Abstract
To obtain an inventory of all human genes that code for α-crystallin–related small heat shock proteins (sHsps), the databases available from the public International Human Genome Sequencing Consortium (IHGSC) and the private Celera human genome project were exhaustively searched. Using the human Hsp27 protein sequence as a query in the protein databases, which are derived from the predicted genes in the human genome, 10 sHsp-like proteins were retrieved, including Hsp27 itself. Repeating the search procedure with all 10 proteins and with a variety of more distantly related animal sHsps, no further human sHsps were detected, as was the case when searches were performed at deoxyribonucleic acid level. The 10 retrieved proteins comprised the 9 earlier recognized human sHsps (Hsp27/HspB1, HspB2, HspB3, αA-crystallin/HspB4, αB-crystallin/HspB5, Hsp20/HspB6, cvHsp/HspB7, H11/HspB8, and HspB9) and a sperm tail protein known since 1993 as outer dense fiber protein 1 (ODF1). Although this latter protein probably serves a structural role and has a high cysteine content (14%), it clearly contains an α-crystallin domain that is characteristic for sHsps. ODF1 can as such be designated as HspB10. The expression of all 10 human sHsp genes was confirmed by expressed sequence tag (EST) searches. For Hsp27/HspB1, 2 retropseudogenes were detected. The HspB1–10 genes are dispersed over 9 chromosomes, reflecting their ancient origin. Two of the genes (HspB3 and HspB9) are intronless, and the others have 1 or 2 introns at various positions. The transcripts of several sHsp genes, notably HspB7, display low levels of alternative splicing, as supported by EST evidence, which may result in minor amounts of isoforms at the protein level.
INTRODUCTION
The small heat shock proteins (sHsps) form a ubiquitous family of molecular chaperones, of which the monomer size typically ranges between 16 and 25 kDa (for recent overviews, see Arrigo and Müller 2002; Narberhaus 2002; van Montfort et al 2002). They are characterized by a conserved C-terminal region, called the α-crystallin domain, a more variable N-terminal sequence, and in most cases a short and variable C-terminal tail. The sHsps occur as homo- or heteromeric complexes, comprising 2 to about 40 subunits. These globular complexes are often polydisperse and dynamic, readily exchanging subunits. Crystallization is therefore generally a problem, and crystal structures are only known for an archaebacterial (Kim et al 1998) and a plant sHsp (van Montfort et al 2001). The α-crystallin domain consists of a ß sandwich of 2 antiparallel ß sheets. The domains of 2 monomers tightly interact to form the dimeric building blocks of the sHsp oligomers. The N-terminal regions are variable in structure and contain α-helical components (eg, van Montfort et al 2001; Kappé et al 2002), whereas the very C-terminal tail flexes freely from the surface (Carver 1999).
Functionally, sHsps prevent the in vitro aggregation of unfolding proteins. Bound proteins can be transferred to adenosine triphosphate–dependent chaperones, such as Hsp70, and refolded (Haslbeck and Buchner 2002). In vivo, sHsps confer protection to a variety of cellular stressors (Latchman 2002; Arrigo et al 2002a). The expression of most sHsps is developmentally regulated and can be upregulated by various forms of stress (Davidson et al 2002; Michaud et al 2002). The sHsps are involved in a variety of cellular processes, notably relating to cytoskeletal rearrangements (eg, Wieske et al 2001; Quinlan 2002) and apoptosis (Charette and Landry 2000; Arrigo et al 2002b). In humans, some sHsps occur at increased levels in neurodegenerative disorders (eg, Krueger-Naug et al 2002) and in certain tumors (eg, Ciocca and Vargas-Roig 2002). Differential serine phosphorylation plays an important role in the functioning of mammalian sHsps (Gaestel 2002; Kato et al 2002).
Although several Eubacteria have no or only a single sHsp (Kappé et al 2002; Narberhaus 2002), most organisms express multiple sHsps in a cell-specific and developmentally regulated pattern. The number of sHsp genes in the known eukaryotic genomes ranges from 2 in yeast to 12 in Drosophila melanogaster (Michaud et al 2002), 16 in Caenorhabditis elegans (Candido 2002), and 19 in Arabidopsis thaliana (Scharf et al 2001). An exhaustive analysis of the A thaliana genome revealed an additional 25 genes that were more distantly related but clearly coded for proteins that contained one or more α-crystallin domains (Scharf et al 2001).
