Abstract
Functional steroid receptor complexes are assembled and maintained by an ordered pathway of interactions involving multiple components of the cellular chaperone machinery. Two of these components, Hop and Hip, serve as co-chaperones to the major heat shock proteins (Hsps), Hsp70 and Hsp90, and participate in intermediate stages of receptor assembly. In an effort to better understand the functions of Hop and Hip in the assembly process, we focused on a region of similarity located near the C-terminus of each co-chaperone. Contained within this region is a repeated sequence motif we have termed the DP repeat. Earlier mutagenesis studies implicated the DP repeat of either Hop or Hip in Hsp70 binding and in normal assembly of the co-chaperones with progesterone receptor (PR) complexes. We report here that the DP repeat lies within a protease-resistant domain that extends to or is near the C-terminus of both co-chaperones. Point mutations in the DP repeats render the C-terminal regions hypersensitive to proteolysis. In addition, a Hop DP mutant displays altered proteolytic digestion patterns, which suggest that the DP-repeat region influences the folding of other Hop domains. Although the respective DP regions of Hop and Hip share sequence and structural similarities, they are not functionally interchangeable. Moreover, a double-point mutation within the second DP-repeat unit of Hop that converts this to the sequence found in Hip disrupts Hop function; however, the corresponding mutation in Hip does not alter its function. We conclude that the DP repeats are important structural elements within a C-terminal domain, which is important for Hop and Hip function.
INTRODUCTION
Steroid receptors require extensive interactions with the heat shock proteins (Hsps) and other components of the cellular chaperone machinery to facilitate functional maturation and to maintain full responsiveness toward hormonal signal (Pratt and Toft 1997; Cheung and Smith 2000). The major cytoplasmic chaperones Hsp90 and Hsp70 are central players in the progesterone receptor (PR) assembly pathway; additionally, there are several Hsp-associated co-chaperones that participate in assembly. Two of these co-chaperones are Hop, which binds to both Hsp90 and Hsp70 (Smith et al 1993), and Hip, which binds to Hsp70 alone (Hohfeld et al 1995; Prapapanich et al 1996a). Through mutual binding to both Hsp70 and Hsp90, Hop functions as an adaptor to promote recruitment of Hsp90 to preexisting Hsp70-receptor complexes (Smith et al 1993; Chen and Smith 1998). Hip binds to Hsp70 and appears in early receptor complexes (Smith 1993; Prapapanich et al 1996a), but its functional role in the assembly process has not been fully established (Prapapanich et al 1998; Kanelakis et al 2000). Both Hop and Hip contain tetratricopeptide repeat (TPR) domains that mediate Hsp binding (Hohfeld et al 1995; Chen et al 1996; Prapapanich et al 1996b; Lassle et al 1997; Scheufler et al 2000). They also share a C-terminal sequence we have termed the DP repeat (Prapapanich et al 1998; Smith 1998). Truncation of the DP-repeat region or substitution of conserved amino acids (aa) within the DP repeat leads to functional defects that arrest maturation of receptor complexes. Hop DP mutants fail to support recruitment of Hsp90 into PR complexes, although these mutants retain binding to Hsp90 and, to a lesser extent, Hsp70 (Chen and Smith 1998). Hip DP mutants bind to Hsp70 in a more stable manner than wild-type Hip; however, whereas these mutants permit binding of Hsp70 to receptor, they block recruitment of Hop and Hsp90 to the receptor complex (Prapapanich et al 1998). In the present study, we extend our characterizations of the DP-repeat region and determine that it is a critical part of a discrete protease-resistant domain in both Hop and Hip.
MATERIALS AND METHODS
Construction of Hip and Hop mutants
The in vitro expression plasmids Hip/pSPUTK (Prapapanich et al 1996a) and Hop/pSPUTK (Chen et al 1996) encoding wild-type human Hip and Hop, respectively, were used as templates for generation of mutant complementary deoxyribonucleic acids (cDNAs). Hip mutant APAV2 and Hop mutant AP2 were constructed previously (Chen and Smith 1998; Prapapanich et al 1998). Mutants HipAM (Glu330 and Val331 changed to Ala and Met) and HopEV (Ala503 and Met504 changed to Glu and Val) were created using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The following mutagenic primers (mutated bases in lowercase) were used: HipAM, CAGGATCCAgcaatgATGGTGGCT; HopEV, GAGTGACCCAGaggTGCGC-CTTATC. Sequences of the mutated cDNAs were verified by automated sequencing. C-terminal truncations of the Hop cDNA also were generated through site-directed mutagenesis by introducing stop codons on the 3-prime side of Ile507 (N507), Gln512 (N512), Ala517 (N517), Leu522 (N522), Gln532 (N532), and Val537 (N537). The sequences of all the mutant cDNAs were confirmed by automated sequencing.
