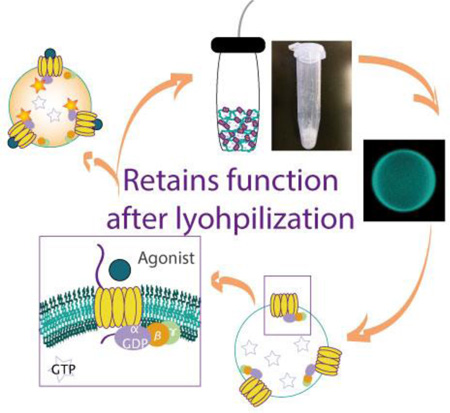
Abstract
We successfully incorporated the human serotonin receptor, G protein coupled receptor (GPCR), 5-HT1AR, in micron scale polymeric giant unilamellar protein-vesicles (pGUPs). By utilizing an agarose rehydration technique for protein incorporation, the GPCR is inserted in biased orientation with the C-terminus cytosolic and N-terminus extracellular as found in the cell plasma membrane. The GPCR is fully functional within the polymeric bilayer exhibiting responses to various ligands. The entire population of incorporated GPCRs displaying activity in pGUPs remains fully functional after lyophilization for 120 hours.
Keywords: block copolymer, GPCR, lyohphilization, polymersome, bilayer vesicle
Graphical Abstract
GPCR is incorporated into polymeric vesicles made up diblock copolymer bilayers. Successfully incorporated GPCRs exhibit correct biased physiological orientation and respond to various ligands. After extended dehydrated storage via lyophilization and subsequent rehydration, diblock copolymer polymersomes retain their shape and incorporated GPCR retains its function.
Since their discovery in 1999 polymersomes have been used as biomimetic platforms to better understand physiological and material properties of cells.[1] Compared to their liposomal counterparts, polymersomes display greater stability and decreased permeability[2, 3] and therefore have been exploited and hybridized with cellular components such as lipids and proteins for drug delivery and research.[4] For example, phase separation has been modeled in lipid-polymer vesicles offering a platform for “windows” of lipid bilayers to be observed in a polymeric framework.[5] The encapsulation of nanometer sized polymersomes within giant unilamellar vesicles (GUVs) further mimics the compartmentalization of living cells.[6] In recent years protein incorporation into lamellar phase polymeric vesicles has been achieved.[7, 8] The incorporation of proteins into polymersomes offers robust platforms for drug discovery and functional screening, though current efforts remain limited. Here, we present a robust platform incorporating G protein-coupled receptors (GPCRs) into diblock copolymer bilayer vesicles that retain protein function after lyophilization and rehydration.
G protein-coupled receptors (GPCRs) are a class of proteins targeted for membrane vesicle incorporation.[9] As druggable targets, there exists over 800 GPCRs with almost half of the therapeutics on the market targeting these proteins.[10] GPCRs are integral membrane protein receptors characterized by seven transmembrane alpha helices. They are associated with cytoplasmic G proteins consisting of α, β, and γ subunits. Binding of an extracellular agonist to the receptor causes a conformational change and dissociation of the G subunits into Gα and βγ. Exchange of GDP for GTP on the Gα subunit results in intracellular signal cascades responsible for many cellular processes such as apoptosis, proliferation and changes in intracellular cyclic adenosine monophosphate (cAMP) levels.[9, 11]
Incorporation of GPCRs into polymersomes via cell free expression has been previously demonstrated. In 2013, May et al. incorporated the dopamine receptor D2 (DRD2) into polymeric vesicles.[12] However, they only observed ligand binding on nanoscale vesicles and were not able to show functionality since the G proteins were not present. Using the same approach de Hoog et al. incorporated the chemokine C-X-C motif receptor 4 (CXCR4) into polymersomes and tracked its binding to antibodies via surface plasmon resonance.[13] While these approaches incorporate GPCRs into polymersomes they are limited by 1) the lack of cognizant G protein subunits, 2) inability to observe and probe receptor activity, and 3) liposomal sized vesicles of 100–150 nm make them inaccessible to common light microscopy. To overcome these limitations we present a robust platform for incorporation of GPCRs into diblock copolymer giant unilamellar polymersomes in the micrometer range that allows for observations of GTP/GDP exchange on G proteins catalyzed by the human serotonin 5-HT1A receptor (5-HT1AR). We further exploit the stability of polymersomes and show that following lyophilization and rehydration, diblock copolymer giant unilamellar protein-vesicles (pGUPs) with integrated GPCRs retain vesicle integrity and protein function.
