Abstract
The α-crystallin–related, small heat shock proteins (sHsps), despite their overall variability in sequence, have discrete regions of conserved sequence that are involved in structural organization, as well as nonconserved regions that may perform similar roles in each protein. Recent X-ray diffraction analyses of an archeal and a plant sHsp have revealed both similarities and differences in how they are organized, suggesting that there is variability, particularly in the oligomeric organization of sHsps. As an adjunct to crystallographic analysis of sHsp structure, we employed the yeast 2-hybrid system to detect interactions between peptide regions of the sHsp of Neurospora crassa, Hsp30. We found that the conserved α-crystallin domain can be divided into N-terminal and C-terminal subdomains that interact strongly with one another. This interaction likely represents the tertiary contacts of the monomer that were visualized in the crystallographic structures of MjHsp16.5 and wheat Hsp16.9. The conserved sHsp monomeric fold is apparently determined by these regions of conserved sequence. We found that the C-terminal portion of the α-crystallin domain also interacts with itself in 2-hybrid assays; however, this interaction requires peptide extension into the semiconserved carboxyl tail. This C-terminal association may represent a principal contact site between dimers that contributes to higher-order assembly, as seen for the crystallized sHsps.
INTRODUCTION
Heat shock proteins (Hsps), belonging to several protein families, are abundantly synthesized by all organisms as a response to heat stress (Parsell and Lindquist 1993). It is now known that most of these Hsps function as chaperones and help to protect cellular proteins from irreversible denaturation (Hendrick and Hartl 1993). The Hsp70 protein family is especially abundant and well represented in organelles and cytoplasm (Hartl 1996) and was the first Hsp identified as a chaperone (Lewis and Pelham 1985). Hsp60, as a 14-subunit double toroid, folds proteins within eukaryotic mitochondria and chloroplasts, as well as in bacteria (Hartl 1996); dimeric Hsp90, assisted by cochaperones, helps to activate signaling proteins (Chadli et al 2000; Donze et al 2001). The small Hsps (sHsps), related to α-crystallin of the vertebrate eye lens, are known to be chaperones, but their specific functions and structural organization have proved more difficult to analyze. The sHps share a diagnostic region of sequence homology to α-crystallin, outside of which they vary extensively in amino acid sequence (Plesofsky-Vig et al 1992; de Jong et al 1998), and although most of them assemble into large multimeric structures, the number of subunits varies depending on the protein species.
sHsps have been found highly expressed in diseased human tissue, such as that associated with Alzheimer's and Parkinson's diseases (Renkawek et al 1999), where their function may be initially protective. However, they also become immune system targets in autoimmune disorders such as multiple sclerosis (Aquino et al 1997), and their association with estrogen-responsive tumors protects these tumors against chemotherapeutic agents (Oesterreich et al 1996). The antiapoptotic role of human sHsps has been described and analyzed in cells experiencing chemical and oxidative stress (Mehlen et al 1996), as well as during stem cell differentiation (Mehlen et al 1997). Several sHsps have been shown to function in vitro as adenosine triphosphate (ATP)-independent chaperones that protect model substrate proteins from irreversible heat denaturation (Lee et al 1997). By binding to nonnative proteins, sHsps maintain them as folding-competent substrates for subsequent renaturation by the ATP-dependent Hsp70 (Ehrnsperger et al 1997). Bacterial survival has been shown to be enhanced by overexpression of sHsps (Yeh et al 1997; Kitagawa et al 2000), with reduced protein aggregation, after exposure to heat shock and other stresses. We found that N crassa cells disrupted in hsp30 survived poorly under stress conditions (Plesofsky-Vig and Brambl 1995), and these cells were impaired in glucose metabolism and hexokinase activity (Plesofsky and Brambl 1999) and in protein import into mitochondria (Plesofsky et al 1999).
The assembled, multimeric form of sHsps has been shown to be the active form of the chaperone (Leroux et al 1997; Rogalla et al 1999), suggesting that the constituent subunits cooperate in chaperone activity, with substrate proteins either coating the surfaces of the oligomer or being sequestered within its interior. The Hsp26 oligomer of Saccharomyces cerevisiae was found to reorganize into a larger multimer that incorporated substrate proteins under heat stress conditions (Haslbeck et al 1999). Different regions of the sHsps have been reported to play distinct roles in formation of higher-order structures, with the nonconserved N-terminal region being required for oligomer formation (Leroux et al 1997) and the semiconserved C-terminal tail helping to solubilize the large oligomer (Smulders et al 1996). Formation of the dimer, the core structural unit of the assembled multimer, does not require the nonconserved, N-terminal region (Feil et al 2001).
The recent crystallization of 2 sHsps has provided new insight into their structural organization. Hsp16.5 of Methanococcus jannaschii forms a 24-subunit hollow sphere with trimeric crystallographic symmetry (Kim et al 1998), in which the monomer is arranged into 2 stacked antiparallel β-sheets. An extensive site of dimerization involves the β6 strand of one monomer entering the β-sheet of a neighboring monomer through contact with its β2 strand. These dimerizing β-strands are within the conserved α-crystallin domain of sHsps, but their sequences are not well conserved. The crystallized Hsp16.9 of wheat assembles into a 12-subunit double disk, each disk consisting of a trimer of dimers (van Montfort et al 2001). But the same monomeric fold, a β-sandwich, exists in wheat Hsp16.9, and its dimeric conformation, involving a projecting β6 strand, is also similar to that of MjHsp16.5. The dimeric structure of αB-crystallin, in contrast, has been proposed to involve contact between symmetrical sites, consisting of a bipartite β-strand that corresponds in position, but not in sequence, to the β6 strand of MjHsp16.5 (Feil et al 2001). The nonconserved N-terminal domain of MjHsp16.5 was disordered and unresolved in the crystal structure but is apparently located within the interior of the sphere (Haley et al 2000). In contrast, only half the N-terminal arms of wheat Hsp16.9 were disordered, with the other 6 seen to help stabilize the oligomer (van Montfort et al 2001). The semiconserved carboxyl tails of both crystallized sHsps were seen to contribute to higher-order assembly, strengthening interdimer contacts and forming associations between tetramers.
These analyses of the crystal structures of 2 sHsps have shown how their monomers, dimers, and oligomers are organized, and they describe the interactions between specific domains that characterize these assemblies. These analyses also reveal divergences in the assembled oligomeric structures of sHsps, which is not unexpected considering that they vary in subunit number. Hsp30 appears to assemble into a 24-subunit particle (unpublished data), like MjHsp16.5 and Hsp26 of S cerevisiae (Haslbeck et al 1999), but it is not known what domain interactions contribute to this higher-order structural organization. In the present study, we have used an alternate approach, based on peptide interactions in yeast, to help identify these interacting domains of Hsp30.
The primary sequence of most sHsps consists of a nonconserved N-terminal domain of 40–100 amino acids, a conserved region of approximately 90 amino acids (the α-crystallin domain), and a semiconserved C-terminal tail of 12–30 amino acids. The conserved region includes 2 strongly conserved subdomains separated by a less conserved sequence (Plesofsky-Vig et al 1992). In the present study, we asked whether we could detect interactions among these distinct regions of a sHsp by dividing Neurospora Hsp30 into peptides, whose interactions with one another we tested by the yeast 2-hybrid assay. By truncating the peptides found to interact with one another in these assays, we have also delineated peptide boundaries that are sufficient for the interaction. We began this analysis by dividing Hsp30 into 2 peptides, one of which includes the N-terminal half of the α-crystallin domain, whereas the other includes the C-terminal half of this domain, thereby separating these 2 strongly conserved subdomains. As reported here, we found that these 2 separate portions of the α-crystallin domain of Hsp30 interact strongly with one another. Furthermore, we found that the C-terminal half interacts with itself and that this reaction requires the semiconserved C-terminal tail. The nonconserved N-terminal region was not found to interact with the other peptide domains of Hsp30.