Tail-swap mutants HipTS (Hip aa 1–317 fused with Hop aa 491–543) and HopTS (Hop aa 1–490 fused to Hip aa 318–369) were generated by first introducing NheI sites immediately upstream of the DP regions. As a consequence, the Hip-coding region was changed from Ile316-Leu317 to Ala316-Ser317. The Hop-coding region was changed from Met490 to Ser490. Using Hip/pSPUTK and Hop/pSPUTK as templates for the respective mutagenesis reactions, the following mutagenic primers (mutated bases in lowercase) were used: Hip-NheI, CTGGAATGCCTGGACTCAATGAAgcTagcAGTGATCCAGAGGTTCTTGCAG and Hop-NheI, GATGTGAAGCGACGAGCtAgcGCCGACCCTGAGGTG (NheI site italicized). In combination with a preexisting NheI cleavage site at the 5′ end of the cDNA, the introduced NheI site was used to exchange the 3′ ends of the Hip and Hop cDNAs. Both exchanges were verified by automated DNA sequencing.
Protease digestion analysis
Hop and Hip forms were subjected to partial proteolytic analysis (Konigsberg 1995) to gain structural insight. Purified protein for limited protease digestions was obtained from intein fusions of Hip and Hop forms (Chen and Smith 1998; Prapapanich et al 1998). The following proteases were used: α-chymotrypsin (Type VII, Sigma, St Louis, MO, USA), trypsin (L-1-tosylamido-2-phenylethylchloromethyl ketone TPCK treated, Sigma), and subtilisin carlsberg (Type VIII, Sigma). Digestion mixtures (60-μL total volume) contained purified Hip or Hop forms (0.5 μg/μL) in 50 mM Tris-HCl, pH 8.0, plus protease (5 ng/μL). Mixtures were incubated at 30°C over a 60-minute time course. Aliquots (10 μL) were removed at different times, and proteolysis was quenched by adjusting the mixtures to 10 mM phenylmethanesulfonyl fluoride and placing them in an acetone–dry ice bath. After addition of sodium dodecyl sulfate (SDS) sample buffer and heating, digested proteins were separated on 4–20% gradient precast SDS–polyacrylamide gel electrophoresis (PAGE) gels (BioRad, Hercules, CA, USA). The gels were stained with Coomassie brilliant blue dye to observe all proteins, or proteins were transferred to Immobilon P polyvinylidene difluoride (PVDF) membrane (Millipore) for immunoblot analysis.
N-terminal protein sequencing
Chymotryptic digestion of Hip and Hop proceeded at 30°C for 15 or 60 minutes, respectively. Protein fragments were then separated by SDS-PAGE and transferred to ProBlott PVDF membrane (BioRad). Thin sample strips were cut from the lanes and immunoblotted to detect C-terminal fragments. The remainder of the membrane was Coomassie stained to detect all protein fragments. The membrane strips were aligned, and the appropriate band plus an adjacent blank membrane region were excised from the Coomassie-stained membrane. The membrane pieces were submitted to the Mayo Protein Core Facility for automated sequencing using a PE Applied Biosystems Procise cLC 492 Capillary Protein Sequencer.
Binding of Hip and Hop forms to Hsp70 and Hsp90, and association with PR complexes
Radiolabeled Hip and Hop forms were generated by in vitro expression (TNT Kit, Promega Corp, Madison, WI, USA) in the presence of [35S]-methionine using pSPUTK plasmids as templates. A 1-μL aliquot of each synthesis reaction was separated by SDS-PAGE and was observed by autoradiography; the labeled product was quantitated by densitometry (Flour-S MultiImager, BioRad). Molar equivalents of each radiolabeled product were added to 200 μL of rabbit reticulocyte lysate ([RL] Green Hectares, Oregon, WI, USA) for immunoprecipitation and receptor assembly trials.
For immunoprecipitations, mouse monoclonal antibodies specific for Hsp70 (BB70) or Hsp90 (H90-10) were preadsorbed to protein G-agarose (Amersham Biosciences, Piscataway, NJ; 1 μg antibody/μL packed resin). For each immunoprecipitation reaction, 200 μL of RL supplemented with 5–10 μL of the radiolabeled synthesis mixture was added to a 10-μL immunoresin pellet. This mixture was incubated at 30°C for 30 minutes with a brief vortex every 5 minutes to resuspend the resin slurry. The resin complexes were washed 4 times in wash buffer and then resuspended in 20 μL of 2× SDS-PAGE sample buffer and separated by SDS-PAGE. The gel was Coomassie stained to visualize the total protein content, then dried, and autoradiographed to observe radiolabeled Hip and Hop forms.
Comparisons of the Hip or Hop forms assembling with PR complexes were done essentially as described previously (Chen and Smith 1998). Briefly, recombinant chick PR A was isolated from Sf9 cell extracts by binding to an immunoaffinity resin, consisting of monoclonal anti-PR antibody PR22 preadsorbed to protein G-agarose. Each assembly reaction contained 10 μL PR resin (representing approximately 1 μg PR), 200 μL RL supplemented with an adenosine triphosphate (ATP)–regenerating system, 5–10 μL of radiolabeled Hip or Hop forms, and geldanamycin (20 μg/mL final concentration) to enhance the recovery of intermediate PR complexes containing Hip and Hop (Smith et al 1995). After a 30-minute incubation at 30°C, the PR resin pellets were washed, and bound proteins were eluted with SDS-PAGE sample buffer.
