Abstract
Nuclear magnetic resonance (NMR) spectroscopy is widely used in metabolomics to perform quantitative profiling of low-molecular weight compounds from biological specimens. The measurement of endogenous metabolites using NMR has proven to be a powerful tool to identify new metabolic biomarkers in physiological and pathological conditions, and to study and evaluate treatment efficiency. In this study we present a rapid approach to indirectly quantify 13C enriched molecules using one-dimensional (1D) 1H NMR. We demonstrate this approach using isotopically labeled [1,6-13C]glucose and in four different cell lines. We confirm the applicability of this approach for treatment follow-up, utilizing a renal cancer cell line with rapamycin as a tool compound to study changes in metabolic profiles. Finally, we validate the applicability of this method to study metabolic biomarkers from ex vivo tumor extracts, after infusion, using isotopically enriched glucose. Given the high throughput and increased sensitivity of direct-detect 1H NMR, this analytical approach provides an avenue for simple and rapid metabolic analysis of biological samples including blood, urine, and biopsies.
Graphical Abstract
Mass spectroscopy (MS) and nuclear magnetic resonance (NMR) have been the major techniques used to perform metabolic profiling, due to their ability to identify multiple compounds in a single sample.1,2 Both approaches provide a diverse set of strengths and weaknesses and this has been explored in comparison as well as in tandem systems (e.g., LC/NMR/MS).3,4 Mass spectrometry using GC-MS or LC-MS is extremely sensitive and has the capability of detecting large numbers of metabolites with little overlap, in comparison to NMR. A few major drawbacks of MS are the long time necessary for analysis, the destructive nature of the analysis and the need for standard curves for each metabolite to be quantitative.5 Moreover, small molecule metabolites of interest can be difficult to resolve since they can ionize similarly and have comparable elution times (e.g., lactate and pyruvate), requiring specialized methods for their detection. In contrast, NMR does not require an initial chromatographic step but can acquire data from the sample intact, which can be used at later time for additional analyses. It is inherently quantitative for the simultaneous measurement of multiple metabolites and also provides high structural specificity.
NMR spectroscopy has proven to be an invaluable tool for the elucidation of crucial structural parameters such as functional groups and stereochemistry. Stable isotopomer-based metabolomics is a powerful NMR atom-based approach applied to study metabolic pathways and fluxes. Information can be deduced from the transformations of stable isotope tracers through metabolic pathways. These transformations are determined by analyzing the isotopic enrichment at different atomic positions of relevant metabolite products (isotopomer analysis).6 Therefore, a major advantage of NMR compared to MS is the determination of positional isotopomer, whereas the MS approach can only gather isotopologues information.7
NMR can be performed on many metabolically relevant nuclei, with the standard for metabolomics being proton (1H) and carbon (13C). Isotopic enrichment using 13C detection has been a staple since its increased chemical shift dispersion makes peak identification and separation easier than 1H NMR. Moreover, proton resonance line shapes are split into well-established patterns when J-coupled to a 13C nucleus. Comparing experiments performed with and without 13C decoupling during acquisition indirectly measures fractional 13C enrichment. Applying this approach to cellular metabolism allows monitoring flux through metabolic intermediates.8
Therefore, the NMR approach can gather more information about a metabolic pathway from relatively small number of 13C metabolites, when compared to MS.
However, 13C NMR requires very long scan times and larger sample sizes, in comparison to 1H NMR and MS methods, making it difficult to apply to samples of limited mass and high-throughput analysis.9,10 Various studies have explored the feasibility of using multidimensional NMR as a metabolic tool.6,11,12 While 2D NMR is a useful approach to elucidate the structure of metabolite mixtures, its intrinsically low sensitivity, complicated acquisition and spectral interpretation limit its application for rapid high throughput metabolites analysis.13
In this work, we demonstrate the application of high-throughput 1H NMR as a rapid quantitative method to analyze the metabolic status both at steady state and with isotopic flux tracing using 13C enriched substrates. Since 13C NMR suffers from lack of sensitivity can be overcome using 1H NMR in combination with resonance deconvolution.14,15 By indirectly measuring the incorporation of 13C into a pool of interest through the loss of the center directly attached 1H resonance, we can increase the sensitivity of our detection to the high nanomole range on a conventional NMR system, utilizing very simple sequences. Using this rapid method, we demonstrate quantitation and separation of both steady state pool sizes and 13C isotopically enriched molecules derived from 13C enriched glucose in a range of cell lines. Moreover, we extend this approach to metabolic changes from mTOR signaling inhibition in vitro. Finally, we apply this method in vivo and demonstrate that with very small tissue masses (<100 mg), this same tracing experiment can yield quantitative results providing a high throughput method for future metabolic investigations.
METHODS
Chemicals
Unless otherwise indicated, all chemicals and solvents were purchase from Sigma-Aldrich (St. Louis, MO, U.S.A.). A total of 99% enriched [1,6-13C]glucose was purchase from Sigma-Aldrich (St. Louis, MO, U.S.A.) and used without further purification.