In humans, 9 α-crystallin–related sHsps have until now been recognized (Kappé et al 2001), for which the formal names HspB1–HspB9 have been proposed in accordance with the guidelines of the HUGO Gene Nomenclature Committee (Wain et al 2002). The best-known representatives are Hsp27/HspB1, αB-crystallin/HspB5, and Hsp20/HspB6. Other low–molecular weight Hsps, like Hsp32 (human heme oxygenase), are sometimes also indicated as sHsps but lacking the α-crystallin domain, they do not belong to this gene family. The 9 genuine human sHsps differ considerably in properties—as far as known—and in distribution, although most of them occur most abundantly in various types of muscle cells. To explore and to eventually understand the whole array of structures and functions of the human sHsps, it is a prerequisite to first know the actual number of sHsp genes that can be expressed in humans. We therefore performed exhaustive BLAST searches for sHsp-coding sequences in the human genome drafts of the Celera Discovery System (CDS) and the National Center for Biotechnology Information (NCBI) public database, which recovered a 10th sHsp, as such designated HspB10. This protein is well known already as outer dense fiber protein 1 (ODF1; Gastmann et al 1993) but has not been recognized earlier in the literature as a member of the sHsp family. It appears that not more than these 10 active sHsp genes can currently be recognized in the human genome.
MATERIALS AND METHODS
Sequences used in searches
The accession numbers of the known human sHsps that were used as queries to search the human genome are listed in Table 1 (under protein accession number). This includes the newly recognized ODF1/HspB10. Also, the mouse or rat orthologs of these 10 sHsps were used as queries (HspB1, accession number P14602; HspB2, Q99PR8; HspB3, Q9QZ57; HspB4, P02490; HspB5, P23927; HspB6, P97541; HspB7, P35385; HspB8, Q9JK92; HspB9, Q9DAM3; HspB10, Q61999) as well as various more distantly related animal sHsp sequences: Xenopus laevis Hsp30C (P30218); C elegans C14F11.5 (Q17992), T27E4.3 (P02513), C14B9.1 (P34328), and C09B8.6 (Q17849); D melanogaster Hsp22 (P02515) and Hsp26 (P02517); Artemia franciscana p26 (AF031367); and Schistosoma mansoni p40 (P12812). Searches were performed both with the complete sequences and with their α-crystallin domains alone.
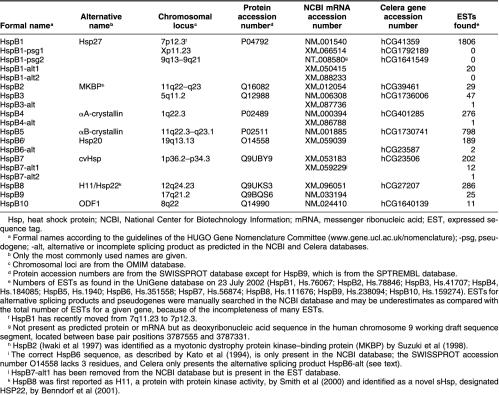
Table 1.
Small Hsp genes, pseudogenes, and putative alternative splicing products as retrieved from the Celera and NCBI human genome databases
Database searches
The human genome assembly constructed by Celera was searched using CDS (Kerlavage et al 2002), and the publicly available human genome database at NCBI (www.ncbi.nlm.nih.gov) was searched with the NCBI BLAST programs. BLASTP searches with our sHsp query sequences were performed on the protein databases that are derived by translation of the predicted genes in these versions of the genome. The entire human genome was also searched for sHsp-related sequences directly at the deoxyribonucleic acid (DNA) level, using translated nucleotide queries (TBLASTN). To find evidence for transcriptional activity of the retrieved genes and for the existence of predicted alternative splicing products, TBLASTN searches for these gene products were performed in the expressed sequence tag (EST) database at the NCBI site. All BLAST searches were performed using the default settings, except for the E value, which was set to 10 instead of 1 to increase the chances to recover more distantly related sHsps. As a control of our search procedures, we tried to find all earlier detected sHsp-related genes in the A thaliana genome (Scharf et al 2001) by BLASTP searches against its derived protein database (www.arabidopsis.org).
It is important to mention that the databases of CDS and NCBI are constantly upgraded; all computationally predicted genes and their translation products are going through a curation process, which means that specific information about a gene is subject to revision. It thus occurred that HspB1/Hsp27 and HspB9 had been curated as being obsolete in CDS databases in November 2001 and at that time were undetectable in the protein, messenger ribonucleic acid (mRNA), or gene databases. Similar inconsistencies were experienced for HspB7/cvHsp (see Results and Discussion) and may unknowingly have affected our results for other sHsps.
Identifying the α-crystallin domain
Demonstrating the presence of the α-crystallin domain is crucial for the identification of a sHsp. For sequences retrieved by CDS, we relied on its built-in protein classification system, which predicts protein domains—including the “Hsp20/α-crystallin domain”—by using the PROSITE, Pfam, PRINTS, ProDom, and SMART databases. The presence of an Hsp20/α-crystallin domain in protein sequences retrieved from NCBI was ascertained using the web-based ProfileScan server at the Swiss Institute for Bioinformatics (SIB; http://hits.isb-sib.ch/cgi-bin/PFSCAN/). This server uses PROSITE profiles and patterns and Pfam hidden Markov models to search for domains.