MATERIALS AND METHODS
Plasmid construction
Deoxyribonucleic acid (DNA) sequence encoding Hsp30 peptides was derived from cloned complementary DNA (cDNA) (Plesofsky-Vig and Brambl 1990), either by restriction enzyme digestion or by polymerase chain reaction (PCR) amplification. The DNA was amplified by Taq polymerase in a Perkin-Elmer DNA Thermal Cycler at 58°C with 0.5 μM oligonucleotide primers from Integrated DNA Technologies (Carolville, IA, USA). The PCR-amplified DNA was purified through a Millipore Ultrafree-MC (30 000 NMWL) filter and ligated into the pGEM-T vector (Promega, Madison, WI, USA), and Escherichia coli was transformed by electroporation. The inserted DNA was excised and ligated to appropriately restricted pAS2-1 or pGADGH 2-hybrid vectors (Clontech, Palo Alto, CA, USA), followed by E coli transformation.
DNA encoding full-length Hsp30 (aa 1–228) was amplified by PCR and cloned into the BamHI and PstI sites of the Gal4-binding domain (BD) vector pAS2-1. For transfer to the Gal4-activation domain (AD) vector pGADGH, hsp30 DNA was excised from pAS2-1 at the upstream XmaI site and the internal EcoRI site (following the native stop codon) and was ligated into the appropriately restricted pGADGH.
The N-terminal half of Hsp30 (aa 1–135) was generated by restricting pGADGH-hsp30 with SalI, which cuts both within hsp30 and at a downstream vector cloning site, followed by recircularization of the plasmid. For transfer to pAS2-1, the N-terminal half of hsp30 was excised by BamHI and SalI restriction from pGADGH and ligated into appropriately restricted pAS2-1.
Two truncated derivatives were generated from N-terminal Hsp30. Peptide 1–33 was generated by cutting pAS-30N at an upstream EcoRI and an internal NruI site, and peptide 1–71 was generated by cutting pAS-30N at EcoRI and an internal Cac8I site; each fragment was ligated into the EcoRI and SmaI sites of pAS2-1. A stop-codon linker (described below) was inserted between the vector BamHI and SalI sites. Other derivatives of the N-terminal half of Hsp30 were generated by PCR amplification of Hsp30 cDNA, using the primers listed below, and cloned into pAS2-1. These include peptides 1–39, 1–49, 1–80, 1–100, 1–111, 1–117, 17–135, and 40–117.
The C-terminal half of Hsp30 was generated by cutting pAS2-hsp30 at an internal AatII site and the upstream XmaI cloning site, followed by recircularizing the plasmid with an AatII-XmaI adaptor. The hsp30 C-terminal half was excised with XmaI and EcoRI, which cuts downstream of the native stop codon, and was ligated to pGADGH (which has its own AatII site).
The C-terminal peptide 129–174 was generated by cutting pGADGH-30C at the upstream BamHI cloning site and at the internal ScaI site and ligating this fragment with pGADGH, restricted with BamHI and SmaI. The C-terminal peptide 173–228 was generated by restricting pGADGH-30C with Cac8I and EcoRI at internal sites and ligating this fragment with pGADGH, restricted with SmaI and EcoRI. The DNA for the other C-terminally derived peptides, 185–216 and 185–228, was generated by PCR amplification, using Hsp30 cDNA and the primers listed below, and ligated into pGADGH.
We used the following PCR primers, with the position of the first encoded amino acid (sense) or the last encoded amino acid (antisense) indicated by number, and the incorporated restriction sites are underlined:
sense 1, 5′CTC GGA TCC TCG CGC TCT TCC CTC GTG GCT T (BamHI);
sense 1, 5′CTC CAT ATG GCG CTC TTC CCT CGT GGC TT (NdeI);
sense 17, 5′CTC CAT ATG AGC TTC ACC AAC CTC TTT CGC CT (NdeI);
sense 40, 5′CTC CAT ATG CCA GAG ACC GGC AGC CGT CGT (NdeI);
sense 185, 5′GTC GGA TCC CGA GTT CTC GCG GAC TTT CAA (BamHI);
antisense 39, 5′CTC GAA TTC TTA GGC GCT GCC TTG AAC CTC GCG A (EcoRI);
antisense 49, 5′CTC GAA TTC TTA CTG GGT ATG ACG ACG GCT GCC GGT (EcoRI);
antisense 80, 5′CGT GGA ATT CTC AAT TGT CAC GAT CAA TGC CGG GA (EcoRI);
antisense 100, 5′CGC TGC AGG TTG CGC TCA ACA CGG CC (PstI);
antisense 111, 5′CTG CTG CAG AGC AAC CTG TGC GGG AGG GGT (PstI);
antisense 117, 5′CTC GAA TTC TCA CTT TTC TGT GAG GAC GCC AGC A (EcoRI);
antisense 135, 5′CTC GAA TTC TCA GTC GAC GTC GTC TTC AAC GGT (EcoRI);
antisense 216, 5′CGT GGA ATT CTT ACT TCT TGG CTT GGG AAC CGT (EcoRI); and
antisense 228, 5′CGT GCT GCA GGA ATT CTT CCG AAC CTT ATC (PstI).
The following linker and adaptor were used:
stop linker (termination codons in 3 frames), with internal NheI site—sense, 5′GAT CCG CTA GCC TAA GGT AG and antisense, 5′TCG ACT ACC TTA GGC TAG CG and
AatII-XmaI adaptor, for 5′-end cloning of DNA for peptide 129–228 into pAS2-1—sense, 5′CCG GGC ACC GTT GAA GAC GAC GT and antisense, 5′CGT CTT CAA CGG TGC.
DNA sequencing
Plasmid DNAs encoding Gal4-peptide fusions were purified with Qiagen Tip-20 columns. Automated sequencing was performed with ABI, using primers for the Gal4-AD or -BD from Clontech. The correct DNA sequence for the following peptides was confirmed—pAS30:1–33; pAS30:1–39; pAS30:1–80; pAS30:1–100; pAS30:1–111; pAS30:17–135; pAS30:40–117; pAS30:129–228; pGAD30:185–216; and pGAD30:185–228.
Transformation of S cerevisiae
Transformation was done according to the protocol of Gietz et al (1995). Briefly, yeast cells were grown to a density of 107/mL, and the cells were washed with water and with 100 mM lithium acetate. For transformation, polyethylene glycol, lithium acetate, single-stranded DNA, and plasmids were added to an aliquot of yeast cells. After incubation at 30°C the cells were exposed to a 42°C heat shock for 25 minutes, collected, resuspended in water, and plated on medium that lacked tryptophan or leucine (or both) to select for transformants with one or both plasmids.