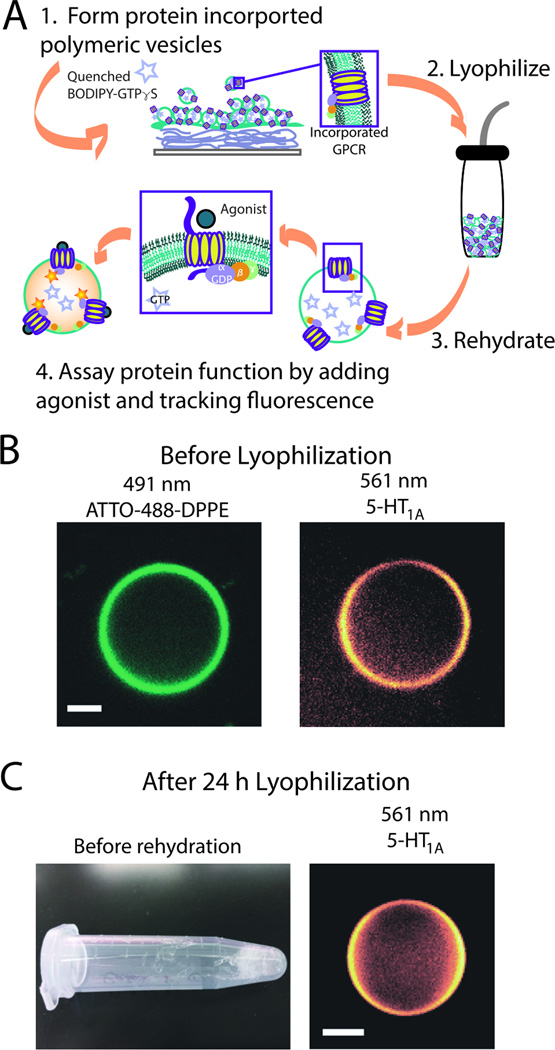
We utilize the agarose hydration method that we have previously reported for the incorporation of 5-HT1AR into giant unilamellar vesicles (Figure S1–S2).[14–16] We incorporate membrane preparations of 5-HT1AR with associated G proteins into polymeric membranes made of polybutadiene-b-poly(ethylene oxide) (PBd(650)-PEO(400)) at a polymer-to-protein molar ratio of 100:1. To detect the functionality of 5-HT1AR in the diblock copolymer bilayers, pGUPs were formed to encapsulate BODIPY-GTPγS, a quenched fluorophore. When an agonist binds to 5-HT1AR on the pGUPs, G protein subunits exchange bound GDP for BODIPY-GTPγS and this exchange unquenches its fluorescence (Figure 1 and S1–S2). Using this system we detect the response of 5-HT1AR in the presence of different antagonists and further show retained protein function after lyophilizing and rehydrating pGUPs.
Figure 1.
GPCR incorporation into diblock copolymer bilayer vesicles, pGUPs. (A) Schematic of pGUP formation and protein incorporation. Films of protein, agarose, and polymer are made on a coverslip and rehydrated with a sucrose buffer solution containing BODIPY-GTPγS. pGUPs formed of diblock copolymer bilayers can be lyophilized and the GPCR retains its function (steps 2–4); for enlarged image see Supporting Information (Figure S1). (B) Confocal micrographs of pGUPs prior to lyophilization. The left micrograph shows the polymer bilayer tagged with ATTO-488-DPPE. The right micrograph shows that rhodamine antibody-tagged 5-HT1AR is evenly distributed throughout the polymer bilayer. (C) The left image shows a pGUP sample after lyophilization. Upon rehydration, pGUPs can still be detected as shown in the right micrograph. All scale bars represent 5 µm.
Figure 1B and 1C show the successful incorporation of 5-HT1AR membrane preparations into diblock copolymer bilayer membranes. The GPCRs, visualized using a rhodamine-labeled anti-5-HT1AR antibody, are evenly distributed throughout the bilayer in both non-lyophilized (Figure 1B, right panel) and lyophilized pGUPs (Figure 1C, right panel). Vesicles can be formed without the GPCR or ATTO-488-DPPE and cannot be formed by ATTO-488-DPPE alone or by ATTO-488-DPPE with the GPCR, confirming the polymeric makeup of the vesicles. (Figure S3). This result is consistent with the diblock copolymer taking on an I-shape or unfolded structure,[17] and forming a bilayer vesicle as depicted in Figure 1A and Figure S1.
