1988; Wadhwa et al 1995). Subsequent studies with a variety of techniques including confocal laser microscopy of the native protein with protein-specific antibodies, localization of the exogenously expressed protein by protein- and tag-specific antibodies, density gradient cell fractionation, and the use of organelle-specific markers assigned mortalin to different subcellular sites (Wadhwa et al 1995; Ran et al 2000). These included mitochondria, endoplasmic reticulum, cytoplasmic vesicles, and cytosol (Domanico et al 1993; Dahlseid et al 1994; Webster et al 1994; Singh et al 1997; Soltys and Gupta 1999; Ran et al 2000). Most recently, 3D reconstruction and deconvolution microscopical analyses confirmed the multiple subcellular sites of mortalin in different human transformed cell lines (Poindexter et al 2002). Mitochondria appeared to be the primary niche that was dependent on the presence of the leader sequence in the N-terminus of the protein, and hence the protein was also called mthsp70 (Dahlseid et al 1994; Webster et al 1994; Bhattacharyya et al 1995; Ran et al 2000). Requirement of the leader sequence for translocation of mortalin in other organelles remains unclear so far. In contrast to the mouse situation, where the 2 mortalin cDNAs, mot-1 and mot-2, were shown to code for differentially distributed proteins (Wadhwa et al 1993c), cloning of human mortalin cDNA from various human transformed cells showed identical sequences; these varied from mouse mot-1 and mot-2 (Fig 1). These data led to the speculation that there are, at least, 2 mechanisms operating for differential distributions of the mortalin protein. One is by distinct mortalin cDNAs, mot-1 and mot-2 found in mouse, and the other by as yet undefined protein modifications or cellular factors found in mouse and human cells.
Fig 1.
Protein sequence comparisons of human and mouse mortalins
MULTIFUNCTIONAL ASPECTS OF MORTALIN
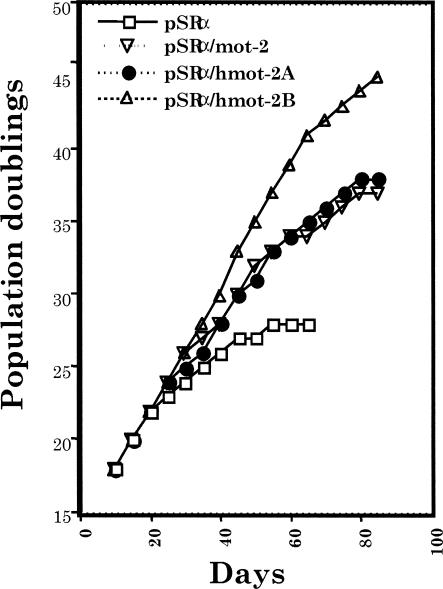
Mortalin is expressed in all cell types and tissues examined so far (Wadhwa et al 1995; Kaul et al 1997) and is expected to perform some essential functions. Expression levels of mortalin correlated with muscle activity, mitochondrial activity, and biogenesis (Ornatsky et al 1995; Ibi et al 1996; Takahashi et al 1998). It was induced by low levels of ionizing radiation (Sadekova et al 1997; Carette et al 2002), glucose deprivation (Merrick et al 1997), calcium ionophore (Resendez et al 1985), ozone (Wu et al 1999), and hyperthyroidism (Craig et al 1998; Schneider and Hood 2000). Many of the human transformed and tumor-derived cells had a high level of mortalin expression (Takahashi et al 1994; Bini et al 1997; Takano et al 1997; Kaul et al 1998; and Kaul and Wadhwa, unpublished observations). In contrast to mot-1, which induced senescence in NIH 3T3 cells (Wadhwa et al 1993c), an overexpression of mot-2 cDNA resulted in malignant transformation of the cells (Kaul et al 1998). Similar to the mouse mot-2 cDNA, human mortalin cDNA induced malignant transformation of NIH 3T3 cells and was thus called hmot-2 (Kaul et al 1998). Human lung fibroblasts (MRC-5) when stably transfected with hmot-2 cDNA underwent extended population doublings in vitro (Fig 2) (Kaul et al 2000b). It was also shown that differentiation of HL-60 promyelocytic leukemia cells was accompanied by a decreased level of hmot-2/mthsp70 expression (Xu et al 1999), whereas overexpression of hmot-2/mthsp70 imparts growth advantage and attenuates their differentiation (Xu et al 1999).
Fig 2.