RESULTS
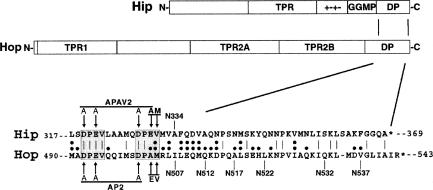
The co-chaperones Hop and Hip have, in addition to Hsp-binding TPR domains, a similar C-terminal region that is marked by a DP-repeat motif (Fig 1). Mutations to this region functionally disrupt either Hop or Hip, but little is known about how the DP-repeat region participates in co-chaperone function. To gain a better understanding of the structural context of the DP-repeat region, we generated partial proteolytic profiles (Konigsberg 1995) of wild-type and DP-mutant proteins.
Fig 1.
Diagram of major Hop and Hip domains and shared C-terminal sequences. The Hop domains tetratricopeptide repeat 1 (TPR1) and TPR2a are important binding sites for heat shock protein 70 (Hsp70) and Hsp90, respectively. A binding partner for TPR2B has not been identified. Hip contains a TPR plus an adjacent charged region (“+−+−”) that form the Hsp70 binding site. Hip also contains a GGMP-repeat motif whose function in unclear. Near the C-terminus of Hop and Hip, there is a region of sequence homology that contains a shared motif we have termed the DP repeat (shaded amino acids in sequence alignment). The notations above and below the sequence alignment signify various mutations used in the present study
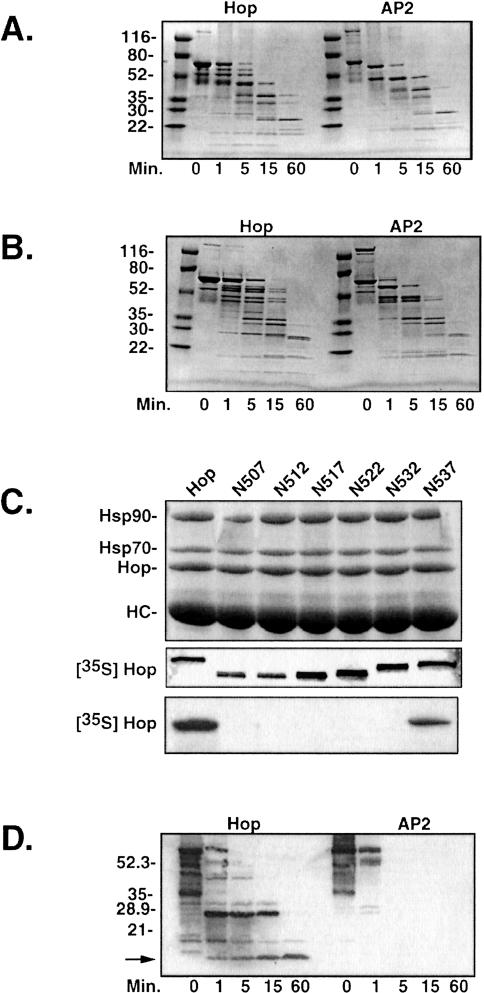
Wild-type Hop and the AP2 mutant, which contains alanine substitutions for the charged DP-repeat residues at positions 492, 494, and 501 (Chen and Smith 1998), were digested over a time course with a limiting concentration of protease. The fragments generated were separated on SDS gels and visualized using Coomassie staining. No differences in fragment patterns were observed with tryptic digests (not shown); however, parallel digestions of Hop and AP2 with subtilisin (Fig 2A) or chymotrypsin (Fig 2B) resulted in fragment patterns that differed at the early proteolytic time points (1 and 5 minutes) but merged into a common profile after 15 minutes. In the subtilisin-digested samples (Fig 2A), several large and small Hop fragments were generated after 1–5 minutes that are absent in the corresponding AP2 lanes. A similar decrease in complexity was observed when comparing Hop and AP2 chymotryptic patterns from early time points (Fig 2B). The reduced complexity of the early AP2 fragment profile, as seen with both subtilisin and chymotrypsin, suggests that partially resistant regions of Hop are more accessible to protease in the DP mutant. Thus, the C-terminal DP repeat may normally interact with other sequences to form a more compactly folded polypeptide.
Fig 2.