Luminescence assays for β-galactosidase
The assay for β-galactosidase was performed according to the procedure of the supplier Tropix (a subsidiary of Applied Biosystems, Foster City, CA, USA), with several modifications. A small inoculum of S cerevisiae, freshly transformed for optimal gene expression, was suspended in 7–8 mL of selective medium and incubated at 30°C with shaking for at least 16 hours. The final OD600 of these cells was generally between 0.1 and 0.5. The volume of cells representing 0.075 OD600 was collected, washed, and resuspended in 25 μL yeast lysis buffer (Tropix) with 0.3 mM dithiothreitol (to stabilize the luminescence signal). The cells were subjected to 3 freeze-thaw cycles (liquid nitrogen and 37°C) to aid in cell lysis. For the β-galactosidase assay, 75 μL of the substrate-reaction buffer (Tropix) were added serially to 10 μL of lysed cells at 1-minute intervals. After 70 minutes of reaction, 100 μL of Accelerate (Tropix) were added at 1-minute intervals to each of the reaction tubes, and photon emission was measured after 10 seconds (for 10 seconds) with a luminometer (Berthold Lumat LB 9501). Duplicate reactions (with different yeast colonies) were performed in parallel for each transformant, and these experiments were repeated at least 3 times with freshly transformed cells for each interaction. The examples described represent typical, reproducible results. Reactions for several different yeast transformants, including those containing a single plasmid, were assayed in parallel for comparison. Dilutions of pure β-galactosidase were also assayed to establish linearity and reproducibility of the reaction (data not shown).
Photomicroscopy
Yeast colonies were visualized with a Zeiss Stemi DRC microscope and photographed with a Nikon Cool Pix 900 digital camera.
In vitro synthesis and coimmunoprecipitation of peptides
PCR was used to add the Myc epitope or the hemagglutinin (HA) epitope to peptides that were originally fused to either the Gal4-BD or the Gal4-AD, respectively, including only a small amount of either Gal4 domain (8 residues of BD or 7 residues of AD). The oligonucleotide primers, supplied by Clontech, also included the bacteriophage T7 promoter for in vitro transcription. The primers for the BD insert, with the Myc-tag sequences boldfaced, were sense, 5′AAA ATT GTA ATA CGA CTC ACT ATA GGG CGA GCC GCC ACC ATG GAG GAG CAG AAG CTG ATC TCA GAG GAG GAC CTG GGT CAA AGA CAG TTG ACT GTA TCG and antisense, 5′TAC CTG AGA AAG CAA CCT GAC CTA CAG G.
The primers for the AD insert, with the HA-tag sequences boldfaced, were sense, 5′AAA ATT GTA ATA CGA CTC ACT ATA GGG CGA GCC GCC ACC ATG TAC CCA TAC GAC GTT CCA GAT TAC GCT CCA CCA AAC CCA AAA AAA GAG and antisense, 5′ACT TGC GGG GTT TTT CAG TAT CTA CGA T.
The PCR products were purified over Microcon YM-50 spin columns (Millipore, Bedford, MA, USA) and were subjected to in vitro transcription and translation in the TnT system of Promega, according to the supplier's protocol. A mixture of protease inhibitors (ethylenediamine-tetraacetic acid–free Complete, Boehringer Mannheim, Indianapolis, IN) was added to some reactions. Tritiated leucine or [35S]methionine was incorporated into these peptides, which contain little or no internal methionine. Using Clontech's procedure for coimmunoprecipitation, we combined the reticulocyte lysates containing the test peptides (60 minutes) and added anti-Myc (monoclonal) or anti-HA (polyclonal) antibody (60 minutes), followed by addition and continuous mixing of the protein A-resin (60 minutes). Antibodies and protein A-resin, as well as oligonucleotide primers, were generously provided to us by Clontech for evaluation purposes. The immune complexes were washed 5–6 times with 25 volumes of buffer 1 and 2–3 times with 25 volumes of buffer 2. Buffer 1 comprises 10 mM N-2-hydroxyethylpiperazine-N′-2-ethane-sulfonic acid (pH 7.7), 500 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 50 mM sucrose, and 0.5% NP-40. Buffer 2 has the same composition but lacks NP-40 and has only 100 mM KCl. These immune complexes were heated to 100°C in 2× Laemmli sample buffer (Laemmli 1970), and the peptides were separated in a denaturing 15% polyacrylamide gel. For comparison, peptides in the reticulocyte lysate were also subjected to electrophoresis. After protein fixation the gels were treated with the fluorographic agent Amplify (Amersham Biosciences, Piscataway, NJ), and dried gels were exposed to XAR-5 film (Kodak) at −70°C.
RESULTS
Interactions between the N-terminal and the C-terminal halves of Hsp30
We used the S cerevisiae 2-hybrid binding assay (Chien et al 1991) to measure the interactions between specific peptide regions of Hsp30. The reporter gene was lacZ, encoding β-galactosidase, whose transcription depended on binding between the test peptides, which were fused to the AD or the BD of the transcriptional activator protein Gal4. The synthesis of β-galactosidase, therefore, provides a measure of the test proteins' interaction. We used a sensitive, quantifiable solution assay of β-galactosidase activity that uses a chemiluminescent substrate, and we measured photon emission with a luminometer. This assay yielded reproducible results and allowed us to evaluate the relative binding strengths of interacting peptides.
Two full-length monomers of Hsp30 were found to interact by this assay, as expected for proteins that dimerize and self-assemble, but this measurable interaction was not strong (data not shown), possibly because of its toxicity. Toxicity was indicated both by the enhanced interaction detected when Hsp30 was expressed from the weakly expressing vectors, pGAD424 and pGBT9, and by the consistent loss of β-galactosidase activity when colonies were subcultured. To understand the basis for self-interaction, we divided Hsp30 into 2 parts, an N-terminal region (residues 1–135) and a C-terminal region (residues 129–228). The division zone is in the midst of a nonconserved sequence that in many sHsps separates the 2 conserved subdomains of the α-crystallin domain (Plesofsky-Vig et al 1992). Conserved sequences are, therefore, contained within both the N-terminal and the C-terminal halves of Hsp30 (Fig 1), although the most highly conserved sequence is within the C-terminal half. These subdomains correspond to consensus regions II and I in plant sHsps (Waters 1995). We tested whether there was an interaction between the N-terminal and the C-terminal halves of Hsp30, and we also tested whether these halves could interact with themselves. We found in these assays that the N-terminal half of Hsp30, fused to the Gal4-BD, interacted very strongly with the protein's C-terminal half, fused to the Gal4-AD (Fig 2a); the reciprocal fusion proteins also interacted moderately (data not shown). We also found that the C-terminal half interacted strongly with itself (Fig 3), but self-interaction was not shown by the N-terminal half (data not shown).
Fig. 1.
The amino acid sequence of Hsp30. Filled circles denote the conserved residues that are similar in 26 out of 35 aligned sHsps (Plesofsky-Vig et al 1992). The predicted amphipathic helix is boxed, and lowercase letters indicate the extensive sequence that separates the 2 subdomains of the conserved α-crystallin domain. The -GVLT- is an iteration of the conserved, signature sequence of sHsps. Numbered residues represent the boundaries of peptides tested in yeast interaction assays. The sequences that correspond most closely to β strands of Methanococcus jannaschii Hsp16.5 (Kim et al 1998) are shaded. sHsps, small heat shock proteins
Fig. 2.