Cell Lines
Normal human renal proximal tubular (HK-2), an aggressive fumarate hydratase mutant renal cancer cell (UOK-262), human pancreatic tumor (PANC-1) and human gastrointestinal stromal tumor (GIST-T1) cell lines were cultured under standard conditions in DMEM supplemented with 10% FBS and penicillin streptomycin. The HK-2 and PANC-1 cell lines were obtained from American Type Culture Collection (ATCC, Manassas, U.S.A.). The UOK-262 and GIST-T1 cell lines were kindly provided by the Zhen Lab (UCSF) and Chi Lab (MSKCC), respectively.
Cell Lysate and Western Blot
For each cell line, 5 × 106 cells were plated in a 10 cm3 plate. Cells were treated with either “vehicle” (6% DMSO) or rapamycin (50 nM) for 24 h. Cells were then washed three times with cold PBS. A 1 mL aliquot of cold PBS was then added to each plate and the cells were scraped and collected in 1.5 mL Eppendorf tube. The cell suspension was centrifuged at 1300 rpm for 5 min. The supernatant was discarded and the cell pellet was resuspended in 500 µL of RIPA buffer (Thermo Scientific Pierce) with protease and phosphatase inhibitors (Thermo Scientific Pierce). The cell were kept in ice for 30 min and centrifuged at 4000 rpm for 20 min. Then the supernatant was collected and BCA assay (Thermo Scientific Pierce) was performed on the cell lysate to accurately measure the protein concentration.
Cell lysates samples (20 µL at 2 mg/mL) were mixed with 5 µL of sample loading buffer (5X: 6% SDS, 15% 2-mercaptoethanol, 30% glycerol, and 0.3 mg/mL bromphenol blue in 188 mM Tris-HCl, pH 6.8), heated at 70 °C for 10 min, and separated by 12% NuPAGE Bis-tris Gel (Invitrogen, Thermo Fisher Scientific). Separated proteins in the gels were electrophoretically transferred onto PVDF membrane 0.2 µm pore size for 90 min (NOVEX, Life Technology). The blotted membrane was blocked with 5% BSA in TBS containing 0.05% Tween 20 (TBS-T buffer) for 15−30 min. After washing the membrane with PBS-T, pS6K, PFK, pPKM2, pLDH-a, and β-actine antibody (1 µg/mL; Cell Signaling Technology), diluted in TBS-T containing 5% BSA, was added and incubated overnight at 4 °C. The bound antibodies were detected by horseradish peroxidase-conjugated antigoat Ig secondary antibody (Santa Cruz Biotechnology) followed by ECL detection system (Thermo Scientific).
In Vitro NMR Treatment Conditions
For each cell line, 1 × 106 cells were plated in a six-well plate. The cells in each well-plate were treated with either vehicle (6% DMSO) or rapamycin (50 nM) for 24 h. Subsequently, the media was exchanged for media containing either 5 mM nonenriched glucose (3 wells) or 5 mM [1,6-13C]glucose enriched media (3 wells), for total pool size or tracing experiment, respectively, and incubated for 3 h. DMSO and rapamycin remained in the media during the entire incubation time. After 3 h, the media was removed and stored at −80 °C for further purification and analysis. The cells were quickly washed with cold PBS. A total of 2 mL of 80% of cold methanol was used to extract the water-soluble component of the cells. The cells were placed at −80 °C overnight to ensure optimal metabolites extraction. Subsequently, the sample was centrifuged at 4000 rpm at 4 °C for 30 min. The supernatant was isolated from the cell pellet and lyophilized.16,17
Sample Preparation for In Vitro NMR
Media samples were filtered at 14000 rpm and 4 °C for 30 min using Amicon Ultra 0.5 mL centrifugal filters (3kD; Merck Millipore Ltd. Darmstadt, Germany) to eliminate the protein component that could interfere with the NMR acquisition. A prewashing step of the filter was performed with 500 µL of water in order to eliminate glycerol from the cartridge. A total of 100 µL of 10× PBS in D2O, containing 0.5 mM of DSS as an internal standard and 10 mM imidazole as a pH indicator, was added to 500 µL of the purified media. The dried water-soluble intracellular content was dissolved in 600 µL of standard and 10 mM of imidazole as pH indicator.