Partial proteolytic digests of wild-type Hop and the AP2 mutant. Purified recombinant Hop forms were subjected to proteolysis with either subtilisin (A) or chymotrypsin (B). After the times indicated below each panel, digestion was stopped, and the resulting fragments were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Coomassie staining. Sizes of the molecular weight standards are indicated to the left of each gel image. (C) A series of C-terminal truncation mutants (the position of the final amino acid is indicated) were expressed in vitro in the presence of [35S]-methionine and tested in immunoprecipitation trials with anti-Hop monoclonal antibody F5. Proteins in the immunoprecipitates from RL were separated by SDS-PAGE and visualized by Coomassie staining (top panel). The bands representing Hop and its major binding partners, heat shock protein 90 (Hsp90) and Hsp70, are indicated to the left along with the band representing the F5 heavy chain. The middle panel is an autoradiograph that illustrates the migration position and relative load for each of the Hop forms used in the F5 immunoprecipitation trial. The bottom panel is an autoradiograph of the gel in the top panel that reveals Hop forms that are recognized by F5. (D) Chymotryptic digests for Hop and AP2 were immunoblotted with F5 to detect proteolytic fragments retaining the F5 epitope. Numbers on the left indicate migration positions for molecular weight markers, and an arrow marks the minimal protease-resistant fragment that retains the F5 epitope
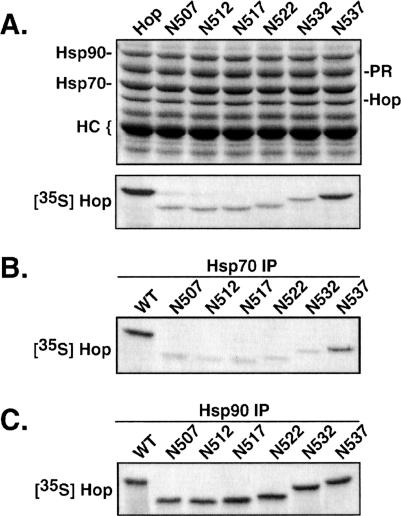
To examine the DP-repeat region more directly, we first established that the epitope for anti-Hop monoclonal antibody F5 (Smith et al 1993) lies near the C-terminus immediately downstream from the DP repeats (Fig 2C). A series of Hop C-terminal truncation mutants, generated by introducing stop codons into Hop/pSPUTK, were individually expressed in rabbit RL to generate radiolabeled products for F5 immunoprecipitation trials. Samples from each synthesis reaction were separated by gel electrophoresis, and radiolabeled Hop forms were detected by autoradiography (middle panel). Equal aliquots from the synthesis mixtures were added to RL, and immunoprecipitation with F5 antibody resin was performed. Washed immunoprecipitates were separated by gel electrophoresis and stained to visualize total protein (upper panel). As indicated, the stained bands represent the endogenous Hop-Hsp90-Hsp70 complex that is abundantly precipitated from RL (Smith et al 1993) and the F5 heavy chain (HC). The stained gel was dried and autoradiographed to detect radiolabeled Hop forms that bind to F5 (bottom panel). Truncations shorter than N537 completely lacked binding to F5, indicating that critical aa in the F5 epitope reside in the C-terminus of Hop, downstream of the DP repeats.
We next probed chymotryptic digests of Hop and AP2 with F5 to detect fragments containing this epitope (Fig 2D). With wild-type Hop, a protease-resistant fragment of approximately 9–10 kDa (indicated by arrow on the left) was observed after 60 minutes of digestion. In contrast, there was only a minimal recovery of F5-reactive bands after the briefest exposure of AP2 to chymotrypsin. The mutations in AP2 do not alter the F5 epitope because the full-length wild-type and mutant proteins are recognized equally by F5 immunostaining (compare 0-minute lanes in Fig 2D). Therefore, the DP-repeat region, which is resistant to proteolysis in wild-type Hop becomes highly sensitive in the AP2 mutant.
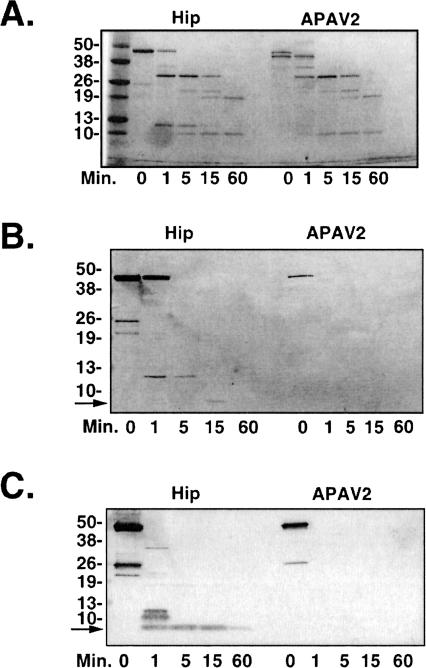
Similar to the Hop studies presented in Figure 2, the DP-repeat region of Hip was probed by limited proteolytic mapping (Fig 3). The Hip mutant APAV2, which contains alanine substitutions for the glutamic and aspartic acid residues in 2 DPEV sequences (see Fig 1) and has altered Hsp70-binding properties (Prapapanich et al 1998), was compared with wild-type Hip in protease assays. The chymotrypsin digestion patterns for Hip and APAV2 (Fig 3A) are mostly similar and closely resemble the Hip chymotryptic pattern recently reported by Velten et al (2002). Two differences are noted when comparing wild-type and mutant Hip-digestion patterns. First, the undigested APAV2 resolves as a doublet, whereas wild-type Hip is a single band. As will be demonstrated below, the lower band of the APAV2 doublet probably represents protein in which the C-terminal DP-repeat region was cleaved away by bacterial proteases before purification of recombinant protein. A second distinction in the Hip fragment pattern is a 12-kDa fragment generated during the brief digestions (1- and 5-minute lanes). Overall, there are fewer differences in the Hip wild-type and mutant protease patterns when compared with the Hop major fragment patterns (Fig 2 A,B). Similar results were obtained when Hip and APAV2 were digested with subtilisin or trypsin (results not shown).
Fig 3.