Lifespan extension of normal human lung fibroblasts by over expression of mouse (mot-2) and human (hmot-2A and hmot2B) mortalins in a mammalian expression vector, pSRα. hmot-2A and hmot-2B are complementary deoxyribonucleic acid isolates from HT1080 (human fibrosarcoma assigned to complementation group A of immortalization) and HeLa (human cervical carcinoma assigned to complementation group B of immortalization) cells, respectively. The level of expression of mortalin was slightly higher in hmot-2B–expressing cells than in the mot-2– or hmot-2A–expressing cells (experimental and technical details described in Kaul et al 2000b)
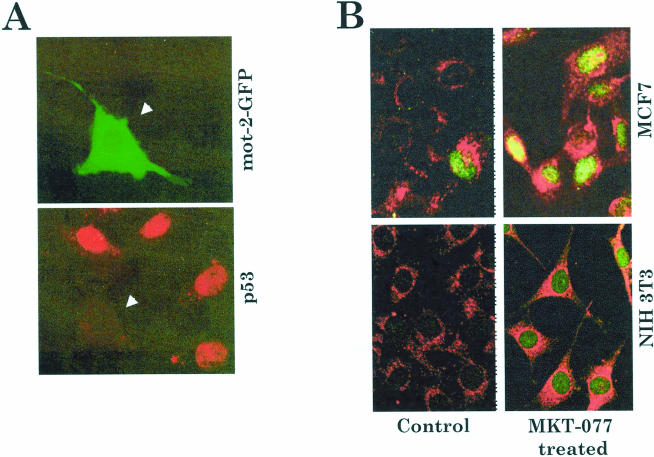
Malignant transformation of NIH 3T3, lifespan extension of MRC-5, and attenuation of differentiation of HL-60 cells by overexpression of mot-2 can be explained, at least in part, by its recently demonstrated p53 inactivation function. mot-2 and p53 were shown to interact in the cytoplasm, resulting in cytoplasmic retention and transcriptional inactivation of the latter (Merrick et al 1996; Wadhwa et al 1998, 1999). Nuclear exclusion of p53 as a possible mechanism of its inactivation was proposed for some tumors (Moll et al 1992, 1996; Takahashi and Suzuki 1994; Takahashi et al 1994; Moll and Schramm 1998). Such nuclear exclusion of p53 was demonstrated in serum-starved NIH 3T3 cells microinjected with green fluorescence protein (GFP)-tagged mot-2 (Fig 3A) (Wadhwa et al 1998). mot-2 (amino-terminus region) was found to bind to the carboxy-terminus region of p53 (Kaul et al 2001; Wadhwa et al 2002); therefore the p53 sequestration activity of mot-2 was consistent with the studies that assigned a carboxy-terminus region of p53 as a cytoplasmic sequestration domain (Moll et al 1996). Significantly, abrogation of mot-p53 interactions by MKT-077 (a water-soluble delocalized lipophilic cationic dye), which binds to mot-2, restored nuclear translocation and activation of p53 function, followed by growth arrest of tumor cells (Fig 3B) (Wadhwa et al 2000). Other potential roles assigned to mortalin include antigen processing, in vivo nephrotoxicity, radioresistance, and cell fate determination (Domanico et al 1993; Dahlseid et al 1994; Merrick et al 1997; Sadekova et al 1997; Carette et al 2002; Rivolta and Holley 2002). Mortalin-p53 complexes were also detected in the mitochondria during p53-induced apoptosis (Marchenko et al 2000), implicating its role in transcriptionally independent apoptotic signaling. Its precise role and mechanism in apoptotic pathways and phenotypes warrant further investigations.
Fig 3.