In Vivo Infusions of 13C-Labeled Glucose
The animal portion of this study was performed under a protocol approved by the Institutional Animal Care and Use Committee at MSK. GIST-T1 xenografted mice were used as a tumor model for the following in vivo tracing study. The 5 × 106 GIST-T1 cells were trypsinized and resuspended in a 1:1 solution of complete media/Matrigel before being injected subcutaneously in the flank of immune compromised NOD.CB17-Prkdcscid mice (Jackson Laboratory, Charles River, U.S.A.). Tracer studies were initiated 4−6 weeks after GIST-T1 inoculation. A total of 24 h before the infusion, mice were treated with 15 mg/kg of rapamycin for comparison to 6% DMSO (vehicle). After treatment, [1,6-13C]glucose was infused as follows: a bolus of 0.4 mg/g body weight (0.3 mL) was infused for 1 min, followed by a continuous infusion of 0.012 mg/g/min at 150 µL/h for 150 min. At the end of the infusions, 100−150 µL of blood was collected from the orbit prior to termination, and this plasma was used to determine the enrichment of the relevant nutrient pool. The tumors were rapidly dissected, weighed, frozen in liquid nitrogen, and stored at −80 °C. Tumors were finely ground in a mortar under liquid nitrogen and prepared for NMR analysis as previously published.10,18
1H NMR Data Acquisition and Processing
NMR spectroscopy was performed on a 14.1 T NMR spectrometer (Bruker Biospin, Billerica, MA) equipped with a cryoprobe (1H and 13C) and automatic sample changer. 1H spectra were acquired using a one-pulse sequence (30 degree pulse), following selective presaturation of the water resonance, with and without composite adiabatic 13C decoupling. Adiabatic 13C decoupling was accomplished by CHIRP-95 pulses (1.5 ms pulse, 3.2 kHz bandwidth) with a composite 20 step phase cycle. The 20 step phase cycle is (0, 150, 60, 150, 0) × 2, (180, 330, 240, 330, 180) × 2, which is a composite of a five-step phase cycle and MLEV4.19
Resonances were identified and quantified using Chenomx NMR Suite 8.0 professional (Chenomx Inc. Edmonton, Canada). Experimental spectra were adjusted for T1 saturation effects by comparison to a sample fully relaxed spectra (180s recycle delay) for each metabolite (Table S1). DSS (0.5 mM) was added to samples to reference chemical shifts and concentrations. Imidazole (10 mM) was added to reference pH.20
Statistical Analysis
All data are expressed as mean ± standard deviation. Significances were calculated using Graph-Pad Prism. The Student’s t-test was used to calculate significance at the 0.05 level.
RESULTS AND DISCUSSION
Quantification of Total Pool Size Metabolites Using 1H NMR
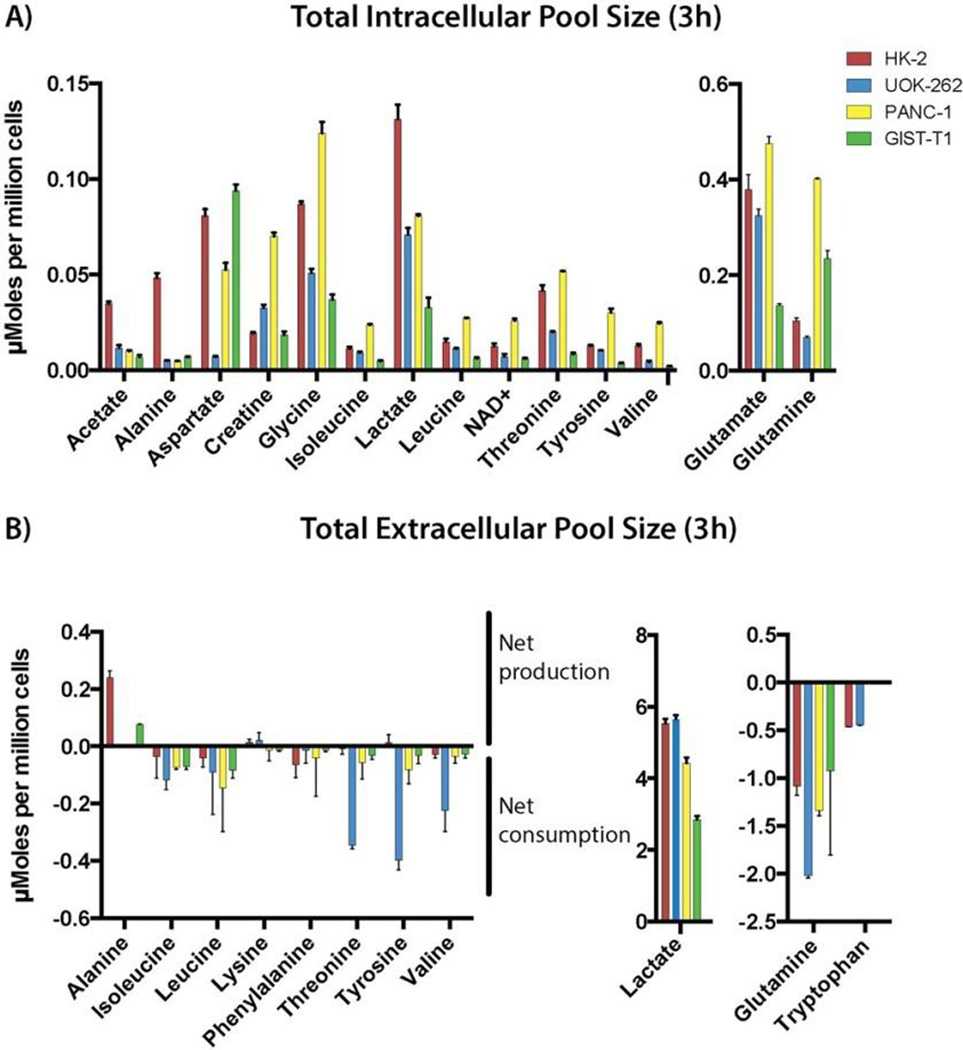
Cell culture media (extracellular water-soluble metabolites) and cell extracts (intracellular water-soluble metabolites) were analyzed from four cell lines (Figure 1). Peak assignments and multiplet information were used to quantify each resonance of interest and using a library of 300 metabolites, 40 metabolites were confidently detected and quantified (Figure S1). Figure 1 highlights 16 intracellular and 11 extracellular metabolites for the four different cell lines. With this method we were able to fit different metabolites derived from different metabolic pathways including glycolysis, TCA cycle, and biosynthesis or catabolism of amino acids.