Partial proteolytic digests of wild-type Hip and the APAV2 mutant. Purified recombinant proteins were digested as in the previous figure. (A) Chymotryptic fragment patterns were generated by gel separation and Coomassie staining of samples. The anti-Hip monoclonal antibody 2G6, whose epitope is localized to the C-terminal region of Hip, was used to immunostain samples digested with chymotrypsin (B) or trypsin (C). Arrows indicate the minimal protease-resistant fragments containing the 2G6 epitope
We identified 2G6, one of our previously developed anti-Hip monoclonal antibodies (Prapapanich et al 1996a), as having an epitope near the C-terminus of Hip. A truncation mutant, N334, which retains the DP repeats but lacks downstream sequences, was not recognized by 2G6 (data not shown). The 2G6 and F5 epitopes are fully contained within the respective C-terminal regions of Hip and Hop because each antibody cross-reacts with the corresponding tail-swap chimera we describe in later experiments (results not shown). We used 2G6 in Western immunoblots to probe digests for proteolytic fragments containing C-terminal Hip sequences. In a chymotryptic time course (Fig 3B), a 12-kDa fragment was immunostained that corresponds to the fragment noted in Figure 3A. The 12-kDa fragment degrades further to a fragment of less than 10 kDa (indicated by arrow) after 15 minutes of digestion. No 2G6-reacting fragments were detected in any of the digested APAV2 samples, indicating that this region is rendered more sensitive to proteolysis by mutation of the DP repeats. Also, note that in the undigested APAV2 lane 2G6 reacts exclusively with the upper band of the doublet, supporting our previous contention that the lower band of the doublet lacks the C-terminal region.
Immunoblots of tryptic digests provide further support for a proteolytically resistant fragment containing the DP repeat (Fig 3C). Although the major tryptic fragments from Hip and APAV2 are similar (Coomassie-stained gel not shown), 2G6 detected a C-terminal Hip fragment of approximately 9–10 kDa size (indicated by arrow), which was detected at the 1-minute– through 60-minute–digestion time points. This fragment was completely absent in the APAV2 digests.
The results in Figures 2 and 3 suggest that the DP repeats of Hop and Hip exist in protease-resistant conformations that could reflect structural domains. To help determine the boundaries of these putative DP-domains, we obtained partial aa sequences from chymotryptic fragments. Samples of Hop and Hip were individually digested with chymotrypsin for 15 or 60 minutes, respectively, after which fragments were separated by gel electrophoresis, transferred to PVDF membrane, and detected by Coomassie staining. Bands corresponding to the minimally sized fragments detected in Figures 2D and 3B were excised and submitted for automated N-terminal sequencing. The partial sequence from the Hop fragment (NRHDSPED) matched the sequence beginning at position 477 that is 15 residues upstream from the initial DPEV. The predicted size of a Hop fragment beginning at 477 and extending to the C-terminus is 7687 Da; this correlates well with the apparent size of the chymotryptic Hop fragment. Whereas the resistant fragment retains the F5 epitope that maps within 10 aa of the C-terminus, the putative DP-domain appears to extend from immediately before the DP repeats to or very near the C-terminus. Partial aa sequence for the protease-resistant Hip chymotryptic fragment (GSFPGGFP) begins at residue 279; this sequence is at the beginning of the GGMP-repeat region (Prapapanich et al 1996a) that immediately precedes the DP repeats. Whether the unusual GGMP repeat is itself resistant to proteolysis or falls within a putative DP-repeat domain is unclear. The predicted size of a Hip fragment extending from residue 279 to the C-terminus is 9217 Da; as in the case of the Hop fragment, this corresponds well with the apparent size of the protease-resistant Hip DP fragment and is consistent with a putative Hip DP-domain that extends to or near the C-terminus.
Truncation of the entire DP-repeat region or DP-point mutation inhibits the ability of Hop to bind to Hsp70 and to support assembly of PR complexes (Chen and Smith 1998). To map out more precisely the extent of C-terminal aa that are functionally important for Hop, we used a series of C-terminal truncation mutants (see Fig 1) to compare coimmunoprecipitation patterns with PR, Hsp70, and Hsp90 (Fig 4). Mutant cDNAs were expressed in vitro to generate radiolabeled products (as in Fig 2C), and equal amounts of each product were added to RL mixtures for receptor assembly (Fig 4A), Hsp70 binding (Fig 4B), or Hsp90 binding (Fig 4C). Removal of 6 aa (N537) had little effect on assembly of Hop in PR complexes or in Hsp interactions. However, truncating 5 additional aa (N532) inhibited assembly with PR complexes. Defective assembly with PR correlates precisely with the loss of Hsp70 binding (Fig 4B); this finding is similar to our previous observations on Hsp70 binding and PR assembly by mutant AP2 (Chen and Smith 1998). As we had observed previously with AP2, the DP region is not required for Hsp90 binding, and all truncation mutants were recovered in Hsp90 complexes at a level identical to that of full-length Hop (Fig 4C). The results in Figure 4 link sequences near the C-terminus with the upstream DP repeats in a common functional role for Hsp70 binding and PR assembly. Thus, the DP repeats contribute to a functional domain that corresponds to the protease-resistant domain encompassing the C-terminus of Hop.
Fig 4.