To determine if 5-HT1AR is correctly oriented in pGUPs, rhodamine tagged anti-5-HT1AR or anti-G protein antibodies were incubated with the membrane preparations of 5-HT1AR prior to incorporation into pGUPs (Figure S2–S8). The monoclonal 5-HT1AR antibody binds to the cytosolic (C-terminal) face of the receptor. The incorporation of the antibody during the preparation results in antibody inside the pGUPs for the correctly oriented GPCRs (C-terminus cytosolic and the N-terminus extracellular), and outside the pGUPs for the incorrect orientation. The rhodamine signal on the vesicle exterior was quenched with the membrane impermeable fluorescence-quenching agent QSY7. Since only fluorophore exterior to the vesicles is quenched, the fraction of quenched fluorescence corresponds to the fraction of receptors oriented with their C-terminus on the outer face of the vesicle. Results of retained fluorescence after quenching are presented in Figure S4. Less than 10% of the receptor-labeled antibody fluorescence is quenched, indicating that 5-HT1AR displays a biased orientation with the C-terminus cytosolic and the N-terminus extracellular. In contrast, the G proteins are anchored to the membrane via a single tether, lacking the complex structure needed for membrane orientation bias. This was confirmed by the quenching experiment, since only ~55% of the G protein-labeling antibody fluorescence was retained after quenching (Figure S4). The results indicate the formation of pGUPs using the agarose technique is likely initiated by the formation of nanoscale liposomes, where the high curvature causes proteins to orient themselves during the agarose rehydration process [18, 19] before the coalescence of vesicles to form giant vesicles on the micrometer scale (Figure S9).
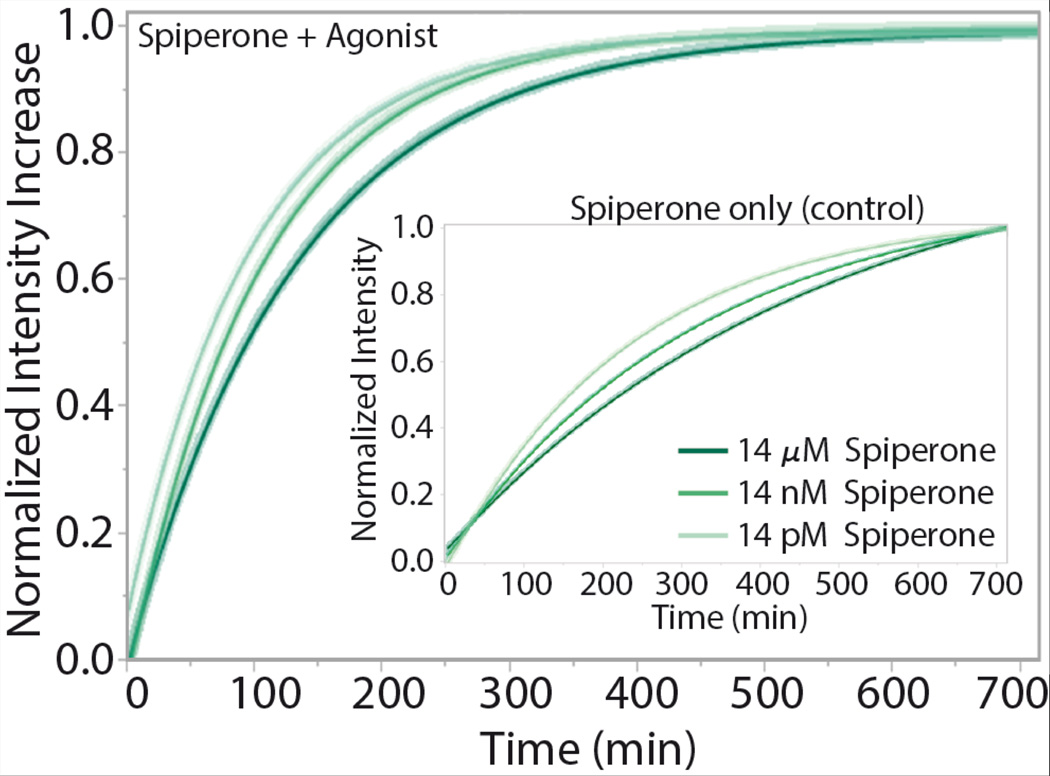
To observe protein function in the synthetic polymer bilayers, pGUPs were formed, settled in glucose and transferred to a 96-microtiter plate. pGUPs were formed in the presence of a receptor antagonist, spiperone (final concentration 14 µM unless otherwise stated), to reduce protein basal activity. Upon initiation of receptor function experiments, the agonist 8-Hydroxy-2-(dipropylamino)tetralin hydrobromide (8-OH-DPAT) was added to the vesicle suspension and the system was incubated at 37°C for 12 hours. Fluorescence unquenching of BODIPY-GTPγS due to G