Figure 1.
(A) Summary of the total intracellular metabolite concentrations for HK-2 (red), UOK-262 (blue), PANC-1 (yellow), and GIST-T1 (green) at 3 h of nonenriched glucose incubation. Acetate, alanine, aspartate, creatine, glycine, isoleucine, lactate, leucine, NAD+, threonine, tyrosine, valine, glutamate, and glutamine for the total intracellular pool size were accurately fitted as quantified. All metabolites are referenced to internal standard DSS (0.5 mM). (B) Summary of the baseline extracellular metabolite concentrations (culture media) for HK-2, UOK-262, PANC-1, and GIST-T1. A positive number of the plot refers to the net production of the metabolites of interest, whereas a negative number reflects the net consumption of the metabolites of interest calculated comparing to the preincubation media. Alanine, isoleucine, lactate, leucine, lysine, phenylalanine, threonine, tyrosine, valine, glutamine, and tryptophan for the extracellular pool size were analyzed.
As shown in Figure 1A, the total intracellular pool size of the metabolites of interest demonstrate different profile patterns between those cell lines reflecting different metabolic needs. Cancer/immortalized cells shift glucose utilization and metabolism from oxidative phosphorylation to aerobic glycolysis (Warburg effect). A total of 80−90% of the glucose is employed for lactate production. Therefore, lactate can be used as a marker to measure glycolytic flux. We observed different intracellular concentrations of lactate in the four cell lines analyzed. Totals of 0.131 ± 0.007, 0.071 ± 0.003, 0.081 ± 0.0007, and 0.033 ± 0.005 µmoles per million cells of lactate were observed for HK-2, UOK-262, PANC-1, and GIST-T1, respectively. Those data may reflect the aerobic glycolytic flux and differential energetic needs of the cell.
In many cells, glutamine can be used as an additional carbon source for metabolism, typically referred to as glutaminolysis. Glutamine is an abundant and versatile nutrient that participates in energy formation, redox homeostasis, macromolecular synthesis, and signaling in cancer cells and glutamine metabolism varies between different cancer type and phenotype. The total pool size of intracellular glutamine and its metabolic intermediate glutamate are 0.401 ± 0.003, 0.141 ± 0.006 and 0.475 ± 0.014, 0.379 ± 0.031 µmoles, million cells respectively for PANC-1 and HK-2. This different grade of glutaminolysis reflect the different glutamine addiction of these two cell lines, PANC-1 being aggressive pancreatic tumor cells and HK-2, which is an immortalized normal renal, tubular cell line. Those values mirror the extracellular pool size of glutamine quantified in the media, where higher glutamine consumption reflects a more aggressive tumor phenotype. For example, extracellular glutamine concentration decreased −1.340 ± 0.053 and −1.083 ± 0.094 µmoles per million cells compared to preincubation media, respectively, for PANC-1 and HK-2. The values represent the net consumption of glutamine in 3 h for each cell type and demonstrate that PANC-1 cells consumed glutamine to a higher degree than HK-2 cells. All quantified cell lines consume glutamine and, presumably, use it to varying degrees for cell proliferation and survival.21–23
We observed a higher intracellular concentration of branched-chain amino acids (leucine, isoleucine, and valine) in PANC-1 cells as compared to the other cell lines. Elevated plasma levels of branched-chain amino acids (BCAAs) have been recently associated with an elevated risk of pancreatic cancer development and are now considerate as an early diagnostic marker for pancreatic ductal adenocarcinoma (PDAC).24 With this method we were able to readily quantify this phenomenon as compared to other cancer cell lines.
Interestingly, we also observed high rates of tryptophan consumption (Figure 1B) in both HK-2 and UOK-262 renal cells, as compared to the other cell lines. A recent study demonstrated the importance of tryptophan metabolism in RCC tumorigenesis. First, the catabolism of tryptophan produces kynurenine, which has the capability to reduced immune response interfering directly with infiltrating tumor T cells. Moreover the catabolism of this amino acid leads to the production of quinolinate and NAD+, a reducing cofactor essential to increase the energetic potential of cancer cells.25 In this study, we confirm tryptophan as an important biomarker for renal cancer cells.