Protein interactions of Hop C-terminal truncation mutants. (A) Radiolabeled Hop forms were synthesized by in vitro expression, and an equal amount of each form was added to reticulocyte lysate (RL) used for assembly of PR complexes. The assembly mixtures were supplemented with the Hsp90-binding drug geldanamycin to enhance recovery of intermediate receptor complexes containing Hop. Samples were separated by gel electrophoresis, and protein components were detected by Coomassie staining (upper panel). Bands representing the major components of the receptor complex and the anti-PR heavy chain are indicated on the left. The dried gel was autoradiographed to detect radiolabeled Hop forms that were recovered in PR complexes (lower panel). (B) Heat shock protein 70 (Hsp70) complexes were immunoprecipitated from RL that was supplemented with radiolabeled Hop forms. The autoradiograph of the resulting gel analysis is shown. (C) Hsp90 complexes were similarly targeted for immunoprecipitation from RL, and the coprecipitating Hop forms were detected by autoradiography
A single Hip C-terminal truncation mutant (N334) was tested for Hsp70 and PR interactions (results not shown). The recovery of N334 in Hsp70 complexes was similar to wild-type Hip, although there was a reduced recovery of N334 in PR complexes. This resembles what we had observed previously with the APAV2 point mutant (Prapapanich et al 1998) and with N303 (Prapapanich et al 1996b, 1998), a more extended C-terminal truncation mutant.
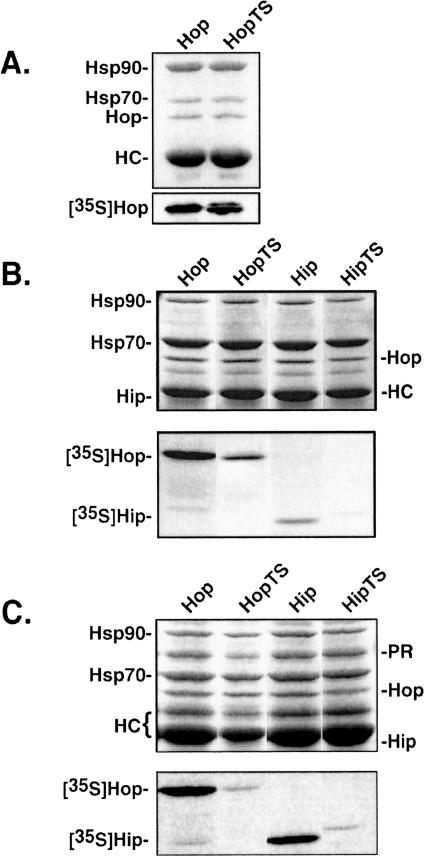
Although the Hop and Hip DP regions have significant sequence similarity and both localize to proteolytically stable fragments, we wondered whether these putative domains would be functionally interchangeable between the 2 proteins. Tail-swap chimeras, as described in the Materials and Methods, were generated and compared with wild-type proteins for recovery in Hsp and PR complexes (Fig 5). To facilitate the generation of the chimeras, first it was necessary to introduce a couple of point mutations in the wild-type cDNAs, as described in the Materials and Methods. It was possible that these intermediate mutations would alter Hip or Hop function, but we observed no difference between wild-type and intermediate mutant interactions with Hsp70, Hsp90, or PR (results not shown). The Hop chimera (HopTS) bound as well as wild-type Hop to Hsp90 (Fig 5A). The Hip forms were not examined in this assay because Hip does not directly bind Hsp90. Both Hop and Hip bind Hsp70, so these were compared alongside the respective tail-swap chimeras for Hsp70 binding (Fig 5B). Relative to wild-type proteins, both chimeras are recovered at a reduced level in Hsp70 complexes. We next compared recovery of Hop, Hip, and tail-swap chimeras with PR complexes assembled in vitro (Fig 5C). Reflecting their reduced interactions with Hsp70, the tail-swap chimeras are impaired in their ability to assemble into PR complexes. Therefore, the DP domains of Hip and Hop are not functionally equivalent despite their structural similarities.
Fig 5.
Interactions by Hop and Hip tail-swap chimeras with heat shock protein 90 (Hsp90) (A), Hsp70 (B), and progesterone receptor complexes (C). Radiolabeled test proteins were added to reticulocyte lysate (RL) before immunoprecipitation reactions. For each set of figures, the upper panel is a Coomassie-stained gel to illustrate total proteins present, and the lower panel is an autoradiograph of the corresponding gel to detect recovery of radiolabeled proteins. Only Hop forms are shown in A because Hip does not bind Hsp90. The abundance of endogenous Hip is low relative to Hop, and Hip closely migrates with antibody heavy chains; thus, unlabeled Hip is poorly resolved in the Coomassie-stained gels
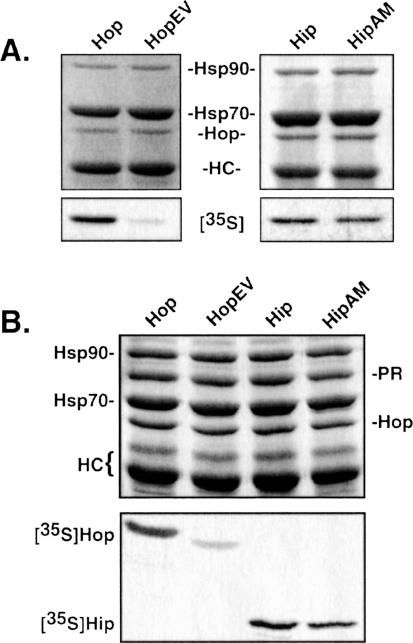
Our final analysis focuses on sequence differences within the DP repeats of Hip and Hop. As illustrated in Figure 1, the first unit of the DP repeat in both proteins consists of DPEV, but the second unit is DPEV in Hip and DPAM in Hop. To address the significance of these differences, we generated mutants in which the EV and AM amino acids of the second unit were exchanged (Fig 6). HopEV and wild-type Hop bound identically to Hsp90 (results not shown); however, HopEV bound Hsp70 much more weakly than wild-type Hop (Fig 6A). Conversely, there was little difference in binding of Hip and HipAM to Hsp70. Paralleling this observation, recovery of HopEV in PR complexes was greatly reduced, whereas HipAM behaved much like its wild-type counterpart (Fig 6B).