protein oligonucleotide exchange was monitored every 5 minutes. Control pGUVs without protein do not display any nonspecific BODIPY-GTPγS unquenching (Figure S10). Tracked fluorescence curves over time were first calculated as percent intensity increase to account for variation in sample size and are averages of six independent replicates of the same experimental protocol. Fluorescence intensity increase curves were then normalized to one for ease in viewing the differences in protein functional rate. Rates are obtained from single exponential fitting on the fluorescence intensity increase vs time curves. Agonist activated fluorescence intensity of the pGUPs displays a much faster rate than the no-agonist control indicating that 5-HT1AR function is agonist induced in the diblock copolymer membranes (Figure 2).
Figure 2.
5-HT1AR in pGUPS display response to increasing antagonist concentration while keeping agonist concentration constant. Fluorescence unquenching due to the irreversible binding of BODIPY-GTPγS to G proteins was tracked for 12 hours for pGUPs formed with increasing amount of the antagonist (spiperone) and constant amount of agonist. Increasing the amount of antagonist in the system decreases the protein functional rate (See Table 1, for change in intensity rates). The inset shows control curves for the pGUPs that were incubated without agonist. 5-HT1AR basal activity is captured in the pGUPs lacking agonist, spiperone only (control).
5-HT1AR in pGUPs displays a dose-dependent response with respect to the antagonist. Reducing the final concentration of spiperone in pGUPs from 14 µM to 14 pM and keeping the concentration of added agonist constant results in an expected increase in the protein functional rate (Table 1, Figure S11). Furthermore, other known 5-HT1AR antagonists, methiothepin maleate (methiothepin), NAN-190, and WAY 100635, that are known to inhibit agonist binding more strongly than spiperone also affect the protein functional rate.[20–22] Thus pGUPs as fabricated are sensitive to spiperone concentration and show a reduction in rate in the presence of other, more tightly binding, anatagonists.
Table 1.
5-HT1AR in pGUP response to changes in antagonist species and concentration.
| Antagonist | Agonist Induced Rate ± Std Er [×10−3 min−1] |
Control Rate ± Std Er [×10−3 min−1] |
|---|---|---|
| 14 pM Spiperone | 10.2 ± 0.3 | 4.2 ± 0.1 |