In Vitro [1,6-13C]Glucose Tracing Studies Using 1H NMR
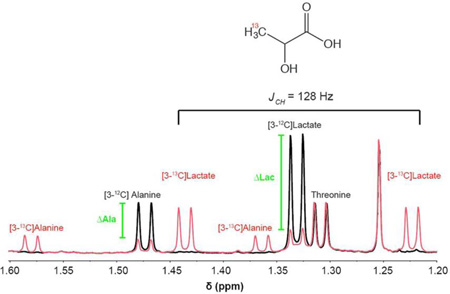
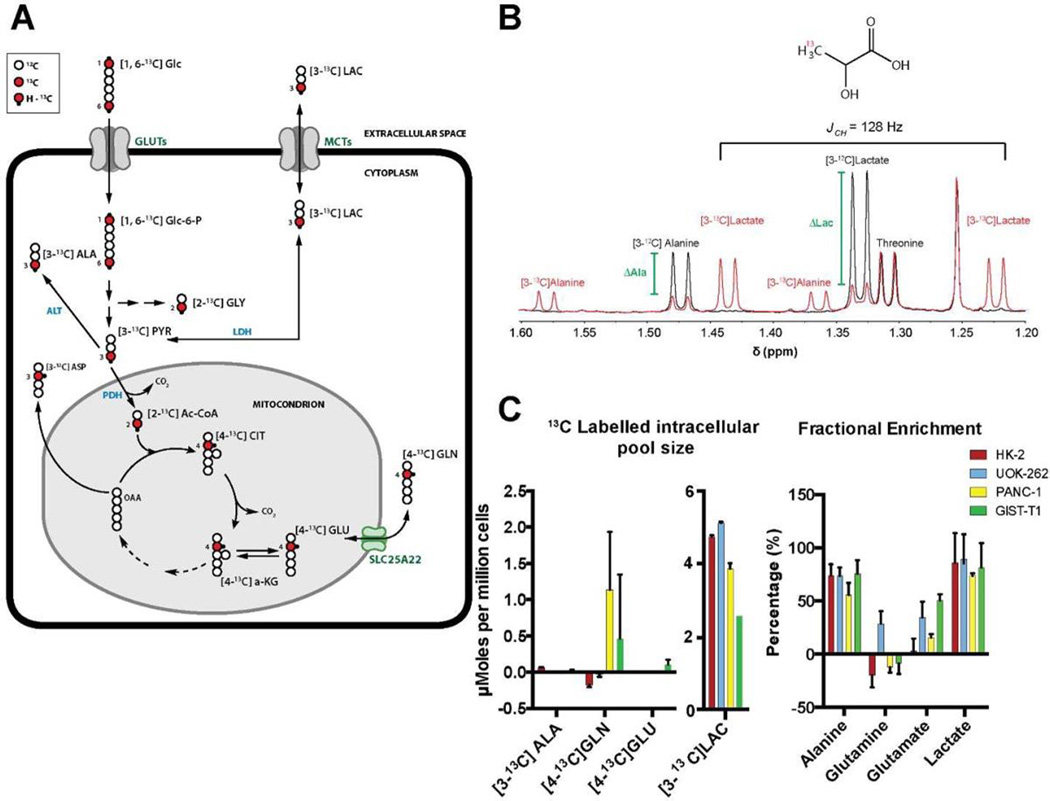
Numerous metabolomics studies, using NMR as a tool to quantify and track metabolites, have been performed with direct 13C measurement. Here, we demonstrate a reduced acquisition time of many orders of magnitude than the time for recording 2D NMR or direct 13C NMR (30 min each sample compared to several h) as well as the acquisition of quantitative data. 13C−1H coupling of 13C enriched metabolites derived from [1,6-13C]glucose has been determined by 1H NMR. Figure 2B shows an example of the effect of the enrichment in [3-13C]lactate derived from incubation of cells with enriched glucose. 1H of the methyl group of unlabeled lactate resonates at 1.32 ppm. Isotopic enrichment of 3-13C lactate was detected by J-splitting (128 Hz) of the methyl resonance (1.32 ppm). Using this phenomenon we are able to easily distinguish between the lactate derived from the tracing study and the lactate from total pool size and accurately measure both components. With 12C and 13C measurements in hand, we can then calculate the Fractional Enrichment (FE) of each compound of interest, here comparing a sample, which is unlabeled to a labeled sample.
Figure 2.
(A) Simplified schematic of [1,6-13C]glucose metabolism in glycolysis and the TCA cycle. Open circles are 12C atoms, filled red circles are 13C atoms, and black dots are protons attached to 13C atoms. Numbers refer to the position of the 13C atom. (B) 1H spectra of UOK-262 demonstrating the effect of 13C enrichment on lactate and alanine. The green bar indicates the change in the 1H−12C resonance for each metabolite, here lactate and alanine, and by fitting it in both instances we derive a ΔMetabolite, which corresponds to the contribution of 13C labeling. (C) Labeled intracellular and (D) extracellular pool size of 13C labeled water-soluble molecules with their respective FE after 3 h incubation with [1,6-13C]glucose. HK-2, UOK-262, PANC-1, and GIST-T1 are represented with red, blue, yellow, and green bars, respectively.
In order to overcome the problem of peak overlap, we identified nonoverlapping parts of a metabolite spin system to quantify the compound concentration and then deconvolution of overlapping peaks for separation of resonances of interest with a minimal residual. A table of identified and quantified compounds (Figure S1A) illustrates the 21 metabolites we assessed in these cell lines. For example, using the 1Hs attached to the C4 of glutamate (2.33−2.36 ppm) we were able to quantify this metabolite. The multiplets of C4 glutamate are in a region of the spectra with less overlap in comparison to other regions, and therefore easy to fit. To confirm fit, along with good quantification, we were able to assess the expected area of another glutamate resonance at 2.0 ppm as a control (Figure S1B). All resonances were fit using the complete spin system in our nonenriched samples.