Fig 6.
Binding of Hop, Hip, and corresponding DP–point mutants to heat shock protein 70 (A) and to PR complexes (B). As in the previous figures, the upper panel in each set is a Coomassie-stained gel of total proteins in the immunoprecipitates, and the lower panel is the corresponding autoradiograph that reveals recovery of radiolabeled proteins
DISCUSSION
Previous studies showed that the DP-repeat motif found in the C-terminal regions of both Hip and Hop plays a functionally critical role in the proper assembly of PR complexes. Here, we used partial proteolytic digestion assays to probe the structure of the DP-repeat segments of Hop and Hip. Taking advantage of monoclonal antibodies with epitopes near the DP repeats, we find that the DP-repeat falls within a discrete, protease-resistant region and that mutation of charged residues within the DP repeats leads to increased protease sensitivity of this region (Fig 2). It appears, then, that the DP-region normally folds into a proteolytically resistant structural domain that is disrupted by point mutations in the DP-repeat motif. We therefore conclude that the DP repeats play an important structural role in maintaining the compact folding of a C-terminal domain. Sequencing of aa of isolated DP-domains helped resolve the N-terminal boundaries of the respective domains in Hop and Hip. The minimal DP-containing Hop fragment contained 16 amino acids upstream from the first DP and appears to continue through to the C-terminus. The minimal Hip fragment began with the GGMP-repeat region that precedes the DP repeats. The GGMP-repeat region does not contain sites for tryptic cleavage, but the region contains multiple potential sites for chymotryptic cleavage. However, we cannot definitively conclude that the GGMP repeat is folded together with the DP domain because the GGMP region could assume a conformation that is protease resistant by itself.
We have previously shown that alanine substitutions of acidic aa in the DP motifs functionally alter full-length Hip and Hop. As shown by the results in Figure 2, the DP domains of these mutants are much more sensitive to protease digestion. We can also glean some information on the potential interaction of the DP domain with other protein regions. Disruption of the Hop DP domain in the AP2 mutant resulted in changes to the overall pattern and rate of early proteolytic steps (Fig 2 A,B), indicating a DP domain influence on the overall conformation of Hop. In contrast, the Hip APAV2 mutant had a global digestion pattern similar to wild-type Hip (Fig 3A), suggesting that the Hip DP domain has less influence on the overall Hip conformation.
Functionally, the Hop DP domain clearly plays a role in Hsp70 binding. As with point mutations in the DP motifs, truncation of 11 aa from the C-terminus similarly inhibited Hsp70 binding and assembly into receptor complexes (Fig 4, N532 lanes). Either the DP domain directly binds to Hsp70, or the DP domain interacts with another region of Hop to form a functional Hsp70 binding site. Arguing against direct interaction is the failure of a GST-Hop DP domain fusion to bind Hsp70 (results not shown), but this does not exclude direct binding of DP to Hsp70. The GST moiety may sterically hinder bindings, or additional Hop sequences may be required for effective binding of DP to Hsp70. The N-terminal TPR1 domain of Hop is required for Hsp70 binding (Prapapanich et al 1996b; Lassle et al 1997); crystallographic (Scheufler et al 2000) and biochemical (Brinker et al 2002) evidence suggest that the highly conserved EEVD sequence terminus of Hsp70 binds in the TPR1 pocket. An interesting possibility we are currently investigating is whether the DP domain interacts with TPR1 to form the full Hsp70 binding site on Hop.
A different picture emerges with the Hip DP domain. Mutations that destabilize the Hip DP domain were previously found to stabilize interactions with Hsp70 while inhibiting PR assembly (Prapapanich et al 1998). Velten et al (2002) recently published that the 2 halves of Hip could each bind to Hsp70 independently. The N-terminal half contained the TPR domain, and the C-terminal half contained the highly charged region plus downstream sequences, presumably including the DP domain. Earlier observations (Prapapanich et al 1996b; Irmer and Hohfeld 1997) showed that mutations in either the TPR domain or the highly charged region could inhibit Hsp70 binding, so these 2 regions may cooperate for high-affinity binding to Hsp70. An intact Hip DP domain may actually lower Hsp70-binding affinity by disrupting interactions between the TPR and the charged regions.