RESULTS AND DISCUSSION
Ten active sHsp genes in the human genome
An initial BLASTP search with the sequence of human HspB1/Hsp27 in the Celera protein database (October 2001) yielded the 9 already known sHsps (Kappé et al 2001), as well as a sperm tail ODF1 (Table 1). The alignment in Figure 1 reveals that ODF1 contains a genuine α-crystallin domain (positions 129–203). It thus belongs to the human sHsp family in which it can be designated as HspB10/ODF1. In addition, 2 hypothetical proteins were retrieved that correspond to the open reading frames of 2 HspB1/Hsp27 pseudogenes (Table 1, and see below). These 12 hits formed the uninterrupted top of the list in the results file, with E values ranging from 10−111 to 10−4. After these sHsps came a dozen hits, with E values quickly rising from 10−2 to 10. Using the protein classification provided by Celera, and the SIB ProfileScan server in cases of doubt, no α-crystallin domains were found among these hits. The same search procedure was repeated with all other human sHsps, including HspB10/ODF1, and with the 2 translated HspB1 pseudogenes. Between the queries, differences were found in the order and number of hits and in the efficiency of finding the other sHsps; especially HspB9 and HspB10 were ineffective queries. No further candidate sHsps, with recognizable α-crystallin domains, were discovered. BLAST searches in the Celera human protein database were also performed with the rat or mouse orthologs of HspB1–10 and with more distantly related sHsps from X laevis, C elegans, D melanogaster, A franciscana, and S mansoni (for specifications, see Materials and Methods). Again, no additional sHsps were found.
Fig 1.
Alignment of the 10 human small heat shock proteins (sHsps). Intron positions and predicted secondary structure elements are indicated by arrowheads above and arrows (for ß strands) below the alignment, respectively. The α-crystallin domain is approximately located between positions 118 and 203. Sequences correspond to the protein accession numbers in Table 1 (but see footnote i for HspB6). The alignment is made with ClustalW and manually edited in GeneDoc. Residues in black are conserved in 8 or more sHsps and those in gray in 5–7. The secondary structure was predicted from the 10 aligned sequences with PHD (Rost and Sander 1994); predicted ß strands are numbered according to the approximate location of the corresponding ß strands in the crystal structures of M jannaschii Hsp16.5 (Kim et al 1998) and wheat Hsp16.9 (van Montfort et al 2001). Numbers above the arrowheads correspond to the HspB1–10 numbering: thus, in the HspB1 and HspB8 genes, 2 introns are present at the same positions, 152 (phase 1) and 175 (phase 2); HspB2 has a single phase 1 intron, at position 66; HspB3 and HspB9 are intronless; HspB4, HspB5, and HspB6 have 2 phase 0 introns at identical positions, between 117 and 118 and between 158 and 159; HspB7 has 2 introns, at positions 91 (phase 1) and between 157 and 158 (phase 0); HspB10 has 1 phase 2 intron, at position 107
Similar searches in the public human genome protein database, using the NCBI website, gave comparable results, and in addition, various predicted alternatively spliced sHsp forms were retrieved (Table 1, and see below). To further investigate the entire human genome directly at DNA level, also a TBLASTN screening of the NCBI and Celera genome databases was performed to search for sHsp-related sequences missed by the automated gene discovery process and therefore not present in the protein databases. These searches using the protein sequences of all 10 sHsp genes and 2 HspB1 pseudogenes as queries did not recover any new entries.
To validate our BLASTP search procedure, we applied precisely the same approach to the genome database of A thaliana. Performing BLASTP searches with a number of A thaliana sHsps (AtHsp17.4, AtAcd31.2, AtAcd28.1, AtAcd25.4, AtAcd21.4, and AtAcd55.2) we could retrieve all 44 sHsps and other “α-crystallin domain (Acd) proteins” as reported by Scharf et al (2001). This indicates that our BLASTP searches in the human genome should have detected such more distantly related α-crystallin domain–containing proteins.
The BLASTP searches on NCBI human database retrieved 2 predicted alternative splicing products for HspB1 and 1 each for HspB3, HspB4/αA-crystallin, HspB6/Hsp20, and HspB7/cvHsp (see Table 1). EST searches provided evidence that most of these predicted alternative splicing products were merely splicing intermediates or mispredictions based on cryptic splice sites in the gene sequences. EST searches further revealed a second alternative splicing product for HspB7/cvHsp (Table 1) and confirmed the absence of transcripts of the 2 HspB1 pseudogenes.
Alignment of the 10 sHsps that are encoded and expressed by the human genome (Fig 1) confirms again that the α-crystallin domain (approximately positions 118–203) is the only recognizably homologous region in all sHsps. In the N- and C-terminal regions, only short sequence motifs (eg, positions 56–67 and 219–223) are more or less conserved in most human sHsps. The structural conservation of the α-crystallin domain is supported by the fact that 6 of the ß strands as present in the crystal structures of Methanococcus jannaschii Hsp16.5 (Kim et al 1998) and wheat Hsp16.9 (van Montfort et al 2001) are correctly predicted from the alignment of the human sHsps, as indicated in Figure 1. However, the inability to predict in the human sHsps the 4 ß strands ß1, ß2, ß6, and ß10 also indicates considerable differences. Notably, the absence of the ß6 strand, which is important for dimer contacts in the sHsp complex (van Montfort et al 2002), suggests that these contacts are different between the human and plant or archaebacterial sHsps.