To study glucose metabolism, we performed tracing experiments using [1,6-13C]glucose. Figure 2A illustrates the fate of [1,6-13C]glucose in cells with the respective labeling position of the 13C enriched metabolites. [1,6-13C]glucose enters the cells via glucose transporters (GLUTs) and is rapidly converted by hexokinase to [1,6-13C]glucose-6-P, which continues through the glycolytic pathway ending in [3-13C]-pyruvate. In the setting of normal cells under typical normoxic conditions, [3-13C]pyruvate continues into the tricarboxylic acid cycle (TCA cycle) with first a decarboxylation by PDH to yield [2-13C]Ac-CoA, which can continue through the cycle to [4-13C]α-KG. [4-13C]α-KG can then continue through the TCA cycle or exchange into the glutamate pool as [4-13C]-glutamate. For this reason, [4-13C] glutamate is typically used as an indicator of the first pass of TCA cycle flux. In a wide range of cells we observe differential flux of glucose into the TCA cycle when comparing the labeled glutamate and the corresponding FE. The FE of glutamate is 3 ± 1%, 34.6 ± 14%, 15.8 ± 3.1%, and 50 ± 5.3% for HK-2, UOK-262, PANC-1, and GIST-T1, respectively, demonstrating a real-time range of glutamate production from glucose and potentially identifying that in each cell line glutamate can be derived from other pathways.
In tumor cells, because of dysregulated metabolism (i.e., Warburg effect), [3-13C]pyruvate is reduced to [3-13C]lactate by lactate dehydrogenase (LDH) and subsequently excreted to the extracellular space via monocarboxylate transporters (MTCs) ending with a strong acidification of the extracellular environment. The acidification of the tumor environment is a classic characteristic of tumor invasiveness.26 Extracellular labeled lactate varies from cell lines and among the 4 cell lines analyzed, the metastatic and aggressive UOK-262 shows the highest production of extracellular lactate (5.115 ± 0.125 µmoles per million cells) in 3 h, demonstrating a correlation between the metabolic profile and the corresponding aggressive phenotype. More importantly, this provides a means for assessing a wide range of quantitative metabolic rates.