(A) Abrogation of nuclear translocation of p53 by mot-2. NIH 3T3 cells were microinjected with green fluorescence protein (GFP)-tagged mot-2 (green fluorescence) and serum-starved for 24 hours to induce nuclear translocation of p53 protein. Note that the cells microinjected with mot-2 (green fluorescence) lacked nuclear p53 (red fluorescence). Similar results were obtained by using N- or C-terminal GFP fusion (experimental and technical details described in Wadhwa et al 1998). (B) Nuclear translocation of p53 (green) in MKT-077–treated human breast carcinoma (MCF7) cells and mouse immortal fibroblasts (NIH 3T3) (Wadhwa et al 2000). Control and MKT-077–treated MCF7 and NIH 3T3 cells were double stained for mortalin (red) and p53 (green). Note that only a negligible number of cells showed nuclear p53 in control cells; MKT-077–treated culture exhibited nuclear p53 in more than 80% of cells
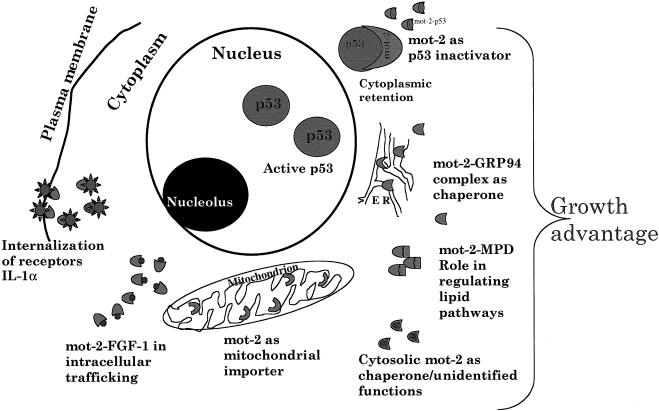
Mortalin was assigned to multiple subcellular sites (Ran et al 2000; Poindexter et al 2002). In support of this, it was shown to bind to residents of different organelles by a variety of protocols. These included yeast and mammalian 2-hybrid interactive screens, affinity binding, and in vitro and in vivo binding assays. It was shown to bind to FGF-1 and aid in its intracellular trafficking (Mizukoshi et al 1999), and this was mediated by its cell cycle–specific phosphorylation (Mizukoshi et al 2001). Adenosine triphosphate (ATP)–sensitive association of mortalin with the interleukin-1 receptor type was also detected and predicted to have a role in receptor internalization (Sacht et al 1999). It was shown to bind to GRP94 (a glucose regulated endoplasmic reticulum chaperone) by 2-hybrid assays and in vitro and in vivo coimmunoprecipiations (Takano et al 2001). Other binding partners of mortalin isolated by yeast interactive screen include mitochondrial reduced form of nicotinamide adenine dinucleotide dehydrogenase (an inner mitochondrial membrane protein), mevalonate pyruvate decarboxylase (a peroxisomal protein), and Tim23 (mitochondrial inner membrane translocase) (Wadhwa, Yaguchi and Kaul, unpublished observations). Their interactions with mortalin and their functional significance remain to be known. The multiple sites and proposed associated functions of mortalin are shown in Figure 4. It remains to be clarified how, when, and where exactly mortalin interacts with these different proteins that have been assigned to different subcellular sites. Multiple subcellular routing and docking of binding partners of mortalin cannot be ruled out. It will be interesting to resolve the functional aspects of such interactions and their physiological relevance to biological phenotypes including cellular senescence, immortalization, stress, and apoptosis. Nevertheless, it can be speculated that mortalin routes through multiple subcellular sites and may interact with different proteins therein. Such interactions in different subcellular organelles may be important for its multiple functions.
Fig 4.
A model showing multiple subcellular sites, binding partners, and functions of mortalin
An overexpression of a mortalin homologue in Caenorhabditis elegans (Hsp70F) resulted in lifespan extension (Yokoyama et al 2002). The yeast homologue of mortalin, SSC1p, is essential for cell viability (Craig et al 1989) and has indispensable functions in mitochondrial import (Voos et al 1999; Krimmer et al 2000; Geissler et al 2001). It binds to Tim-44, an inner mitochondrial membrane anchor, and is an essential component of mitochondrial import machinery (Voisine et al 1999; Krimmer et al 2000; Strub et al 2001). Mutations in Tim-44 that result in inefficient recruitment of mthsp70/SSC1 are lethal in Saccharomyces cerevisiae (Merlin et al 1999; Schulke et al 1999). On the basis of the studies in yeast at least 3 kinds of activities can be hypothesized for mortalin. These include (1) unfolding of proteins outside mitochondria, (2) unidirectional translocation across mitochondrial membranes initiated by membrane potential MΔΨ, and (3) completion of import by acting as an ATP-driven motor. It was shown to bind to endonuclease, Endo. SceI, and it confers on it broader sequence specificity, greater stability, and higher activity (Mizumura et al 1999). It is also required for degradation of misfolded peptides by m-AAA and PIM1 proteases in mitochondria (Lim et al 2001; Liu et al 2001). Evidence suggests that it cooperates with mthsp60 and CPN-10 chaperones for folding of imported proteins to functionally competent forms in mitochondria and for yet undefined roles of mthsp60 at extra-mitochondrial sites (Cechetto et al 2000; Strub et al 2001). Taken together, these studies have shown that overexpression of mot-2 confers proliferation-growth advantage to cells in vitro and in vivo.