The dispersal of the 10 sHsp genes over 9 different chromosomes (Table 1) is a witness for the ancient duplications that have generated the human sHsp family. Even the head-to-head located genes for HspB2 and HspB5/αB-crystallin on chromosome 11 are highly divergent in sequence (Iwaki et al 1997). It is therefore difficult, if not impossible, to reliably resolve the gene phylogeny of all 10 human sHsps. Using sophisticated, likelihood-based methods for phylogenetic inference (Whelan et al 2001), we only found consistent support for the grouping of HspB4/α-crystallin, HspB5/αB-crystallin, and HspB6/Hsp20 as the most closely related ones among the 10 human sHsps (data not shown). This relationship actually is also clear from some uniquely shared amino acid replacements (eg, positions 38W, 41R, 151H, 171H, 175R in Fig 1) and seems to be confirmed by the common intron-exon structures of HspB4, HspB5, and HspB6, as indicated in Figure 1. However, the genes for HspB1 and HspB8 also precisely share the positions of their 2 introns but do not group together on the basis of their sequence resemblance. Just like the absence of introns in the genes for HspB3 and HspB9, common intron positions are difficult to interpret phylogenetically in view of the ongoing introns early/late debate (De Souza et al 1998).
The human sHsps and their genes for which new features have been revealed are further discussed in the following paragraphs.
HspB1/Hsp27
One active and 2 pseudogenes for HspB1/Hsp27 were found in the human genome databases (Table 1). The sequences of the active gene and pseudogene 1, located on chromosomes 7 and X, respectively, correspond to those reported by Hickey et al (1986). Pseudogene 2 is located on chromosome 9, in agreement with the assignment of Hsp27 sequences to this chromosome by McGuire et al (1989). As already described by Hickey et al (1986), pseudogene 1 is a classical intronless retropseudogene. It has 3 deletions and 16 nonsynonymous and 26 synonymous point mutations as compared with the coding region of the original HspB1 gene (Fig 2). Despite these deletions and base substitutions, it has an uninterrupted open reading frame. Pseudogene 2 is a 5′-truncated semiprocessed retropseudogene (Fig 2). It lacks exon 1, starts at the 3′ end of intron 1, and further comprises exons 2 and 3. It also has an open reading frame, starting at the first ATG in exon 2. As mentioned already, no transcripts of these pseudogenes are found in the EST database. Both pseudogenes 1 and 2 are flanked by a poly-A stretch at the 3′ end, as well as at the 5′ end. HspB1 is the only human sHsp gene for which pseudogenes have been found. Orthologous HspB1 retropseudogenes could not be detected in the mouse genome database of Celera, suggesting their relatively recent origin.
Fig 2.
Alignment of the open reading frames and flanking intron sequences of the HspB1/Hsp27 gene and its 2 processed pseudogenes. The alignment was made with ClustalW and edited with GeneDoc. The 2 introns in the HspB1 gene are in gray. Sequence differences are highlighted in black; the black nucleotides from positions 390 to 425 in pseudogene 2 demarcate its 5′ boundary. Start and stop codons are underlined (the potential start codon for pseudogene 2 is at positions 468–470). The GA and AG sequences in bold at positions 343–344 and 615–616 indicate the alternative splice sites in exon 1 and exon 3 that are predicted to give rise to HspB1-alt2 (see Table 1 and text)
The NCBI protein database contains 2 computationally predicted proteins that could result from incomplete or alternative splicing of the HspB1 pre-mRNA (Table 1). In HspB1-alt1, intron 1 is not spliced out of the pre-mRNA, and on translation, the first in-frame stop codon in intron 1 would be used. This could yield the normal HspB1 protein sequence up to the middle of the α-crystallin domain (position 151 in Fig 1), followed by a completely different C-terminal region of 65 amino acids. Twenty ESTs were found for HspB1-alt1, representing only about 1% of all ESTs for HspB1. Given this low abundance of ESTs, and the questionable viability of the partially out-of-frame translation product, this gene transcript is unlikely to have any physiological relevance. HspB1-alt2 is based on the presence of assumed cryptic splice sites: a nonconsensus GA splice donor site in exon 1 and a consensus AG splice acceptor site in exon 3 (indicated in Fig 2). No ESTs were found for HspB1-alt2 nor for any splicing product combining a cryptic with a regular splice site, suggesting that the cryptic splice sites are not used.
Heat shock protein B3
Although the HspB3 gene is intronless (Fig 1), the NCBI protein database contains a predicted protein (HspB3-alt in Table 1) that would result from splicing of a 117-bp pseudointron, bordered by a nonconsensus intron-exon boundary, GC-AG. This spliced mRNA would be translated in an HspB3 protein missing the 39 residues corresponding to positions 37–79 in Figure 1. Because we found only a single EST for this sequence, among the 47 for HspB3, the removal of this pseudointron must be a rare event.