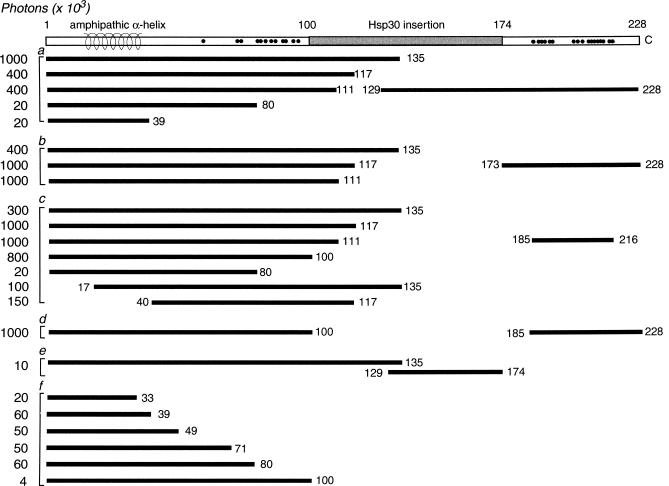
Photon measurements of β-galactosidase activity resulting from interaction between the N-terminal half and the C-terminal half of Hsp30. On the left are the [BD] N-terminal half and its truncated peptide derivatives, which were each assayed for interaction with the (a) [AD] C-terminal half and (b–e) its peptide derivatives, shown opposite on the right. At the bottom are (f) measurements for [BD] N-terminal peptides of Hsp30 in the absence of Gal4 AD-peptides. Photons (×103) measured by a luminometer are at the far left. We assume that measurement variation in the high range of 400–1000 (×103) likely reflects experimental variables rather than differences in peptide-binding affinity. Single Gal4 BD- or AD-peptide fusions (other than those shown) produced photon measurements of 1–6 (×103). Measurements of 20 (×103) or less are considered background. BD, binding domain; AD, activation domain
Fig. 3.
Photon measurements of β-galactosidase activity resulting from interaction between the 2 C-terminal halves. Interactions between the [BD] C-terminal half of Hsp30 and the [AD] full-length or truncated peptide derivatives of the C-terminal half are shown. Measured photons (×103) are on the left. Single Gal4 BD- or AD-peptide fusions produced photon measurements of 1–6 (×103). BD, binding domain; AD, activation domain
We sought to determine what sequences within the N- and C-terminal halves were involved in these interactions by engineering the peptide derivatives of each half whose interaction we assayed. We first made truncations at each end of the N-terminal half. The approximate boundaries of conserved sequence in the N-terminal half of Hsp30 are aa 51 through 100 (Fig 1). We truncated the C-end of the N-terminal half, creating peptides 1–117, 1–111 (removing -GVLT-, which repeats the conserved sequence within the C-terminal half), 1–100, and 1–80. We found that the first 3 of these peptides bound strongly to the C-terminal domain of Hsp30, whereas peptide 1–80 was inactive in the interaction assay (Fig 2). This indicates that the peptide sequence between aa 80 and 100 is required for binding, but the nonconserved sequence between aa 101 and 135 is not required. Approaching from the other direction, deletion of the extreme N-terminus of Hsp30, creating peptide 17–135, lessened binding to the C-terminal domain but still maintained it at a strong level (Fig 2c). Between aa 18 and 35, Hsp30 has a predicted amphipathic helix, a motif that is shared by only a few other sHsps (Plesofsky-Vig and Brambl 1990). Deletion extending through this helical motif, creating peptide 40–117, also yielded a strong, albeit reduced, level of binding to the C-terminal domain (Fig 2c). We believe that this reduction in binding is likely due to steric constraints or other indirect causes, rather than loss of a binding site, because the peptides representing these deleted regions, such as peptides 1–33 (data not shown) and 1–39 (Fig 2a) showed no binding to the C-terminal domain. These truncation experiments indicate that N-terminal sequences sufficient for binding to the C-terminal domain are within aa 40–100 of Hsp30, which corresponds approximately to the conserved region within the N-terminal half.
Although peptide 1–80 and smaller peptides did not bind to the Hsp30 C-terminal domain, several of them induced a low amount of β-galactosidase activity by themselves, in the absence of a Gal4-AD fusion protein, suggesting that these Gal4-BD fusion peptides may interact with transcriptional activating proteins to induce lacZ transcription. The peptides from the N-terminal half that were sufficient to induce β-galactosidase activity include peptides 1–39, 1–49, 1–71, and 1–80 (Fig 2f). Peptide 1–33 induced 30% of this activity, at most, and peptide 1–100 (and longer) did not induce this activity (Fig 2f). The smallest peptide that shows this general protein-binding activity, peptide 1–39, includes the amphipathic helix (aa 18–35), suggesting that this motif may be responsible for protein binding and activation of lacZ transcription. Extension of the peptide to residue 100 presumably masks or alters this protein-binding motif.
To find the regions within the C-terminal half of Hsp30 (residues 129–228) that are responsible for binding to the N-terminal half, we divided the C-terminal half into peptide 129–174, containing nonconserved sequence, and peptide 173–228, which includes sequence that is highly conserved among sHsps (Fig 1). Peptide 173–228 was found to interact strongly with the N-terminal half of Hsp30 (Fig 2b), whereas peptide 129–174 was inactive (Fig 2e). We trimmed away the less conserved sequence at both ends of peptide 173–228 to create peptide 185–216, which corresponds to the most conserved region within sHsps (Fig 1). This tightly conserved peptide strongly interacted with the N-terminal half and its active peptide derivatives, whereas it did not interact with its inactive derivatives (Fig 2c).
Interaction of the C-terminal half of Hsp30 with itself
In addition to binding to the N-terminal half, the C-terminal half of Hsp30 binds to itself (Fig 3). We assayed whether the C-terminal half would interact with the C-terminal derivatives we generated. As found for the N-terminal half, there was no interaction with the nonconserved peptide 129–174, but the C-terminal half strongly interacted with peptide 173–228 (Fig 3). Unlike the N-terminal half, however, the C-terminal half did not bind to the trimmed, conserved peptide 185–216, but it did bind to peptide 185–228 (Fig 3). This indicates that peptide extension toward the carboxyl end of Hsp30 is required for this self-interaction, whereas residues 173–184 are not necessary. Therefore, the semiconserved tail of Hsp30, as an extension of the tightly conserved domain, appears to be required for self-interaction of the C-terminal region.