DIFFERENTIAL SUBCELLULAR DISTRIBUTION IN NORMAL AND IMMORTAL CELLS
More than 60 different cell lines have been analyzed for mortalin staining patterns; none of these have shown pancytoplasmic staining. In sharp contrast, normal cells have invariably shown widely distributed mortalin staining in the cytoplasm (Wadhwa et al 1995; Yaguchi, Kaul and Wadhwa, unpublished observations). Induction of senescence in transformed cells by introduction of a single chromosome (Nakabayashi et al 1999), chromosome-fragments, and genes (Bertram et al 1999) or chemicals (Michishita et al 1999) was accompanied by reversion of subcellular distribution of mortalin from the nonpancytoplasmic type to the pancytoplasmic type. MKT-077 is a rhodacyanine dye that is selectively toxic to cancer cells. MKT-077–induced growth arrest of cancer cells was also accompanied by a change in mortalin staining pattern from the nonpancytoplasmic type (perinuclear) to the pancytoplasmic type, characteristic of normal cells (Fig 3) (Wadhwa et al 2000). This was accompanied by nuclear translocation and activation of p53 as a result of abrogation of its interactions with mortalin (Wadhwa et al 2000). Heat shock treatment translocated mortalin from the pancytoplasmic locale to the perinuclear one in normal cells, which is typical in immortal cells (Kaul et al 1993). Of the multiple subcellular sites of mortalin, mitochondria are a dominant localization (Bruschi and Lindsay 1994; Webster et al 1994; Ran et al 2000). Interestingly, the distribution of mitochondria was reported to change in response to heat shock treatment (Collier et al 1993) in chicken embryo fibroblasts and in response to virus infection in mammalian cells (Rojo et al 1998; Murata et al 2000). At normal temperature, chicken embryo fibroblasts exhibit evenly distributed mitochondria as elongated, tubular, and dynamic organelles in the cell cytoplasm, but upon heat shock they move to the perinuclear region and form a tight ring of short swollen and in some cases fused vesicles (Collier et al 1993). Rojo et al (1998) and Murata et al (2000) reported that African swine fever virus– and herpes simplex virus–infected cells show clustering of mitochondria in the perinuclear sites. This migration of mitochondria from the cytoplasm to the perinuclear region requires microtubules because it is blocked in the presence of the microtubule-disassembling drug nocodazole. Recently, it was shown that functional inactivation of mthsp70 causes mitochondrial aggregation in yeast (Kawai et al 2001). This was shown to be independent of the defects in mitochondrial protein import or mitochondrial translocases (Kawai et al 2001). It was proposed that mthsp70 is essential for optimizing the functions of, yet unidentified, heat-labile protein(s) in the mitochondrial matrix in controlling mitochondrial morphology (Kawai et al 2001). In light of these studies and the prominent mitochondrial residence of mortalin protein (Bruschi and Lindsay 1994; Webster et al 1994; Ran et al 2000), it is possible that the differential subcellular distribution of hmot-2/mthsp70 in different human transformed cells represents altered mitochondrial morphology, at least in part. This would imply that changes in mitochondrial morphology and distribution are consistent alterations that occur with change of the cellular divisional phenotype from mortal to immortal. Because mitochondria play essential roles in supply of energy, regulation of calcium levels, and control of apoptotic cell death, it is plausible that alterations in their morphology, localization, and function would occur with cellular immortality. The mechanism of such differential localizations, role of cytoskeleton elements, and downstream signal transduction and its role in regulation of proliferation warrant further studies.
PROSPECTIVES
Mortalin (mot-2/mthsp70/PBP74/GRP75) is an essential protein belonging to the Hsp70 family of chaperones. It sojourns in multiple subcellular sites although it resides predominantly in mitochondria and performs multiple functions including mitochondrial import, intracellular trafficking, receptor internalization, and inactivation of tumor suppressor protein p53. Differential staining patterns of mortalin may predict its different functions in normal and transformed cells. Some of these functions such as inactivation of p53 can be employed as an advantage in immortalization of human cells in vitro. Targeting of other functions such as chaperoning mitochondrial biogenesis and intracellular trafficking may provide novel tools for tumor therapy.