HspB4/αA-crystallin
A predicted case of incomplete splicing of HspB4/αA-crystallin was found in the NCBI protein database that is comparable with that of HspB1-alt1, ie, in which intron 1 is still present (HspB4-alt in Table 1). Translation of the mRNA would yield the normal N-terminal region of HspB4/αA-crystallin, up to position 118 in Figure 1, followed by a completely different C-terminal region of 72 residues. With only 1 detected EST among the 276 for HspB4, it is likely to represent a pre-mRNA or splicing intermediate. Interestingly, no ESTs were found for the well-known alternative splice variant αAins-crystallin, in which an optional 69-bp pseudoexon is spliced in between the regular exons 1 and 2 because it is flanked by a nonconsensus GC 5′ donor splice site (King and Piatigorsky 1983). EST searches were specifically performed with the sequence of the human optional exon (Jaworski and Piatigorsky 1989). αAins-crystallin is quite abundantly expressed in rodents and in some other mammals (Hendriks et al 1988) but has not been observed at protein level in humans, probably because of a frame-shift mutation in the optional exon. However, the human optional exon is flanked by the same splice site sequences as in rodents and the alternatively spliced αAins-crystallin mRNA could therefore be expected to be present, unless the regulation of alternative splicing is different in humans and rodents. Alternatively, any possible human αAins-crystallin mRNA might be rapidly degraded because insertion of the optional exon causes out-of-frame translation of exon 2, up to a stop codon 81 nucleotides upstream of intron 2. This will lead to nonsense-mediated mRNA decay, which occurs when translation terminates more than 50–55 nucleotides upstream of the 3′-most exon-exon junction (Maquat 2002). The same decay mechanism may actually also operate in the case of HspB1-alt1 and HspB4-alt.
HspB6/Hsp20
The HspB6/Hsp20 sequence from the NCBI protein database as given in Figure 1 corresponds to the protein sequence as originally determined by Kato et al (1994) but differs from the SwissProt entry (O14558). The latter lacks 3 residues, VAQ at positions 115–117 in Figure 1, corresponding to the 3′ end of exon 1. Also, because all HspB6 sequences in the NCBI human EST database have the VAQ insert, we conclude that the HspB6 sequence in Figure 1 is the correct one. The Celera protein database predicts only an alternative form of HspB6 and not HspB6 itself. This HspB6-alt (Table 1) has the sequence of HspB6 as shown up to position 201 in Figure 1, including the VAQ insert, but the 15 C-terminal residues have been replaced by a completely different 51-residue sequence. It would result from the removal of an optional 59-bp intron (including the normal stop codon) from the third exon by the use of a cryptic splice site pair GC-GG. One could imagine that this yields a viable C-terminally elongated form of HspB6. However, only 2 of the 189 HspB6 ESTs could be confirmed to correspond to HspB6-alt (Table 1).
HspB7/cvHsp
TBLASTN searches in the Celera database and in the NCBI working draft of February 2002 retrieved 2 linked and highly similar copies of the HspB7 gene, located on chromosome 1. In both databases, 1 of the copies was incomplete at the 5′ coding end, but the order and distance of the 2 copies differed. In a newer version of the NCBI working draft (May 2002), only the incomplete HspB7 gene was present and at a different position. Genome assembly in this part of chromosome 1 is clearly still in progress.
HspB7-alt1 was predicted in the NCBI protein database. It would result from the use of a start codon in intron 1, 45 nucleotides upstream of the regular 3′ splice site of this intron. The deduced protein would therefore have the normal HspB7 sequence from position 91 onward (cf Fig 1), preceded by a deviating N-terminal region of 15 residues: MQGLLHASLTAAHPT. HspB7-alt1 is represented by 12 ESTs, as compared with a total of 202 ESTs for HspB7, which makes it an intriguing gene product, especially because the alt-1 ESTs are predominantly present in muscle rhabdomyosarcoma.
EST searches revealed a single entry of a second alternatively spliced variant of HspB7, HspB7-alt2 (Table 1). It results from the use of an alternative 3′ splice site AG, located 12 bp upstream of the regular 3′ splice site of intron 1, and would cause an insert of 4 amino acids (AHPT) between positions 90 and 91 in Figure 1. Krief et al (1999), who first described HspB7/cvHsp, reported a similarly located insert in a complementary DNA (cDNA) clone, although with the sequence AAHPT.
On basis of EST and cDNA analyses, Krief et al (1999) proposed an HspB7 gene organization with 4 introns. We inferred from the genomic data the presence of 2 introns (Fig 1), of which the first one, at position 91, was correctly predicted by Krief et al (1999).