Given the similar sequences and positioning of the C-terminal DP domains, we tested whether the DP domains are functionally interchangeable in Hip and Hop (Fig 5). However, functional defects in both DP swap mutants suggest that there are functionally important, context-sensitive interactions that are specific for each DP domain (Fig 5). In an attempt to refine differences in the DP domains, we generated double-point mutants that interconverted the second DP motifs (Fig 6). The HipAM mutant largely retained its ability to bind to Hsp70 and assemble with PR complexes, but the converse HopEV mutant had greatly reduced binding to Hsp70 and consequently was recovered at low levels in PR complexes.
Now that we know that the DP motifs are important components within what is likely a C-terminal structural-functional domain, we are pursuing studies to understand how the DP domains impact Hsp70 interactions and whether the DP domains play a role in the progressive assembly of steroid receptor complexes. One possibility is that direct, intramolecular interaction between the Hop DP domain and the N-terminal TPR domain is the key to maximal Hsp70 binding. If this is true, it might explain apparent changes in the overall Hop fold that result from DP domain disruption. Along this line, we further postulate that the Hip DP domain, when presented in an Hsp70-containing PR complex, may compete or cooperate with the similar Hop DP domain to link Hip and Hop actions and promote rearrangement of components during the progressive assembly of PR complexes.
Acknowledgments
We thank other members of the Smith Lab for their technical assistance and critical comments on the manuscript. We also thank the Mayo Protein Core Facility and the Mayo Molecular Biology Core Facility for their expert services. We were supported by NIH R01 DK-44923 and the Mayo Foundation.
REFERENCES
- Brinker A, Scheufler C, and Von Der Mulbe F. et al. 2002 Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 × Hop × Hsp90 complexes. J Biol Chem. 277:19265–19275. [DOI] [PubMed] [Google Scholar]
- Chen S, Prapapanich V, Rimerman RA, Honore B, Smith DF. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins hsp90 and hsp70. Mol Endocrinol. 1996;10:682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- Cheung J, Smith DF. Molecular chaperone interactions with steroid receptors: an update. Mol Endocrinol. 2000;14:939–946. doi: 10.1210/mend.14.7.0489. [DOI] [PubMed] [Google Scholar]
- Hohfeld J, Minami Y, Hartl FU. Hip, a novel co-chaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- Irmer H, Hohfeld J. Characterization of functional domains of the eukaryotic co-chaperone Hip. J Biol Chem. 1997;272:2230–2235. doi: 10.1074/jbc.272.4.2230. [DOI] [PubMed] [Google Scholar]
- Kanelakis KC, Murphy PJ, and Galigniana MD. et al. 2000 Hsp70 interacting protein Hip does not affect glucocorticoid receptor folding by the hsp90-based chaperone machinery except to oppose the effect of BAG-1. Biochemistry. 39:14314–14321. [DOI] [PubMed] [Google Scholar]
- Konigsberg WH. Limited proteolysis of DNA polymerases as probe of functional domains. Methods Enzymol. 1995;262:331–346. doi: 10.1016/0076-6879(95)62028-1. [DOI] [PubMed] [Google Scholar]
- Lassle M, Blatch GL, Kundra V, Takatori T, Zetter BR. Stress-inducible, murine protein mSTI1. Characterization of binding domains for heat shock proteins and in vitro phosphorylation by different kinases. J Biol Chem. 1997;272:1876–1884. doi: 10.1074/jbc.272.3.1876. [DOI] [PubMed] [Google Scholar]
- Prapapanich V, Chen S, Nair SC, Rimerman RA, Smith DF. Molecular cloning of human p48, a transient component of progesterone receptor complexes and an Hsp70-binding protein. Mol Endocrinol. 1996a;10:420–431. doi: 10.1210/mend.10.4.8721986. [DOI] [PubMed] [Google Scholar]
- Prapapanich V, Chen S, Smith DF. Mutation of Hip's carboxy-terminal region inhibits a transitional stage of progesterone receptor assembly. Mol Cell Biol. 1998;18:944–952. doi: 10.1128/mcb.18.2.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prapapanich V, Chen S, Toran EJ, Rimerman RA, Smith DF. Mutational analysis of the hsp70-interacting protein Hip. Mol Cell Biol. 1996b;16:6200–6207. doi: 10.1128/mcb.16.11.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, and Bourenkov G. et al. 2000 Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 101:199–210. [DOI] [PubMed] [Google Scholar]
- Smith D, Sullivan W, and Marion T. et al. 1993 Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol Cell Biol. 13:869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF. Dynamics of heat shock protein 90-progesterone receptor binding and the disactivation loop model for steroid receptor complexes. Mol Endocrinol. 1993;7:1418–1429. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- Smith DF. Sequence motifs shared between chaperone components participating in the assembly of progesterone receptor complexes. Biol Chem. 1998;379:283–288. doi: 10.1515/bchm.1998.379.3.283. [DOI] [PubMed] [Google Scholar]
- Smith DF, Whitesell L, Nair SC, Chen S, Prapapanich V, Rimerman RA. Progesterone receptor structure and function altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol. 1995;15:6804–6812. doi: 10.1128/mcb.15.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten M, Gomez-Vrielynck N, Chaffotte A, Ladjimi MM. Domain structure of the HSC70 co-chaperone, HIP. J Biol Chem. 2002;277:259–266. doi: 10.1074/jbc.M106881200. [DOI] [PubMed] [Google Scholar]