REFERENCES
- Bertram MJ, Berube NG, Swanson XH, Pereira-Smith OM. Assembly of a BAC contig of the complementation group B cell senescence gene candidate region at 4q33-q34.1 and identification of expressed sequences. Genomics. 1999;56:353–354. doi: 10.1006/geno.1998.5726. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya T, Karnezis AN, Murphy SP, Hoang T, Freeman BC, Phillips B, Morimoto RI. Cloning and subcellular localization of human mitochondrial hsp70. J Biol Chem. 1995;270:1705–1710. doi: 10.1074/jbc.270.4.1705. [DOI] [PubMed] [Google Scholar]
- Bini L, Magi B, and Marzocchi B. et al. 1997 Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis. 18:2832–2841. [DOI] [PubMed] [Google Scholar]
- Bruschi SA, Lindsay JG. Mitochondrial stress protein actions during chemically induced renal proximal tubule cell death. Biochem Cell Biol. 1994;72:663–667. doi: 10.1139/o94-087. [DOI] [PubMed] [Google Scholar]
- Carette J, Lehnert S, Chow TYK. Implication of PBP74/mortalin/GRP75 in the radio-adaptive response. Int J Radiat Biol. 2002;78:183–190. doi: 10.1080/09553000110097208. [DOI] [PubMed] [Google Scholar]
- Cechetto JD, Soltys BJ, Gupta RS. Localization of mitochondrial 60-kD heat shock chaperonin protein (Hsp60) in pituitary growth hormone secretory granules and pancreatic zymogen granules. J Histochem Cytochem. 2000;48:45–56. doi: 10.1177/002215540004800105. [DOI] [PubMed] [Google Scholar]
- Collier NC, Sheetz MP, Schlesinger MJ. Concomitant changes in mitochondria and intermediate filaments during heat shock and recovery of chicken embryo fibroblasts. J Cell Biochem. 1993;52:297–307. doi: 10.1002/jcb.240520306. [DOI] [PubMed] [Google Scholar]
- Craig EE, Chesley A, and Hood DA 1998 Thyroid hormone modifies mitochondrial phenotype by increasing protein import without altering degradation. Am J Physiol. 275 C. 1508–C1515. [DOI] [PubMed] [Google Scholar]
- Craig EA, Kramer J, Shilling J, Werner-Washburne M, Holmes S, Kosic-Smithers J, Nicolet CM. SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol Cell Biol. 1989;9:3000–3008. doi: 10.1128/mcb.9.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlseid JN, Lill R, Green JM, Xu X, Qiu Y, Pierce SK. PBP74, a new member of the mammalian 70-kDa heat shock protein family, is a mitochondrial protein. Mol Biol Cell. 1994;5:1265–1275. doi: 10.1091/mbc.5.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanico SZ, DeNagel DC, Dahlseid JN, Green JM, Pierce SK. Cloning of the gene encoding peptide-binding protein 74 shows that it is a new member of the heat shock protein 70 family. Mol Cell Biol. 1993;13:3598–3610. doi: 10.1128/mcb.13.6.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler A, Rassow J, Pfanner N, Voos W. Mitochondrial import driving forces: enhanced trapping by matrix hsp70 stimulates translocation and reduces the membrane potential dependence of loosely folded preproteins. Mol Cell Biol. 2001;21:7097–7104. doi: 10.1128/MCB.21.20.7097-7104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibi T, Sahashi K, Ling J, Marui K, Mitsuma T. [Immunostaining of mitochondrial heat shock proteins (mtHSPs) in skeletal muscle fibers of mitochondrial cytopathy]. Rinsho Shinkeigaku. Clin Neurol. 1996;36:61–64. [PubMed] [Google Scholar]
- Kaul SC, Duncan EL, Englezou A, Takano S, Reddel RR, Mitsui Y, Wadhwa R. Malignant transformation of NIH3T3 cells by overexpression of mot-2 protein. Oncogene. 1998;17:907–911. doi: 10.1038/sj.onc.1202017. [DOI] [PubMed] [Google Scholar]
- Kaul SC, Duncan EL, Sugihara T, Reddel RR, Mitsui Y, Wadhwa R. Structurally and functionally distinct mouse hsp70 family members mot-1 and mot-2 proteins are encoded by two alleles. DNA Res. 2000a;7:229–231. doi: 10.1093/dnares/7.3.229. [DOI] [PubMed] [Google Scholar]
- Kaul SC, Matsui M, Takano S, Sugihara T, Mitsui Y, Wadhwa R. Expression analysis of mortalin, a unique member of the Hsp70 family of proteins, in rat tissues. Exp Cell Res. 1997;232:56–63. doi: 10.1006/excr.1997.3503. [DOI] [PubMed] [Google Scholar]
- Kaul SC, Reddel RR, Mitsui Y, Wadhwa R. An N-terminal region of mot-2 binds to p53 in vitro. Neoplasia. 2001;3:110–114. doi: 10.1038/sj.neo.7900139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul S, Reddel RR, Sugihara T, Mitsui Y, Wadhwa R. Inactivation of p53 and life span extension of human diploid fibroblasts by mot-2. FEBS Lett. 2000b;474:159–164. doi: 10.1016/s0014-5793(00)01594-5. [DOI] [PubMed] [Google Scholar]