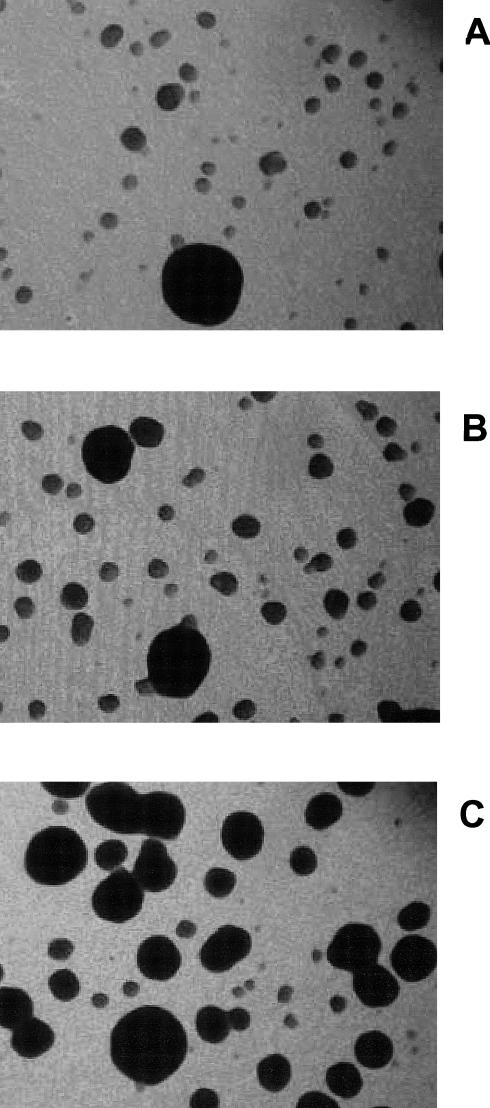
The interaction we observed between the N- and C-terminal halves of Hsp30 was associated with moderate toxicity to the transformed yeast, as indicated by the small size of their colonies (Fig 4). These small colonies had high β-galactosidase activity, whereas the few interspersed large colonies lacked this activity (data not shown) and had probably stopped expressing the fusion proteins. This toxicity was not evident for transformed yeast expressing only the N-terminal or the C-terminal half (or truncated derivatives). With each truncation of the C-terminal half, there was a small increase in colony size, suggesting a slight reduction in toxicity of the interaction, but this toxicity was completely lost in strains coexpressing the Hsp30 N-terminal half and the shortest C-terminal peptide 185–216, which lacks the carboxyl tail (Fig 4). When we assayed cells that coexpressed the N-terminal peptide 17–135 and the C-terminal half, it proved difficult with these slow-growing cells to measure binding. However, there was no difficulty in detecting the interaction of peptide 17–135 with peptide 185–216 (Fig 2c), and the coexpressing yeast cells grew as uniform colonies on solid medium (Fig 4). Removal of the carboxyl tail should block self-interaction of the C-terminal peptide. That this truncation correlates with the disappearance of toxicity and the restoration of normal yeast growth suggests that cell injury may derive from the simultaneous interactions of the C-terminal half with itself and with the N-terminal half of Hsp30, when these are synthesized as separate fusion proteins.
Fig. 4.
Photomicrographs of yeast colonies resulting from cotransformation with interacting N-terminal and C-terminal Hsp30 peptides. (A) Colonies on solid medium cotransformed with peptides BD: 17–135 and AD: 129–228. These colonies are extremely small, with the occasional large colony. (B) Colonies on solid medium cotransformed with peptides BD: 17–135 and AD: 185–228. The small colonies are somewhat larger than those in (A), but there are only a few large colonies. (C) Colonies on solid medium cotransformed with peptides BD: 17–135 and AD: 185–216. Most colonies are of a uniformly large size. The field is 3 mm across. BD, binding domain; AD, activation domain
Interactions of Hsp30 peptides synthesized in vitro
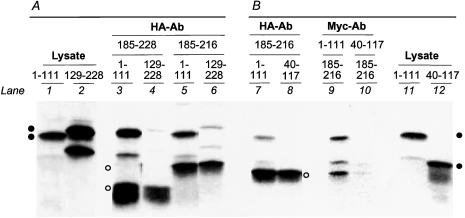
To test peptide interactions by an alternate method, we synthesized in vitro select peptides of Hsp30 that were tagged with either the Myc (BD) or the HA (AD) epitope for immunoprecipitation. These peptides also differed from the fusion proteins made in yeast by their lack of most of the Gal4 domain sequences. When we combined the Myc-tagged peptide 1–111 with peptide 185–216 or 185–228, tagged with the HA epitope, anti-HA antibody coimmunoprecipitated peptide 1–111 along with peptide 185–216 or 185–228 (Fig 5). In the reciprocal experiment, anti-Myc antibody similarly coprecipitated peptide 185–216 along with Myc-tagged peptide 1–111 (Fig 5). Therefore, the interaction identified by 2-hybrid assay between the N-terminal half of Hsp30 and the C-terminal peptide is supported by in vitro biochemical evidence. Other interactions detected in yeast were not evident when tested in vitro. Myc-peptide 40–117, which interacted with peptide 185–216 in the 2-hybrid assay, was not coprecipitated with it by anti-HA antibody, and the amount of peptide 185–216 that was coprecipitated with peptide 40–117 by anti-Myc antibody was not clearly above the background levels of nonspecific precipitation. Similarly, the Myc-tagged peptide 129–228, encompassing the C-terminal half of Hsp30, was coprecipitated at only background levels by antibody against the HA epitope, when mixed with either HA-peptide 185–228 or HA-peptide 185–216 (Fig 5). Anti-Myc antibody did not coprecipitate peptide 185–228 or peptide 185–216 above background levels from the same mixture (data not shown).
Fig. 5.
Coimmunoprecipitation of in vitro–synthesized peptides of Hsp30. Proteins that were radiolabeled with (A) [35S]methionine or with (B) [3H]leucine were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Closed circles denote Myc-tagged peptides, and open circles denote HA-tagged peptides. Lanes 1 and 2: Myc-peptide 1–111 and Myc-peptide 129–228; lanes 11 and 12: Myc-peptide 1–111 and Myc-peptide 40–117, respectively, synthesized in rabbit reticulocyte lysate. Lanes 3 through 8: proteins coprecipitated by anti-HA antibody; lanes 9 and 10: proteins coprecipitated by anti-Myc antibody. Lanes 3 and 4: HA-peptide 185–228 was mixed with Myc-peptide 1–111 or Myc-peptide 129–228; lanes 5 and 6: HA-peptide 185–216 was mixed with Myc-peptide 1–111 or Myc-peptide 129–228. Lanes 7 and 9: HA-peptide 185–216 was mixed with Myc-peptide 1–111; lanes 8 and 10: HA-peptide 185–216 was mixed with Myc-peptide 40–117. The results show that Myc-peptide 1–111 coimmunoprecipitated with HA-185–228 and with HA-185–216, whereas there was no or background coprecipitation of Myc-peptide 40–117 and Myc-peptide 129–228 by the HA-tagged peptides. Although little Myc-peptide 40–117 is immunoprecipitated by anti-Myc antibody in this experiment (lane 10), the use of a higher amount of Myc antibody in another experiment (not shown) precipitated a higher amount of peptide 40–117, with possible coprecipitation of peptide 185–216. However, the reciprocal experiments with anti-HA antibody did not indicate coprecipitation. Peptide 185–216 has extra C-terminal amino acids derived from the pGADGH vector, making it longer than peptide 185–228. HA, hemagglutinin
There are several possible reasons for the lack of detectable peptide interactions in vitro. The biochemical assay is probably less sensitive than the 2-hybrid assay; the interaction that was detected by coimmunoprecipitation gave a very strong signal in the 2-hybrid assay, higher than that of the other interactions. Alternatively, the lack of N-terminal Gal4 sequence in the peptides made in vitro may prevent peptides that represent internal sequence (peptides 40–117 and 129–228) from assuming a binding conformation, whereas the N-terminal Hsp30 peptide 1–111 may assume its native conformation for binding to the C-terminal peptides. Further, the Myc tag on peptide 40–117 may be less accessible for antibody binding, or the protein may be unstable during prolonged incubation.