In Vitro Application of 1H NMR Metabolomics on Rapamycin-Treated UOK-262
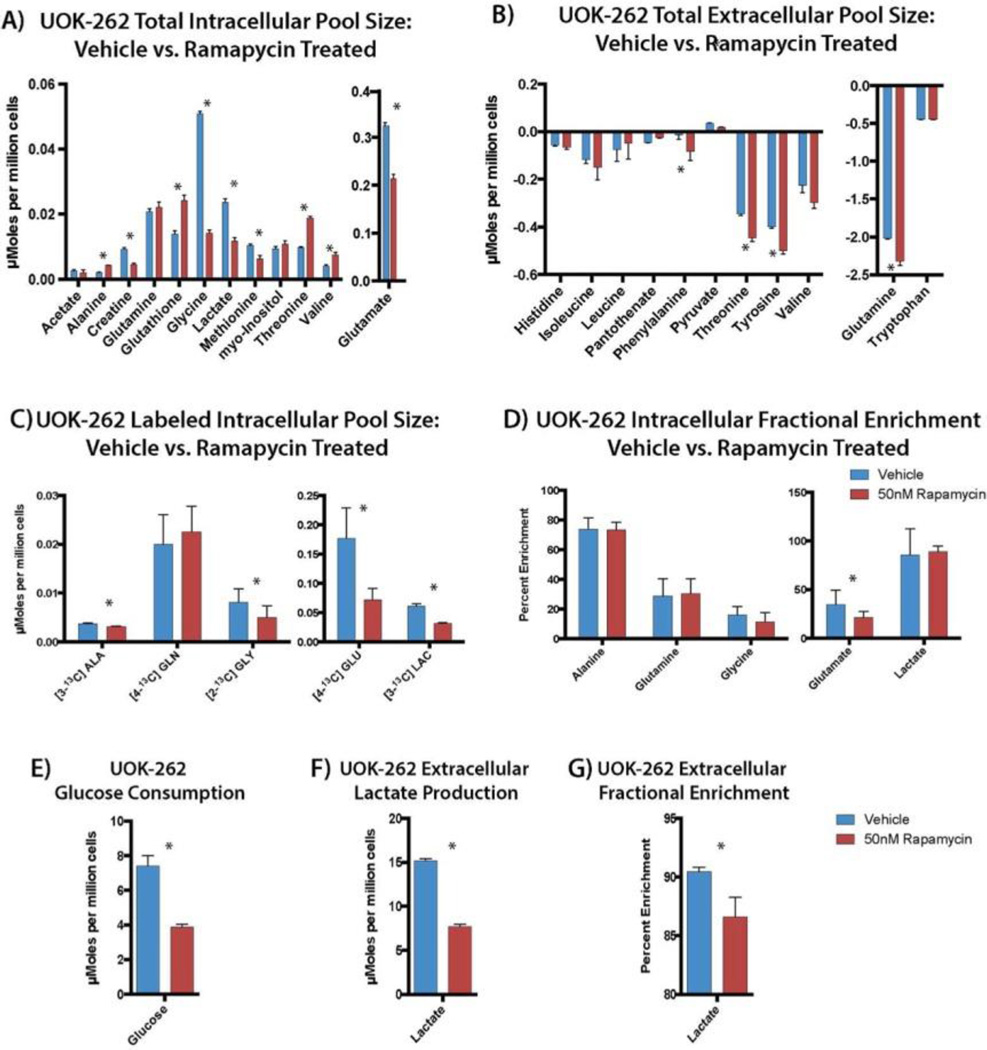
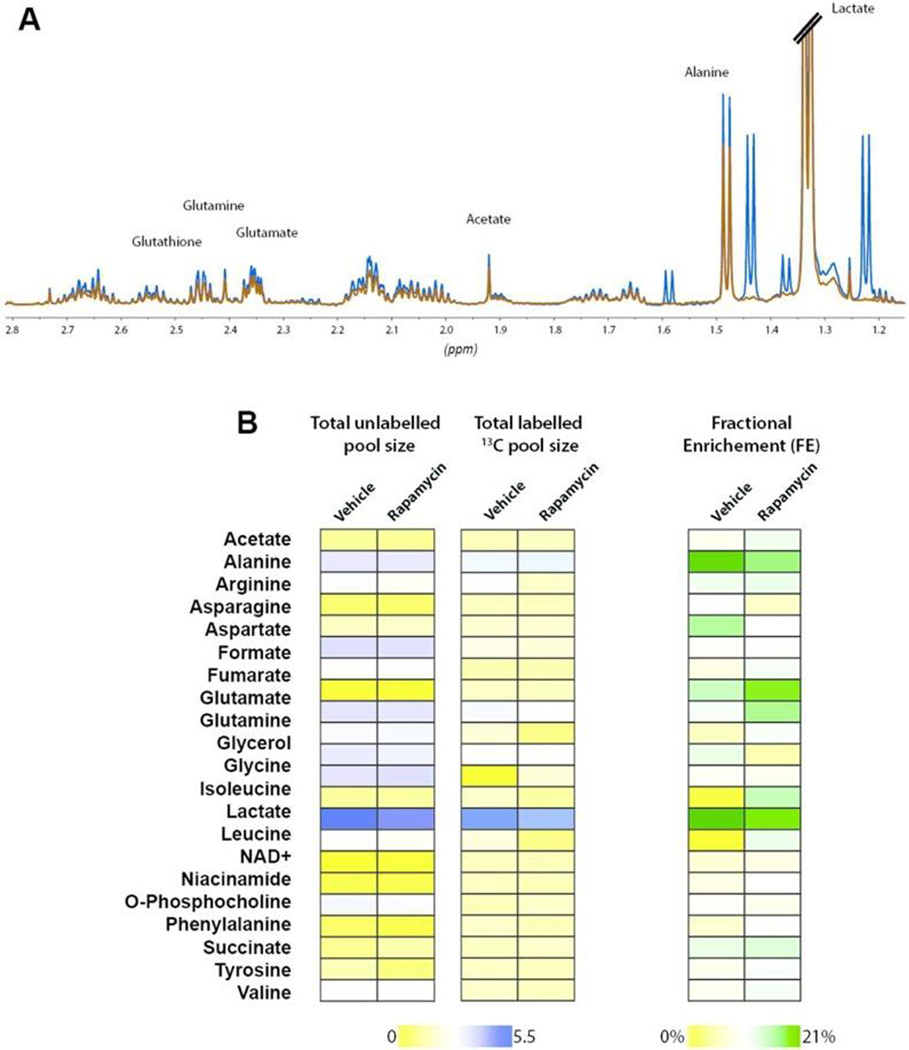
The mammalian target of rapamycin (mTOR) pathway plays a key role in regulating glucose metabolism and promoting cell proliferation. Using rapamycin as a tool compound, we tested the in vitro effect of this inhibitor in UOK-262 cancer cells and this approaches ability to assess quantitative changes in metabolism. Incubation of cells with 50 nM rapamycin inhibits the phosphorylation of p70S6 kinase (S6K) the target of mTORC1 (Figure S3). The total pool size of intracellular lactate and [3-13C]lactate significantly decreased by 50% and 51.9%, respectively (p-value < 0.001, for both), with 24 h of mTOR inhibition using rapamycin, reflecting a strong inhibition of the glycolytic flux. Moreover, the decrease of intracellular lactate production mirrors the decrease of the extracellular lactate concentration, in line with previously published studies of rapamycin.27–29 Another important inhibitory effect of rapamycin is the downregulation of amino acid metabolism. As shown in Figure 3A, glycine and methionine pool sizes are significantly reduced with rapamycin treatment as well as alanine and creatine. We observed a significant decrease in total extracellular pool size of glutamine consumption by 7% (Figure 3B) in rapamycin-treated cells compared to vehicle. In addition we observed a significant decrease of total intracellular pool size glutamate (0.324 ± 0.006 vs 0.214 ± 0.004, p-value < 0.001 for vehicle and rapamycin treated cells, respectively; Figure 3A). The accentuated decrease of intracellular glutamate concentration is most likely derived from inhibition of glycolysis and the TCA cycle together with an inhibition of glutaminolysis. The pool size of glutathione is also significantly decreased, as expected, due to the inhibition of glutamate and glycine metabolism.