- Kaul SC, Wadhwa R, Komatsu Y, Sugimoto Y, Mitsui Y. On the cytosolic and perinuclear mortalin: an insight by heat shock. Biochem Biophys Res Commun. 1993;193:348–355. doi: 10.1006/bbrc.1993.1630. [DOI] [PubMed] [Google Scholar]
- Kaul SC, Wadhwa R, Matsuda Y, Hensler PJ, Pereira-Smith OM, Komatsu Y, Mitsui Y. Mouse and human chromosomal assignments of mortalin, a novel member of the murine hsp70 family of proteins. FEBS Lett. 1995;361:269–272. doi: 10.1016/0014-5793(95)00177-b. [DOI] [PubMed] [Google Scholar]
- Kawai A, Nishikawa Si S, Hirata A, Endo T. Loss of the mitochondrial Hsp70 functions causes aggregation of mitochondria in yeast cells. J Cell Sci. 2001;114:3565–3574. doi: 10.1242/jcs.114.19.3565. [DOI] [PubMed] [Google Scholar]
- Krimmer T, Rassow J, Kunau WH, Voos W, Pfanner N. Mitochondrial protein import motor: the ATPase domain of matrix hsp70 is crucial for binding to Tim44, while the peptide binding domain and the carboxy-terminal segment play a stimulatory role. Mol Cell Biol. 2000;20:5879–5887. doi: 10.1128/mcb.20.16.5879-5887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Martin F, Guiard B, Pfanner N, Voos W. The mitochondrial Hsp70-dependent import system actively unfolds preproteins and shortens the lag phase of translocation. EMBO J. 2001;20:941–950. doi: 10.1093/emboj/20.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Krzewska J, Liberek K, Craig EA. Mitochondrial hsp70 Ssc1: role in protein folding. J Biol Chem. 2001;276:6112–6118. doi: 10.1074/jbc.M009519200. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- Merlin A, Voos W, Maarse AC, Meijer M, Pfanner N, Rassow J. The J-related segment of tim44 is essential for cell viability: a mutant Tim44 remains in the mitochondrial import site, but inefficiently recruits mtHsp70 and impairs protein translocation. J Cell Biol. 1999;145:961–972. doi: 10.1083/jcb.145.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick BA, He C, Witcher LL, Patterson RM, Reid JJ, Pence-Pawlowski PM, Selkirk JK. HSP binding and mitochondrial localization of p53 protein in human HT1080 and mouse C3H10T1/2 cell lines. Biochim Biophys Acta. 1996;13:57–68. doi: 10.1016/0167-4838(96)00089-1. [DOI] [PubMed] [Google Scholar]
- Merrick BA, Walker VR, He C, Patterson RM, Selkirk JK. Induction of novel Grp75 isoforms by 2-deoxyglucose in human and murine fibroblasts. Cancer Lett. 1997;119:185–190. doi: 10.1016/s0304-3835(97)00270-x. [DOI] [PubMed] [Google Scholar]
- Michishita E, Nakabayashi K, Suzuki T, Kaul SC, Ogino H, Fujii M, Mitsui Y, Ayusawa D. 5-Bromodeoxyuridine induces senescence-like phenomena in mammalian cells regardless of cell type or species. J Biochem. 1999;126:1052–1059. doi: 10.1093/oxfordjournals.jbchem.a022549. [DOI] [PubMed] [Google Scholar]
- Mizukoshi E, Suzuki M, Misono T, Loupatov A, Munekata E, Kaul SC, Wadhwa R, Imamura T. Cell-cycle dependent tyrosine phosphorylation on mortalin regulates its interaction with fibroblast growth factor-1. Biochem Biophys Res Commun. 2001;280:1203–1209. doi: 10.1006/bbrc.2001.4225. [DOI] [PubMed] [Google Scholar]
- Mizukoshi E, Suzuki M, and Loupatov A. et al. 1999 Fibroblast growth factor-1 interacts with the glucose-regulated protein GRP75/mortalin. Biochem J. 343:461–466. [PMC free article] [PubMed] [Google Scholar]
- Mizumura H, Shibata T, Morishima N. Stable association of 70-kDa heat shock protein induces latent multisite specificity of a unisite-specific endonuclease in yeast mitochondria. J Biol Chem. 1999;274:25682–25690. doi: 10.1074/jbc.274.36.25682. [DOI] [PubMed] [Google Scholar]
- Moll UM, Ostermeyer AG, Haladay R, Winkfield B, Frazier M, Zambetti G. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol Cell Biol. 1996;16:1126–1137. doi: 10.1128/mcb.16.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Riou G, Levine AJ. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc Natl Acad Sci U S A. 1992;89:7262–7266. doi: 10.1073/pnas.89.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Schramm LM. p53–an acrobat in tumorigenesis. Crit Rev Oral Biol Med. 1998;9:23–37. doi: 10.1177/10454411980090010101. [DOI] [PubMed] [Google Scholar]