HspB10/ODF1
Like HspB9 (Kappé et al 2001), the newly recognized HspB10/ODF1 has a testis-specific expression (Gastmann et al 1993), which was confirmed by our present EST analyses. With a length of 250 residues and a mass of 28.4 kDa, HspB10/ODF1 is the largest and most deviating among the human sHsps. Most conspicuous are the 13 PCX repeats in the C-terminal extension, where X is mostly S or N (positions 203–242 in Fig 1). This region is predicted to form a coiled coil (Shao and van der Hoorn 1996). The other regions of the protein are rich in cysteines too; thereby, the total cysteine amounts to 14%. ODF1 is a major component of ODF of the sperm tail, which is assembled along the axoneme during development of the sperm and functions as a passive elastic structure, providing elastic recoil for the sperm tail (Baltz et al 1990). ODF1 is a highly insoluble protein, also because of the presence of disulfide bonds, and thus will be unable to act as a soluble chaperone-like protein, like most other sHsps. ODF1 can self-interact in the yeast 2-hybrid system and forms multimers in vitro, probably assisted by a putative leucine zipper dimerization motif in the N-terminal region (Shao and van der Hoorn 1996). A 27-bp deletion polymorphism in the PCX repeat region has been described in humans but is not associated with fertility problems (Hofferbert et al 1993).
Although human and mouse HspB9 show the highest protein sequence divergence among the orthologous sHsps in these 2 species (Kappé et al 2001), human and mouse HspB10/ODF1 are almost identical (96%), except for 4 additional PCX repeats and a deletion of 15 amino acids right before the intron (positions 92–107 in Fig 1) in the mouse sequence.
CONCLUSIONS
On the basis of our analyses of the current genome databases from Celera and NCBI, we could not detect more than 10 different sHsps as being encoded and expressed by the human genome. HspB9 and HspB10/ODF1 are specifically expressed in the testis and HspB4/αA-crystallin in the eye lens. The other 7 sHsps are more ubiquitous in their expression, although most of them typically occur in muscle and heart. Our searches revealed a considerable number of predicted alternative and intermediate splicing products, although most of these were not detected at appreciable levels in the EST databases. However, heat shock is known since long to interfere with the splicing of pre-mRNA (eg, Bond 1988). It may therefore be possible that some of the alternative splicing products are more abundant under heat shock conditions.
Considering that orthologs of the 10 human sHsps are present in mouse and rat, it is to be expected that the same 10 orthologs will be present in all mammalian species, and probably in other higher vertebrates (birds and reptiles) as well. In view of the exhaustive search procedures used in the present study, including the EST databases, it seems unlikely that additional genuine sHsp genes will still be detected as encoded and expressed by the human genome, although the presence of genes with more marginal homology can never be excluded. Thus, it is interesting to note that no apparent orthologs of the typical lower vertebrate Hsp30 subfamily of sHsps, as found in multiple copies in X laevis (Abdulle et al 2002) and the fish Poeciliopsis lucida (Norris and Hightower 2002), have been traced in the human genome. Only HspB9 has the tendency to cluster with these Hsp30 sequences in our phylogenetic analyses (data not shown), but whether this reflects genuine orthology may be difficult to establish.
The greatest challenge now is to explore and understand the functional characteristics and physiological properties of this broad array of vertebrate sHsps.
Acknowledgments
We thank Ole Madsen for performing phylogenetic analyses on the human sHsp sequences.
REFERENCES
- Abdulle R, Mohindra A, Fernando P, Heikkila JJ. Xenopus small heat shock proteins, Hsp30C and Hsp30D, maintain heat- and chemically denatured luciferase in a folding-competent state. Cell Stress Chaperones. 2002;7:6–16. doi: 10.1379/1466-1268(2002)007<0006:xshsph>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo AP, Müller WEG. (eds). 2002 Small Stress Proteins,. Springer Verlag, Berlin. [Google Scholar]
- Arrigo AP, Paul C, Ducasse C, Sauvageot O, Kretz-Remy C. Small stress proteins: modulation of intracellular redox state and protection against oxidative stress. Prog Mol Subcell Biol. 2002a;28:171–184. doi: 10.1007/978-3-642-56348-5_9. [DOI] [PubMed] [Google Scholar]