| 14 nM Spiperone | 9.6 ± 0.3 | 3.2 ± 0.1 |
| 14 µM Spiperone | 7.7 ± 0.1 | 3.8 ± 0.1 |
| 14 µM Methiothepin | 8.5 ± 0.2 | 3.1 ± 0.1 |
| 14 µM NAN-190 | 5.7 ± 0.1 | 3.6 ± 0.1 |
| 14 µM WAY 100635 | 5.9 ± 0.1 | 3.4 ± 0.1 |
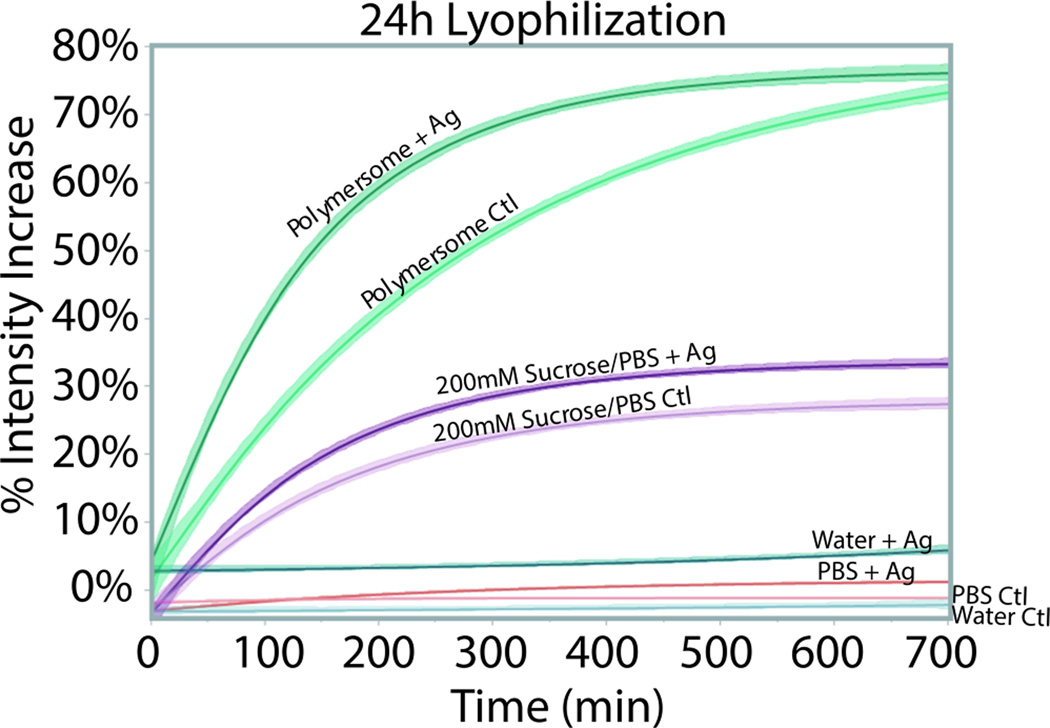
A remarkable feature of the 5-HT1AR pGUPs is their stability after lyophilization and rehydration. Freeze drying of proteins often renders them nonfunctional and larger lipid vesicles (>5 µm) typically display fracturing upon lyophilization.[23, 24] Since polymersomes are known for increased stability, we formed pGUPs as described above and subjected them to flash freezing for five minutes in liquid nitrogen followed by overnight vacuum at 0.5 torr and −35°C to completely lyophilize the samples. pGUPs were kept frozen with desiccant at −20°C for extended storage. At 24 hours (h) and 120 hours (h), lyophilized samples were rehydrated with deionized water (37°C) for 20 minutes. Rehydrated samples were observed using epifluorescence microscopy and analyzed via fluorescence microtiter plate assay as previously described. 24 h and 120 h pGUPs were still vesicular and retained their size (Figure S12). Furthermore 24 h and 120 h lyophilized pGUPs display agonist induced functional rates comparable to that of pGUPs which were not lyophilized (the 0 h samples in Table 2 are tested prior to lyophilization), 7.3 ± 0.2 and 6.5 ± 0.1 vs 7.7±0.1 (× 10−3 min−1) respectively (Table 2). A Tukey-Kramer pair wise comparison of means (α=0.05) shows that the differences in rates are not statistically significant. The percent intensity increase of the pGUPs accounts for varying amounts of pGUPs in individual microtiter wells and is also indicative of the population of functional receptors. Table 1 shows that the percent intensity increase does not vary significantly across all samples and thus the functional protein populations in pGUPs do not decrease upon lyophilization. pGUPs formed without spiperone and subjected to lyophilization, retained activity (Figure S13) and non-lyophilized pGUPs stored at −20°C for 24 h and 120 h display decreased activity (Figure S14). These results indicate that our polymeric bilayers protect protein integrity during lyophilization and extended dehydrated storage, and that lyophilization is necessary for extended storage of functional proteins in pGUPs.
Table 2.