- Murata T, Goshima F, Daikoku T, Inagaki-Ohara K, Takakuwa H, Kato K, Nishiyama Y. Mitochondrial distribution and function in herpes simplex virus-infected cells. J Gen Virol. 2000;2:401–406. doi: 10.1099/0022-1317-81-2-401. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Ogino H, Michishita E, Satoh N, Ayusawa D. Introduction of chromosome 7 suppresses telomerase with shortening of telomeres in a human mesothelial cell line. Exp Cell Res. 1999;252:376–382. doi: 10.1006/excr.1999.4619. [DOI] [PubMed] [Google Scholar]
- Ohashi M, Oyanagi M, Hatakeyama K, Inoue M, Kominami R. The gene encoding PBP74/CSA/motalin-1, a novel mouse hsp70, maps to mouse chromosome 18. Genomics. 1995;30:406–407. [PubMed] [Google Scholar]
- Ornatsky OI, Connor MK, Hood DA. Expression of stress proteins and mitochondrial chaperonins in chronically stimulated skeletal muscle. Biochem J. 1995;311:119–123. doi: 10.1042/bj3110119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Smith OM, Smith JR. Genetic analysis of indefinite division in human cells: identification of four complementation groups. Proc Natl Acad Sci U S A. 1988;85:6042–6046. doi: 10.1073/pnas.85.16.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter BJ, Pereira-Smith OM, Wadhwa R, Buja LM, Bick RJ. 3D reconstruction and localization of mortalin by deconvolution microscopy. Microsc Anal. 2002;89:21–23. [Google Scholar]
- Ran Q, Wadhwa R, Kawaki R, Kaul SC, Sifers RN, Bick RJ, Smith JR, Pereira-Smith OM. Extra-mitochondrial localization of mortalin/mthsp70/PBP 74/GRP75. Biochem Biophys Res Commun. 2000;275:174–179. doi: 10.1006/bbrc.2000.3237. [DOI] [PubMed] [Google Scholar]
- Resendez E Jr,, Attenello JW, Grafsky A, Chang CS, Lee AS. Calcium ionophore A23187 induces expression of glucose-regulated genes and their heterologous fusion genes. Mol Cell Biol. 1985;5:1212–1219. doi: 10.1128/mcb.5.6.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta MN, Holley MC. Asymmetric segregation of mitochondria and mortalin correlates with the multi-lineage potential of inner ear sensory cell progenitors in vitro. Dev Brain Res. 2002;133:49–56. doi: 10.1016/s0165-3806(01)00321-2. [DOI] [PubMed] [Google Scholar]
- Rojo G, Chamorro M, Salas ML, Vinuela E, Cuezva JM, Salas J. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J Virol. 1998;72:7583–7588. doi: 10.1128/jvi.72.9.7583-7588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacht G, Brigelius-Flohe R, Kiess M, Sztajer H, Flohe L. ATP-sensitive association of mortalin with the IL-1 receptor type I. Biofactors. 1999;9:49–60. doi: 10.1002/biof.5520090107. [DOI] [PubMed] [Google Scholar]
- Sadekova S, Lehnert S, Chow TY. Induction of PBP74/mortalin/Grp75, a member of the hsp70 family, by low doses of ionizing radiation: a possible role in induced radioresistance. Int J Radiat Biol. 1997;72:653–660. doi: 10.1080/095530097142807. [DOI] [PubMed] [Google Scholar]
- Schneider JJ, Hood DA. Effect of thyroid hormone on mtHsp70 expression, mitochondrial import and processing in cardiac muscle. J Endocrinol. 2000;165:9–17. doi: 10.1677/joe.0.1650009. [DOI] [PubMed] [Google Scholar]
- Schulke N, Sepuri NB, Gordon DM, Saxena S, Dancis A, Pain D. A multisubunit complex of outer and inner mitochondrial membrane protein translocases stabilized in vivo by translocation intermediates. J Biol Chem. 1999;274:22847–22854. doi: 10.1074/jbc.274.32.22847. [DOI] [PubMed] [Google Scholar]
- Singh B, Soltys BJ, Wu ZC, Patel HV, Freeman KB, Gupta RS. Cloning and some novel characteristics of mitochondrial Hsp70 from Chinese hamster cells. Exp Cell Res. 1997;234:205–216. doi: 10.1006/excr.1997.3609. [DOI] [PubMed] [Google Scholar]
- Soltys BJ, Gupta RS. Mitochondrial-matrix proteins at unexpected locations: are they exported? Trends Biochem Sci. 1999;24:174–177. doi: 10.1016/s0968-0004(99)01390-0. [DOI] [PubMed] [Google Scholar]