DISCUSSION
The group of α-crystallin–related Hsps, including Hsp30, share a region of conserved sequence and assemble into large oligomers, but overall they vary extensively in amino acid sequence, and the multimers they form are of different sizes. Their nonconserved N-terminal region has been shown to be required for sHsp assembly into large oligomeric structures. sHsps that are naturally or experimentally truncated at their N-termini assemble into only dimers and tetramers (Leroux et al 1997; Kokke et al 1998; Feil et al 2001), while also showing reduced capacity to chaperone other proteins. This nonconserved region, therefore, may account for much of the variation in the oligomeric structure of different sHsps and for the observation that sHsps of different classes do not coassemble into oligomers (Helm et al 1997; Studer and Narberhaus 2000). In contrast, the conserved α-crystallin domain is sufficient for dimer formation by truncated proteins.
Although some species of sHsps vary in oligomeric size and are polydisperse (Haley et al 2000), the crystal structures of the more uniform Hsp16.5 of M jannaschii (Kim et al 1998) and Hsp16.9 of wheat (van Montfort et al 2001) have recently been solved. Excluding the disordered N-terminal region, MjHsp16.5 was seen to assemble into a 24-subunit hollow sphere consisting of 3 octamers, each comprising 4 dimers. The most extensive dimerization contact was between a β6 strand of one monomer interacting with a β2 strand within the β-sheet of the other monomer. Both these β-strands are within the conserved, α-crystallin–related region of MjHsp16.5, supporting the experimental sufficiency of this region for dimerization of sHsps. Contacts between dimers comprising the MjHsp16.5 octamer are made by the carboxyl tail of one dimer subunit making contact with the α-crystallin domain of another dimer subunit.
Wheat Hsp16.9 assembles into a double disk consisting of only 12 subunits (van Montfort et al 2001), but it shares many features with the 24-subunit archeal sHsp. In addition to a similar monomeric structure, the major dimerization site involves the same β6 strand, which loops out to enter the β-sheet of the partner monomer. Although the β6 strand sequence is not conserved in the sHsps of animals, it is moderately conserved in the archeal, plant, and fungal sHsps, including a glutamate residue involved in dimerization, which is only absent from chloroplast and mitochondrial sHsps (Plesofsky-Vig et al 1992; Scharf et al 2001). However, the glutamate in wheat Hsp16.9 is involved in an ionic interaction, whereas in MjHsp16.5 it forms a backbone hydrogen bond. These differences suggest that there is some permissiveness in the mechanisms for dimer formation among sHsps, even when the overall structural result is similar. The animal sHsps, as exemplified by αB-crystallin, appear to diverge further. In addition to lacking a conserved β6 strand, the animal proteins lack the adjacent, variable hydrophilic region that separates the conserved α-crystallin subdomains of sHsps (Plesofsky-Vig et al 1992), and they tend to have longer C-terminal extensions (de Jong et al 1998). According to physical analyses, the dimeric structure of N-terminally truncated αB-crystallin differs from that of MjHsp16.5 (Feil et al 2001).
In the present study, we tested the interactions between defined peptides of a sHsp, using the yeast 2-hybrid interaction assay (Chien et al 1991), as an indirect approach for analyzing the contribution of specific domains to the structural organization of a sHsp. Our approach and the results of our 2-hybrid analysis of domain interactions in Hsp30 are distinct from those of other sHsp 2-hybrid studies (Lambert et al 1999; Liu and Welsh 1999), but they are compatible with them. A major difference of our study is that we divided the conserved α-crystallin domain into 2 separate subdomains, one within the N- and the other within the C-terminal half, whose interaction with one another we assayed. We believe that the interactions we detected between these peptides, as part of fusion proteins, likely reflect the interactions that occur in the native Hsp30. It seems unlikely to us that the strong interactions we detected by this technique are artifactual. First, we tested the binding of Hsp30's N-terminal and C-terminal halves to several other proteins in the 2-hybrid assay without detecting any interaction. Furthermore, the interactions we detected were based on testing a series of Hsp30-derived peptides, rather than only 1 peptide, and the results obtained were consistent and supported one another. In addition, we used an independent approach to confirm the interaction of the N-terminal half of Hsp30 with the C-terminal conserved peptide, by synthesizing the peptides in vitro and assaying their coimmunoprecipitation.
When Hsp30 was divided into 2 halves for 2-hybrid analysis, we detected a strong interaction between its N-terminal half (aa 1–135) and its C-terminal half (aa 129–228). We found that the Hsp30 C-terminal half also interacts with itself. The N-terminal half includes both the nonconserved sequence (aa 1–50) and a portion of the conserved α-crystallin domain (aa 51–100). The C-terminal half includes the other part of the conserved α-crystallin domain (aa 174–216) and a semiconserved carboxyl extension (aa 217–228). The intervening region (aa 101–173) consists of a sequence that appears to be inserted into Hsp30, relative to the aligned sHsps of animals (Plesofsky-Vig et al 1992). Plant and microbial sHsps also have an extra, variable sequence at this location, but it is much shorter than that of Hsp30, typically being no more than 10 residues.
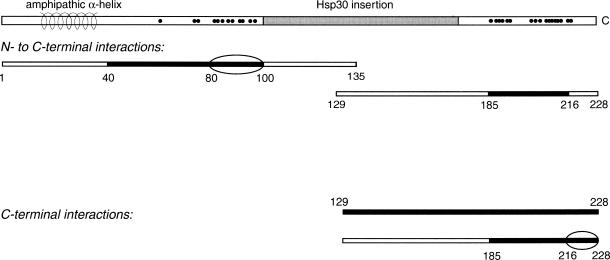
We found that the conserved portion (aa 41–100) of the N-terminal half contains the sequence responsible for binding to the C-terminal half and that aa 81–100 are specifically required for binding (Fig 6). The extreme N-terminus (aa 1–16) and the potential amphipathic helix (aa 18–35) are both dispensable for this interaction, although the N-terminus, especially, appears to facilitate or strengthen the interaction. The conserved subdomain within the N-terminal half corresponds to β strands 1 through 5 of the MjHsp16.5 crystal structure (Fig 1) or to α3 and β2–β5 of wheat Hsp16.9, with aa 81–100 corresponding to β4 and β5 (Kim et al 1998). This subdomain interacts with the tightly conserved sequence (aa 185–216) within the C-terminal subdomain of Hsp30, which corresponds to the β7–β9 strands of MjHsp16.5 but excludes the β6 strand and the variable carboxyl tail (Fig 1). In the MjHsp16.5 monomer, β1, β4, and β5 associate with β7 in one β sheet, whereas β2 and β3 associate with β9 and β8 in the other β sheet. Because the tertiary structures of sHsps, including αB-crystallin (Feil et al 2001), appear to assume the same fold, the interaction between aa 40–100 and 185–216 of Hsp30 likely represents contacts within the β sheets of the monomer. These β strands, therefore, would be able to associate with one another even when incorporated into separate peptides, which is consistent with the physical separation that exists between α-crystallin subdomains in the native protein. We propose that these 2-hybrid interactions between the N- and C-terminal conserved subdomains represent the tertiary contacts seen within the sHsp crystal structures. This further suggests that the N-terminal and C-terminal nonconserved sequences, as well as the presumed central dimerization loop, are not required for the characteristic monomeric fold assumed by the most conserved regions of the α-crystallin domain.
Fig. 6.