Substrate exchange rates and increase in fluorescence intensity of pGUP after lyophilization. Values and standard errors are reported for samples with added agonist (+Ag) and control samples (Ctl) which represent the basal activity levels. 0 h samples are not lyophilized. Increase in fluorescence intensity is indicative of active 5-HT1AR population. A Tukey-Kramer pair wise comparison of means (α=0.05) shows that the differences in rates of grouped samples are not statistically significant.
| Lyophilization Time [h] |
Sample | Rate×10−3 [min−1] |
Increase in Fluorescence Intensity |
|---|---|---|---|
| 0 (not lyophilized) | Ctl | 2.8 ± 0.1 | 85% ± 11% |
| 24 | Ctl | 3.0 ± 0.1 | 73% ± 8% |
| 120 | Ctl | 2.5 ± 0.2 | 83% ± 12% |
| 0 (not lyophilized) | + Ag | 7.7 ± 0.1 | 80% ± 10% |
| 24 | + Ag | 7.3 ± 0.2 | 77% ± 7% |
| 120 | + Ag | 6.5 ± 0.1 | 85% ± 9% |
Proteins may be stabilized by ligands and sugars, which can aid in keeping their functional integrity during lyophilization by decreasing aggregation and providing hydrogen bonding.[25–28] The membrane fragments used here are suspended in 20% sucrose, which may also assist in its shelf life (see SI). It has been previously shown that increasing sucrose content increases the physical stability of proteins.[28] Furthermore interactions between buffer species and proteins can replace water molecules during lyophilzation which further stabilizes the protein in a dehydrated state.[26] To determine if the stabilization in our systems is due in part from sucrose and the buffer used for pGUP formation (200 mM sucrose in PBS), membrane fragments of 5-HT1AR were bound to spiperone and then diluted in deionized water (DI water), PBS (pH 7.4), or 200 mM sucrose in PBS (pH 7.4) with protein concentrations similar to that in our pGUP system. The work, therefore, investigates the stability of the GPCR in membrane preparation alone versus in pGUPs. After 24 h of lyophilization, membrane preparation samples were rehydrated and assessed for protein function. 5-HT1AR membrane fragments alone diluted in DI water or PBS did not retain their function (Figure 3). In 200 mM sucrose in PBS, the protein displays a similar functional rate for both control and agonist treated samples. Despite displaying protein function, the rates do not discriminate between control and agonist exposed samples suggesting that the protein in membrane fragments has lost its agonist binding ability (Figure S14). Furthermore the percent intensity increase of these samples were well below that of the pGUP samples, indicating that only a small population of proteins retained some function (Figure 3). Thus, while these results suggest that sucrose stabilizes 5-HT1AR to some extent during lyophilization, its protective ability is much lower than the overall stability and protection offered by our pGUPs, which not only retain vesicle shape and size, but also retain protein functional integrity (a summary of all results can be found in Table S1).
Figure 3.
Functional rates of 5-HT1AR in polymersomes (pGUPs) versus various solutions. Controls (Ctl), no agonist pGUPs, are plotted alongside agonist-exposed samples (+Ag). The percent intensity increase of the samples indicates the population of functional protein. In DI water and PBS, the 5-HT1AR displays no fluorescence activity. In 200 mM sucrose in PBS (pH 7.4) 5-HT1AR displays weaker fluorescence intensity increase compared to pGUPs. Furthermore there is no difference in rate between the Ctl and +Ag protein in 200 mM sucrose in PBS.
Using an agarose rehydration technique we not only show successful incorporation of GPCR 5-HT1AR into diblock copolymer bilayer vesicles in the form of pGUPs but also show increased protein stability during lyophilization and extended dehydrated storage. Successfully reconstituted 5-HT1AR in diblock copolymer pGUP vesicles on the micrometer scale exhibits expected responses to different antagonists and at various concentrations. Rehydration of pGUPs after 24 h and 120 h of lyophilization retains vesicle size and consistent protein function and offers increased stability as compared to buffered sugar solutions and pGUPs that are not lyophilized. Thus we offer a simple platform to investigate protein function in polymer vesicles in the form of pGUPs. Extension of this work to other types of GPCRs is currently being conducted.
Experimental Section
For pGUP formation, a film of polymer was drop casted on a plasma treated coverslip. A film of protein and agarose (3% w/w, 45°C) was formed on top of the polymer and swollen with 200 mM sucrose in PBS (pH 7.4) with spiperone and BODIPY-γ-FL-GTP (70 nM final concentration) for 20 minutes. For activity assessment, vesicles were harvested and sedimented in an isoosmotic glucose solution. pGUPs were transferred to 96-well plates and agonist 8-OH-DPAT was added. Samples were read at 37°C every 5 minutes for twelve hours. Fluorescence reading of BODIPY-GTPγS unquenching was done on a Biotek Synergy H4 Microplate Reader equipped with a xenon flash lamp (excitation: 485/20 nm and emission: 528/20). For lyophilization, samples were flash-frozen in liquid nitrogen and placed under a vacuum at 0.5 torr overnight.