- Strub A, Lim JH, Pfanner N, Voos W. The mitochondrial protein import motor. Biol Chem. 2001;381:943–949. doi: 10.1515/BC.2000.115. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Chesley A, Freyssenet D, and Hood DA 1998 Contractile activity-induced adaptations in the mitochondrial protein import system. Am J Physiol. 274 C. 1380–C1387. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Mikami T, and Watanabe Y. et al. 1994 Correlation of heat shock protein 70 expression with estrogen receptor levels in invasive human breast cancer. Am J Clin Pathol. 101:519–525. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Suzuki K. DNA synthesis-associated nuclear exclusion of p53 in normal human breast epithelial cells in culture. Oncogene. 1994;9:183–188. [PubMed] [Google Scholar]
- Takano S, Wadhwa R, Mitsui Y, Kaul SC. Identification and characterization of molecular interactions between glucose-regulated proteins (GRPs) mortalin/GRP75/peptide-binding protein 74 (PBP74) and GRP94. Biochem J. 2001;357:393–398. doi: 10.1042/0264-6021:3570393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano S, Wadhwa R, Yoshii Y, Nose T, Kaul SC, Mitsui Y. Elevated levels of mortalin expression in human brain tumors. Exp Cell Res. 1997;237:38–45. doi: 10.1006/excr.1997.3754. [DOI] [PubMed] [Google Scholar]
- Voisine C, Craig EA, Zufall N, von Ahsen O, Pfanner N, Voos W. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix hsp70. Cell. 1999;97:565–574. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- Voos W, Martin H, Krimmer T, Pfanner N. Mechanisms of protein translocation into mitochondria. Biochim Biophys Acta. 1999;16:235–254. doi: 10.1016/s0304-4157(99)00007-6. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Kaul SC, Ikawa Y, Sugimoto Y. Identification of a novel member of mouse hsp70 family. Its association with cellular mortal phenotype. J Biol Chem. 1993a;268:6615–6621. [PubMed] [Google Scholar]
- Wadhwa R, Kaul SC, Mitsui Y, Sugimoto Y. Differential subcellular distribution of mortalin in mortal and immortal mouse and human fibroblasts. Exp Cell Res. 1993b;207:442–448. doi: 10.1006/excr.1993.1213. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Kaul SC, Sugimoto Y, Mitsui Y. Induction of cellular senescence by transfection of cytosolic mortalin cDNA in NIH 3T3 cells. J Biol Chem. 1993c;268:22239–22242. [PubMed] [Google Scholar]
- Wadhwa R, Pereira-Smith OM, Reddel RR, Sugimoto Y, Mitsui Y, Kaul SC. Correlation between complementation group for immortality and the cellular distribution of mortalin. Exp Cell Res. 1995;216:101–106. doi: 10.1006/excr.1995.1013. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Takano S, Robert M, Yoshida A, Reddel RR, Nomura H, Mitsui Y, Kaul SC. Inactivation of tumor suppressor p53 by mot-2, an hsp70 family member. J Biol Chem. 1998;273:29586–29591. doi: 10.1074/jbc.273.45.29586. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Sugihara T, Yoshida A, Nomura H, Reddel RR, Simpson R, Maruta H, Kaul SC. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 2000;60:6818–6821. [PubMed] [Google Scholar]
- Wadhwa R, Takano S, Mitsui Y, Kaul SC. NIH 3T3 cells malignantly transformed by mot-2 show inactivation and cytoplasmic sequestration of the p53 protein. Cell Res. 1999;9:261–269. doi: 10.1038/sj.cr.7290025. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Yaguchi T, Hasan MK, Mitsui Y, Reddel RR, Kaul SC. Hsp70 family member, mot-2/mthsp70/GRP75, binds to the cytoplasmic sequestration domain of the p53 protein. Exp Cell Res. 2002;274:246–253. doi: 10.1006/excr.2002.5468. [DOI] [PubMed] [Google Scholar]
- Webster TJ, Naylor DJ, Hartman DJ, Hoj PB, Hoogenraad NJ. cDNA cloning and efficient mitochondrial import of pre-mtHSP70 from rat liver. DNA Cell Biol. 1994;13:1213–1220. doi: 10.1089/dna.1994.13.1213. [DOI] [PubMed] [Google Scholar]
- Wu R, Zhao YH, and Plopper CG. et al. 1999 Differential expression of stress proteins in nonhuman primate lung and conducting airway after ozone exposure. Am J Physiol. 277 L. 511–L522. [DOI] [PubMed] [Google Scholar]
- Xu J, Xiao HH, Sartorelli AC. Attenuation of the induced differentiation of HL-60 leukemia cells by mitochondrial chaperone HSP70. Oncol Res. 1999;11:429–435. [PubMed] [Google Scholar]
- Yokoyama K, Fukumoto K, Murakami T, Harada S, Hosono R, Wadhwa R, Mitsui Y, Ohkuma S. Extended longevity of C. elegance by knocking in extra copies of hsp70F, a homologue of mot-2 (mortalin)/mthsp70/Grp75. FEBS Lett. 2002;516:53–57. doi: 10.1016/s0014-5793(02)02470-5. [DOI] [PubMed] [Google Scholar]