Summary of the 2 interactions revealed by this study, between the N- and C-terminal halves of Hsp30 and between 2 C-terminal halves. The minimal peptide regions sufficient for interaction are drawn in black, and an oval surrounds the portions known to be necessary for the interaction. Amino acids 217–228 are required for peptide interaction with the C-terminus but not for interaction with the N-terminus
An overlapping, but distinct, region within the C-terminal half of Hsp30 was found to be responsible for C-terminal self-interaction. Although the conserved peptide 185–216, which interacts with the N-terminal half, was not sufficient for interaction with the C-terminal half, extension of this peptide to include the carboxyl tail, creating peptide 185–228, resulted in strong C-terminal interaction (Fig 6). This result implies several alternative possibilities: the nonconserved tails on the 2 peptides may bind to one another; there may be reciprocal binding between the sequences contained within peptide 185–228; or the tail in peptide 185–228 may interact with the C-terminal sequence that is not included in peptide 185–216, such as the variable-inserted sequence or the β6 strand counterpart (or both) (Fig 1). We believe it most likely that there is reciprocal binding between sequences within peptide 185–228. One explanation for the moderate toxicity generated when yeast coexpresses the N-terminal peptide and the extended (but not the unextended) C-terminal peptide is that peptide 185–228 interacts with itself (Fig 4) as well as with the N-terminal peptide, leading to protein aggregates.
In MjHsp16.5 the C-terminal tail of one dimer subunit interacts with the β4 and β8 strands of another dimer subunit, providing a basis for association between dimers in the octamer (Kim et al 1998). In wheat Hsp16.9, different C-terminal tails strengthen either interdimer or intertetramer associations (van Montfort et al 2001), binding in a hydrophobic groove between the β4 and β8 strands of the interacting monomer, thereby contributing to oligomeric assembly. Peptide 185–228 of Hsp30 contains the β7 and β8 strands, but β4 is not included even within the C-terminal half. Therefore, it appears that sequences within the carboxyl tail of Hsp30 may be capable of binding to β8, in the absence of β4, as long as there is reciprocal binding. Binding residues have been identified in the carboxyl tails of MjHsp16.5 and wheat Hsp16.9 that comprise a short motif, Ile-X-Ile/Val, that is conserved in sHsps (de Jong et al 1998), including Neurospora Hsp30 (Fig 1). The sHsp crystal structures show that these C-terminal extensions, although otherwise nonconserved, have an important role in the stabilization of oligomers.
Our 2-hybrid assays also suggest that the N-terminal 39 residues of Hsp30 have a general affinity for proteins. The smallest peptide showing this activity includes a region with the potential to form an amphipathic helix, a motif frequently involved in protein-protein interactions. This region may be part of a nonconserved domain in Hsp30 that binds substrates or other proteins. The Hsp22 of Drosophila melanogaster also has an amphipathic helix-forming region at its N-terminus, but it has a net positive charge characteristic of a mitochondrial targeting sequence, and Hsp22 was shown to be an intramitochondrial protein (Morrow et al 2000). Hsp30, in contrast, appears to associate with the outer membrane of mitochondria (Plesofsky-Vig and Brambl 1990), and we have found that it coimmunoprecipitates with import receptors of the outer membrane (Plesofsky et al 1999). One of these receptors was Tom20, which has been shown to recognize and bind to amphipathic helices regardless of their charge (Abe et al 2000), with positive charge required only for later steps in protein import.
When one compares the various assembly levels for sHsps, it appears that the higher the structural order the less conserved are the amino acid sequences involved and the less predictable are the specific domain contacts. Whereas the conserved tertiary structure of the monomer depends on the most highly conserved regions of the α-crystallin domain, dimerization involves the loosely conserved or nonconserved β6 strand and the adjacent, variable sequence. The nonconserved carboxyl tail of sHsps, which contains a small conserved binding motif, has been implicated in establishing and strengthening the associations between dimers and further oligomeric assembly. The exact connections, however, depend on the oligomeric organization of the particular sHsp. In wheat Hsp16.9, 6 of the carboxyl tails interact with dimers in the same disk, whereas the other 6 tails interact with dimers in the other disk. Complete assembly of sHsps into the oligomer appears to be driven by their nonconserved N-terminal domains (Leroux et al 1997; van Montfort et al 2001).
This 2-hybrid analysis of domain interactions in Hsp30 has verified, for a noncrystallized sHsp, some of the interactions seen for the 2 crystallized sHsps. The conserved monomeric fold, a β sandwich, is reflected in the tight binding of the conserved subdomains of the α-crystallin domain. It should not be surprising that the 2 α-crystallin subdomains of Hsp30, incorporated into separate fusion proteins, can maintain β strand interactions, because these 2 subdomains appear to be separated in the native protein by a loop of variable sequence that contains the dimerizing β6 strand. Furthermore, the importance of the C-terminal tail for dimer interactions and oligomeric assembly, seen for both crystallized proteins, is likely embodied in the self-interaction of the C-terminal peptide, which requires this semiconserved extension. This 2-hybrid analysis offers the additional advantage of suggesting what the minimal unit of interaction might be. At least some of the tertiary interactions within the monomer can occur independently of the nonconserved flanking sequence. It is also likely that the C-tail can interact with the C-terminal α-crystallin subdomain, presumably with the β8 strand, in the absence of the β4-containing N-terminal subdomain. That the 2-hybrid assays did not detect any interaction involving the dimerizing β6 strand may be due to the way sequences were distributed between peptides.
Because such interactions can be detected between peptides in isolation from other protein domains, it should be possible to test the importance of specific domains of sHsps for influencing their structure and chaperone function. For example, the nonconserved N-terminal and C-terminal domains can be exchanged between sHsps, specifically between Hsp30 and plant sHsps, and the properties of these chimeric proteins can be assayed in vitro and in N crassa cells that are disrupted in hsp30 (Plesofsky-Vig and Brambl 1995). The nonconserved N-terminal region of sHsps is reported to be important both for chaperone activity and for oligomerization, and our experiments suggest that the N-terminal 39 aa of Hsp30 may be involved in substrate binding. Because Hsp30 assembles into an approximately 24-subunit multimer (unpublished data), we can distinguish its size from that of the 12-subunit plant sHsp. By creating chimeric proteins we can determine whether the amphipathic helix of Hsp30 is involved in specific substrate interactions or membrane localization (or both), and we can test to what extent this region contributes to the specificity and extent of oligomerization. The carboxyl tail, which was earlier reported to keep the assembled α-crystallin oligomer solubilized (Smulders et al 1996), is now seen to help bind together dimers and tetramers. Although this tail contains a short conserved binding motif, it is otherwise variable. By exchanging the carboxyl tail between sHsps, we can assess its ability to stabilize oligomers of the chimeric sHsp.
Acknowledgments
This research was supported by grants from the National Research Initiative of the USDA Competitive Research Grants Office (94-37100-0290) and the National Institute of General Medical Sciences (GM-19398).