- Arrigo AP, Paul C, and Ducasse C. et al. 2002b Small stress proteins: novel negative modulators of apoptosis induced independently of reactive oxygen species. Prog Mol Subcell Biol. 28:185–204. [DOI] [PubMed] [Google Scholar]
- Baltz JM, Williams PO, Cone RA. Dense fibers protect mammalian sperm against damage. Biol Reprod. 1990;43:485–491. doi: 10.1095/biolreprod43.3.485. [DOI] [PubMed] [Google Scholar]
- Benndorf R, Sun X, and Gilmont RR. et al. 2001 HSP22, a new member of the small heat shock protein superfamily, interacts with mimic of phosphorylated HSP27 (3DHSP27). J Biol Chem. 276:26753–26761. [DOI] [PubMed] [Google Scholar]
- Bond U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988;7:3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido EPM. The small heat shock proteins of the nematode Caenorhabditis elegans: structure, regulation and biology. Prog Mol Subcell Biol. 2002;28:61–78. doi: 10.1007/978-3-642-56348-5_4. [DOI] [PubMed] [Google Scholar]
- Carver JA. Probing the structure and interactions of crystallin proteins by NMR spectroscopy. Prog Ret Eye Res. 1999;18:431–462. doi: 10.1016/s1350-9462(98)00027-5. [DOI] [PubMed] [Google Scholar]
- Charette SJ, Landry J. The interaction of HSP27 with Daxx identifies a potential regulatory role of HSP27 in Fas-induced apoptosis. Ann N Y Acad Sci. 2000;926:126–131. doi: 10.1111/j.1749-6632.2000.tb05606.x. [DOI] [PubMed] [Google Scholar]
- Ciocca DR, Vargas-Roig LM. Hsp27 as a prognostic and predictive factor in cancer. Prog Mol Subcell Biol. 2002;28:205–218. doi: 10.1007/978-3-642-56348-5_11. [DOI] [PubMed] [Google Scholar]
- Davidson SM, Loones M-T, Duverger O, Morange M. The developmental expression of small HSP. Prog Mol Subcell Biol. 2002;28:103–128. doi: 10.1007/978-3-642-56348-5_6. [DOI] [PubMed] [Google Scholar]
- De Souza SJ, Long M, Klein RJ, Roy S, Gilbert W. Toward a resolution of the introns early/late debate: only phase zero introns are correlated with the structure of ancient proteins. PNAS. 1998;95:5094–5099. doi: 10.1073/pnas.95.9.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaestel M. sHsp-phosphorylation: enzymes, signalling pathways and functional implications. Prog Mol Subcell Biol. 2002;28:151–169. doi: 10.1007/978-3-642-56348-5_8. [DOI] [PubMed] [Google Scholar]
- Gastmann O, Burfeind P, Guenther E, Hamerster H, Szpirer C, Hoyer-Fender S. Sequence, expression, and chromosomal assignment of a human sperm outer dense fiber gene. Mol Reprod Dev. 1993;36:407–418. doi: 10.1002/mrd.1080360402. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Buchner J. Chaperone function of sHsps. Prog Mol Subcell Biol. 2002;28:37–59. doi: 10.1007/978-3-642-56348-5_3. [DOI] [PubMed] [Google Scholar]
- Hendriks W, Sanders J, de Leij L, Ramaekers F, Bloemendal H, de Jong WW. Monoclonal antibodies reveal evolutionary conservation of alternative splicing of the αA-crystallin primary transcript. Eur J Biochem. 1988;174:133–137. doi: 10.1111/j.1432-1033.1988.tb14072.x. [DOI] [PubMed] [Google Scholar]
- Hickey E, Brandon SE, Potter R, Stein G, Stein J, Weber LA. Sequence and organization of genes encoding the human 27 kDa heat shock protein. Nucleic Acids Res. 1986;14:4127–4145. doi: 10.1093/nar/14.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofferbert S, Burfeind P, Hoyer-Fender S, Lange R, Haidl G, Engel W. A homozygous deletion of 27 basepairs in the coding region of the human outer dense fiber protein does not result in a pathogenic phenotype. Hum Mol Genet. 1993;2:2167–2170. doi: 10.1093/hmg/2.12.2167. [DOI] [PubMed] [Google Scholar]
- Iwaki A, Nagano T, Nakagawa M, Iwaki T, Fukumaki Y. Identification and characterization of the gene encoding a new member of the α-crystallin/small hsp family, closely linked to the αB-crystallin gene in a head-to-head manner. Genomics. 1997;45:386–394. doi: 10.1006/geno.1997.4956. [DOI] [PubMed] [Google Scholar]
- Jaworski CJ, Piatigorsky J. A pseudo-exon in the functional human αA-crystallin gene. Nature. 1989;337:752–754. doi: 10.1038/337752a0. [DOI] [PubMed] [Google Scholar]
- Kappé G, Leunissen JAM, de Jong WW. Evolution and diversity of prokaryotic small heat shock proteins. Prog Mol Subcell Biol. 2002;28:1–17. doi: 10.1007/978-3-642-56348-5_1. [DOI] [PubMed] [Google Scholar]
- Kappé G, Verschuure P, Philipsen RL, Staalduinen AA, van de Boogaart P, Boelens WC, de Jong WW. Characterization of two novel human small heat shock proteins: protein kinase-related HspB8 and testes-specific HspB9. Biochim Biophys Acta. 2001;1520:1–6. doi: 10.1016/s0167-4781(01)00237-8. [DOI] [PubMed] [Google Scholar]