Supplementary Material
Acknowledgments
MGG was funded by a Viterbi Fellowship, Oakley Fellowship, and ARCS Scholarship. JP was funded by the USC SURE program. This work was supported by the National Institutes of Health (award 1R01GM093279). All microtiter plate reader results were obtained using equipment provided by the USC NanoBioPhysics Core Facility.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Discher BM. Science. 1999;284:1143. doi: 10.1126/science.284.5417.1143. [DOI] [PubMed] [Google Scholar]
- 2.Discher DE, Eisenberg A. Science. 2002;297:967. doi: 10.1126/science.1074972. [DOI] [PubMed] [Google Scholar]
- 3.Rodríguez-García R, et al. Soft matter. 2011;7:1532. [Google Scholar]
- 4.Jain JP, Ayen WY, Kumar N. Current Pharmaceutical Design. 2011;17:65. doi: 10.2174/138161211795049822. [DOI] [PubMed] [Google Scholar]
- 5.Le Meins JF, et al. Materials Today. 2013;16:397. [Google Scholar]
- 6.Peters RJ, et al. Angewandte Chemie International Edition. 2014;53:146. [Google Scholar]
- 7.Kumar M, et al. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20719. doi: 10.1073/pnas.0708762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amado E, et al. ACS Macro Letters. 2012;1:1016. doi: 10.1021/mz300304u. [DOI] [PubMed] [Google Scholar]
- 9.Katritch V, Cherezov V, Stevens RC. Annu. Rev. Pharmacol. Toxicol. 2013;53:531. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drews J. Science. 2000;287:1960. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 11.Lefkowitz RJ. Trends in pharmacological sciences. 2004;25:413. doi: 10.1016/j.tips.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 12.May S, et al. Angewandte Chemie International Edition. 2013;52:749. [Google Scholar]
- 13.de Hoog HP, et al. PLoS One. 2014;9:e110847. doi: 10.1371/journal.pone.0110847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutierrez MG, Malmstadt N. Journal of the American Chemical Society. 2014;136:13530. doi: 10.1021/ja507221m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen JS, et al. Journal of the American Chemical Society. 2013;135:17294. doi: 10.1021/ja409708e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez MG, Mansfield K, Malmstadt N. Biophysical journal. 2016;110:2486. doi: 10.1016/j.bpj.2016.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LoPresti C, et al. Journal of Materials Chemistry. 2009;19:3576. [Google Scholar]
- 18.Horger KS, et al. Journal of the American Chemical Society. 2009;131:1810. doi: 10.1021/ja805625u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bordignon E. Topics in Current Chemistry. 2012;321:121. doi: 10.1007/128_2011_243. [DOI] [PubMed] [Google Scholar]
- 20.Newman-tancredi A, et al. Molecular pharmacology. 2002;62:590. doi: 10.1124/mol.62.3.590. [DOI] [PubMed] [Google Scholar]
- 21.Bojarski AJ, et al. Molecules. 2004;9:170. doi: 10.3390/90300170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman-Tancredi A, et al. Naunyn-Schmiedeberg's Archives of Pharmacology. 1998;357:205. doi: 10.1007/pl00005159. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, et al. Journal of Controlled Release. 2010;142:299. doi: 10.1016/j.jconrel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 24.MacDonald RC, Jones FD, Qiu R. Biochimica et biophysica acta. 1994;1191:362. doi: 10.1016/0005-2736(94)90187-2. [DOI] [PubMed] [Google Scholar]
- 25.Tonnis WF, et al. Molecular Pharmaceutics. 2015;12:684. doi: 10.1021/mp500423z. [DOI] [PubMed] [Google Scholar]
- 26.Allison SD, et al. Archives of Biochemistry and Biophysics. 1999;365:289. doi: 10.1006/abbi.1999.1175. [DOI] [PubMed] [Google Scholar]
- 27.Randolph TW. Journal of Pharmaceutical Sciences. 1997;86:1198. doi: 10.1021/js970135b. [DOI] [PubMed] [Google Scholar]
- 28.Wang B, et al. Journal of Pharmaceutical Sciences. 2009;98:3145. doi: 10.1002/jps.21622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.