REFERENCES
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom 20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- Aquino DA, Capello E, and Weisstein J. et al. 1997 Multiple sclerosis: altered expression of 70- and 27-kDa heat shock proteins in lesions and myelin. J Neuropathol Exp Neurol. 56:664–672. [PubMed] [Google Scholar]
- Chadli A, Bouhouche I, Sullivan W, Stensgard B, McMahon N, Catelli MG, Toft DO. Dimerization and N-terminal domain proximity underlie the function of the molecular chaperone heat shock protein 90. Proc Natl Acad Sci U S A. 2000;97:12524–12529. doi: 10.1073/pnas.220430297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C-T, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong WW, Caspers G-J, Leunissen JAM. Genealogy of the α-crystallin-small heat-shock protein superfamily. Int J Biol Macromol. 1998;22:151–162. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- Donze O, Abbas-Terki T, Picard D. The Hsp90 chaperone complex is both a facilitator and a repressor of the dsRNA-dependent kinase PKR. EMBO J. 2001;20:3771–3780. doi: 10.1093/emboj/20.14.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil IK, Malfois M, Hendle J, van der Zandt H, Svergun DI. A novel quaternary structure of the dimeric α-crystallin domain with chaperone-like activity. J Biol Chem. 2001;276:12024–12029. doi: 10.1074/jbc.M010856200. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Haley DA, Bova MP, Huang Q-L, Mchaourab HS, Stewart PL. Small heat-shock protein structures reveal a continuum from symmetric to variable assemblies. J Mol Biol. 2000;298:261–272. doi: 10.1006/jmbi.2000.3657. [DOI] [PubMed] [Google Scholar]
- Hartl F-U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Walke S, Stromer T, Ehrnsperger M, White HE, Chen S, Saibil HR, Buchner J. Hsp26: a temperature-regulated chaperone. EMBO J. 1999;18:6744–6751. doi: 10.1093/emboj/18.23.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm KW, Lee GJ, Vierling E. Expression and native structure of cytosolic class II small heat-shock proteins. Plant Physiol. 1997;114:1477–1485. doi: 10.1104/pp.114.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick JP, Hartl F-U. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Kim KK, Kim R, Kim S-H. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Matsumura Y, Tsuchido T. Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol Lett. 2000;184:165–171. doi: 10.1111/j.1574-6968.2000.tb09009.x. [DOI] [PubMed] [Google Scholar]
- Kokke BPA, Leroux MR, Candido EPM, Boelens WC, de Jong WW. Caenorhabditis elegans small heat-shock proteins Hsp12.2 and Hsp12.3 form tetramers and have no chaperone-like activity. FEBS Lett. 1998;433:228–232. doi: 10.1016/s0014-5793(98)00917-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lambert H, Charette SJ, Bernier AF, Guimond A, Landry J. HSP27 multimerization mediated by phosphorylation-sensitive intermolecular interactions at the amino terminus. J Biol Chem. 1999;274:9378–9385. doi: 10.1074/jbc.274.14.9378. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux MR, Melki R, Gordon B, Batelier G, Candido EPM. Structure-function studies on small heat shock protein oligomeric assembly and interaction with unfolded polypeptides. J Biol Chem. 1997;272:24646–24656. doi: 10.1074/jbc.272.39.24646. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HRB. Involvement of ATP in the nuclear and nucleolar functions of the 70 kd heat shock protein. EMBO J. 1985;4:3137–3143. doi: 10.1002/j.1460-2075.1985.tb04056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Welsh MJ. Identification of a site of Hsp27 binding with Hsp27 and αB-crystallin as indicated by the yeast two-hybrid system. Biochem Biophys Res Comm. 1999;255:256–261. doi: 10.1006/bbrc.1999.0174. [DOI] [PubMed] [Google Scholar]
- Mehlen P, Mehlen A, Godet J, Arrigo A-P. Hsp27 as a switch between differentiation and apoptosis in murine embryonic stem cells. J Biol Chem. 1997;272:31657–31665. doi: 10.1074/jbc.272.50.31657. [DOI] [PubMed] [Google Scholar]
- Mehlen P, Schulze-Osthoff K, Arrigo A-P. Small stress proteins as novel regulators of apoptosis: heat shock protein 27 blocks Fas/APO-1- and staurosporine-induced cell death. J Biol Chem. 1996;271:16510–16514. doi: 10.1074/jbc.271.28.16510. [DOI] [PubMed] [Google Scholar]
- Morrow G, Inaguma Y, Kato K, Tanguay RM. The small heat shock protein Hsp22 of Drosophila melanogaster is a mitochondrial protein displaying oligomeric organization. J Biol Chem. 2000;275:31204–31210. doi: 10.1074/jbc.M002960200. [DOI] [PubMed] [Google Scholar]
- Oesterreich S, Hickey E, Weber LA, Fuqua SAW. Basal regulatory promoter elements of the hsp27 gene in human breast cancer cells. Biochem Biophys Res Commun. 1996;222:155–163. doi: 10.1006/bbrc.1996.0714. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Plesofsky N, Brambl R. Glucose metabolism in Neurospora is altered by heat shock and by disruption of HSP30. Biochim Biophys Acta. 1999;1449:73–82. doi: 10.1016/s0167-4889(98)00172-4. [DOI] [PubMed] [Google Scholar]
- Plesofsky N, Gardner N, Lill R, Brambl R. Disruption of the gene for Hsp30, an α-crystallin-related heat shock protein of Neurospora crassa, causes defects in import of proteins into mitochondria. Biol Chem. 1999;380:1231–1236. doi: 10.1515/BC.1999.156. [DOI] [PubMed] [Google Scholar]
- Plesofsky-Vig N, Brambl R. Gene sequence and analysis of hsp30, a small heat shock protein of Neurospora crassa which associates with mitochondria. J Biol Chem. 1990;265:15432–15440. [PubMed] [Google Scholar]
- Plesofsky-Vig N, Brambl R. Disruption of the gene for hsp30, an α-crystallin-related heat shock protein of Neurospora crassa, causes defects in thermotolerance. Proc Natl Acad Sci U S A. 1995;92:5032–5036. doi: 10.1073/pnas.92.11.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesofsky-Vig N, Vig J, Brambl R. Phylogeny of the α-crystallin-related heat shock proteins. J Mol Evol. 1992;35:537–545. doi: 10.1007/BF00160214. [DOI] [PubMed] [Google Scholar]
- Renkawek K, Stege GJ, Bosman GJ. Dementia, gliosis and expression of the small heat shock proteins hsp27 and αB-crystallin in Parkinson's disease. Neuroreport. 1999;10:2273–2276. doi: 10.1097/00001756-199908020-00009. [DOI] [PubMed] [Google Scholar]
- Rogalla T, Ehrnsperger M, and Preville X. et al. 1999 Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor α by phosphorylation. J Biol Chem. 274:18947–18956. [DOI] [PubMed] [Google Scholar]
- Scharf K-D, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (Acd proteins) Cell Stress Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulders RHPH, Carver JA, Lindner RA, van Boekel MAM, Bloemendal H, de Jong WW. Immobilization of the C-terminal extension of bovine αA-crystallin reduces chaperone-like activity. J Biol Chem. 1996;271:29060–29066. doi: 10.1074/jbc.271.46.29060. [DOI] [PubMed] [Google Scholar]
- Studer S, Narberhaus F. Chaperone activity and homo- and hetero-oligomer formation of bacterial small heat shock proteins. J Biol Chem. 2000;275:37212–37218. doi: 10.1074/jbc.M004701200. [DOI] [PubMed] [Google Scholar]
- van Montfort RLM, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- Waters ER. The molecular evolution of the small heat-shock proteins in plants. Genetics. 1995;141:785–795. doi: 10.1093/genetics/141.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C-H, Chang P-FL, Yeh K-W, Lin W-C, Chen Y-M, Lin C-Y. Expression of a gene encoding a 16.9-kDa heat-shock protein, Oshsp16.9, in Escherichia coli enhances thermotolerance. Proc Natl Acad Sci U S A. 1997;94:10967–10972. doi: 10.1073/pnas.94.20.10967. [DOI] [PMC free article] [PubMed] [Google Scholar]