- Kato K, Goto S, Inaguma Y, Hasegawa K, Morishita R, Asano T. Purification and characterization of a 20-kDa protein that is highly homologous to αB crystallin. J Biol Chem. 1994;269:15302–15309. [PubMed] [Google Scholar]
- Kato K, Ito H, Inaguma Y. Expression and phosphorylation of mammalian small heat shock proteins. Prog Mol Subcell Biol. 2002;28:129–150. doi: 10.1007/978-3-642-56348-5_7. [DOI] [PubMed] [Google Scholar]
- Kerlavage A, Bonazzi V, and di Tommaso M. et al. 2002 The Celera Discovery System™. Nucleic Acids Res. 30:129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Kim R, Kim S-H. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- King CR, Piatigorsky J. Alternative RNA splicing of the murine αA-crystallin gene: protein-coding information within an intron. Cell. 1983;32:707–712. doi: 10.1016/0092-8674(83)90056-9. [DOI] [PubMed] [Google Scholar]
- Krief S, Faivre JF, and Robert P. et al. 1999 Identification and characterization of cvHsp. A novel human small stress protein selectively expressed in cardiovascular and insulin-sensitive tissues. J Biol Chem. 274:36592–36600. [DOI] [PubMed] [Google Scholar]
- Krueger-Naug AM, Plumier JC, Hopkins DA, Currie RW. Hsp27 in the nervous system: expression in pathophysiology and in the aging brain. Prog Mol Subcell Biol. 2002;28:235–251. doi: 10.1007/978-3-642-56348-5_13. [DOI] [PubMed] [Google Scholar]
- Latchman DS. Protection of neuronal and cardiac cells by HSP27. Prog Mol Subcell Biol. 2002;28:253–265. doi: 10.1007/978-3-642-56348-5_14. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay. Curr Biol. 2002;12:R196–R197. doi: 10.1016/s0960-9822(02)00747-9. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Fuqua SAW, Naylor SL, Helin-Davis DA, McGuire WL. Chromosomal assignments of human 27-kDa heat shock protein gene family. Somat Cell Mol Genet. 1989;15:167–171. doi: 10.1007/BF01535078. [DOI] [PubMed] [Google Scholar]
- Michaud S, Morrow G, Marchand J, Tanguay RM. Drosophila small heat shock proteins: cell and organelle-specific chaperones. Prog Mol Subcell Biol. 2002;28:79–101. doi: 10.1007/978-3-642-56348-5_5. [DOI] [PubMed] [Google Scholar]
- Narberhaus F. α-Crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev. 2002;66:64–93. doi: 10.1128/MMBR.66.1.64-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris CE, Hightower LE. Discovery of two distinct small heat shock protein (HSP) families in the desert fish Poeciliopsis. Prog Mol Subcell Biol. 2002;28:19–35. doi: 10.1007/978-3-642-56348-5_2. [DOI] [PubMed] [Google Scholar]
- Quinlan R. Cytoskeletal competence requires protein chaperones. Prog Mol Subcell Biol. 2002;28:219–234. doi: 10.1007/978-3-642-56348-5_12. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Scharf K-D, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (Acd proteins) Cell Stress Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, van der Hoorn FA. Self-interaction of the major 27-kilodalton outer dense fiber protein is in part mediated by a leucine zipper domain in the rat. Biol Reprod. 1996;55:1343–1350. doi: 10.1095/biolreprod55.6.1343. [DOI] [PubMed] [Google Scholar]
- Smith CC, Yu YX, Kulka M, Aurelian L. A novel human gene similar to the protein kinase (PK) coding domain of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) codes for a serine-threonine PK and is expressed in melanoma cells. J Biol Chem. 2000;275:25690–25699. doi: 10.1074/jbc.M002140200. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Sugiyama Y, and Hayashi Y. et al. 1998 MKBP, a novel member of the small heat shock protein family, binds and activates the myotonic dystrophy protein kinase. J Cell Biol. 140:1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfort RL, Slingsby C, Vierling E. Structure and function of the small heat shock protein/α-crystallin family of molecular chaperones. Adv Protein Chem. 2002;59:105–156. doi: 10.1016/s0065-3233(01)59004-x. [DOI] [PubMed] [Google Scholar]
- van Montfort RLM, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- Wain HM, Bruford EA, Lovering RC, Lush MJ, Wright MW, Povey S. Guidelines for human gene nomenclature. Genomics. 2002;79:464–470. doi: 10.1006/geno.2002.6748. [DOI] [PubMed] [Google Scholar]
- Whelan S, Liò P, Goldman N. Molecular phylogenetics: state-of-the-art methods for looking into the past. Trends Genet. 2001;17:262–272. doi: 10.1016/s0168-9525(01)02272-7. [DOI] [PubMed] [Google Scholar]
- Wieske M, Benndorf R, Behlke J, Dolling R, Grelle G, Bielka H, Lutsch G. Defined sequence segments of the small heat shock proteins HSP25 and αB-crystallin inhibit actin polymerization. Eur J Biochem. 2001;268:2083–2090. doi: 10.1046/j.1432-1327.2001.02082.x. [DOI] [PubMed] [Google Scholar]