Figure 3.
(A) Metabolic profile of the total intracellular and (B) extracellular pool size of vehicle vs rapamycin treated UOK-262. (C) Labeled intracellular pool size and (D) fractional enrichment (FE) of relevant metabolites in vehicle and rapamycin-treated UOK-262 cells. (E) Glucose consumption, (F) labeled extracellular, and (G) FE of lactate production in UOK-262 in vehicle and rapamycin treated cells. Blue bar represents vehicle-treated cell and red bars represent rapamycin-treated cells. *P < 0.05.
Figure 3C shows a smaller panel of labeled metabolites that change after 3 h of [1,6-13C]glucose incubation in UOK262 cells. There is a significant decrease in labeled intracellular and extracellular [3-13C]lactate (38%, p < 0.0001) production in rapamycin treated cells supporting that changes in lactate pool size are derived from glucose consumption in these cells, and that inhibition of mTOR results in decreased glycolysis. Labeling of C2 glycine from glucose is also significantly decreased as a result of mTOR inhibition. This readily demonstrates the feeding of glucose into 1 carbon metabolism and it is shunting upon rapamycin treatment. A strong inhibition of [4-13C]glutamate (59%, p = 0.0024) production is observed in treated cells. The glutamate fractional enrichment (FE) significantly decreased (34.65 ± 14.72% and 21.33 ± 6.43%, respectively, for vehicle versus rapamycin-treated cells, p-value < 0.005), indicating that glutamate production derived not only from glycolysis, but from other metabolic pathways.
In Vivo Tracing Experiment in Xenografted Mice
Utilizing [1,6-13C]glucose infusions in mice harboring xenografted tumors, we extended this method to the analysis of in vivo metabolism. 13C glucose enrichment of the plasma was approximately 70% at the end of the experiment, which is in line with previously published studies.10 In order to overcome the need for unlabeled samples to measure the total pool sizes of metabolites, we took advantage of 13C decoupling. This approach is robust with high signal-to-noise and Figure 4A demonstrates an example of coupled and decoupled 1H NMR spectra of the tumor extract. Figure 4B shows color maps for the percent change of metabolites expression for intracellular unlabeled and 13C-labeled pool size in vehicle versus rapamycin-treated tumor. With normalization for tissue mass, robust data is acquired, supporting the feasibility of this approach for in vivo flux tracing. High signal-to-noise is achieved in these data from minimal tissue (100 mg), readily demonstrating that an extract of 5−10 mg would be achievable in this setting, opening the door to fine needle aspirates and other low volume biological samples.
Figure 4.
(A) 1H NMR spectra (with and without 13C decoupling) of tumor extract after in vivo infusion of [1,6-13C]glucose. Lactate, alanine, glutamate, and glutamine are shown. Red and blue indicates decoupled and coupled spectra, respectively. (B) Color maps of intracellular 12C, 13C, and the fractional enrichment (FE) of water-soluble compounds extracted from rapamycin- and vehicle-treated GIST-T1 tumors after in vivo [1,6-13C]glucose infusion.
CONCLUSIONS
Herein we present an approach to perform rapid, easy, and quantitative metabolomics and flux tracing based on 1H NMR. We are able to readily detect 40 intracellular metabolites (using less than 1 × 106 cells), 13C enrichment of these pools from glucose using only two rapid acquisitions (<30 min) and profile an array of cell types. We demonstrate that this can then be used to rapidly assess metabolic changes in the setting of treatment response. With mTOR inhibition using a tool compound, rapamycin, and quantitative changes in labeling of multiple pools are reproducibly observed allowing for a method to screen these effects across other cells and nutrient sources.
This method is simultaneously quantitative and high throughput, giving it the potential to be translated easily in studies with multiple cell lines, treatments and probes. Since this method can detect changes in the high nanomolar to micromolar range, it can easily be extended to other substrates that target the pathway of interest, providing knowledge of the expected labeling patterns. Additionally, this approach could be used dynamically in order to accurately measure labeling flux rates for high concentration intermediates. Utilizing this as a first step in metabolomics investigations will then allow for more targeted and higher sensitivity assay with other approaches such as GC and LC/MS on the same sample post-NMR processing.
As shown in the Results and Discussion, we have performed analysis from purified mouse plasma and 100 mg of dissected tumor. This approach could not only be applied in basic research, but could be extended to analyzing biological liquids, such as urine, blood, or cerebrospinal fluid, as well as fine needle aspirates of patients who were infused with isotopically enriched substrates. Moreover, by the application of the cryoprobe technology, robotic sample changers and performing experiments with high field magnets the acquisition time and the overlap can be reduced markedly with increased sensitivity. The extract analyzed does not require further purification; therefore, the loss of compounds is minimized and the same sample could easily be applied in tandem with standard 1H metabolomics or mass spectrometry.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Amy Freund who provided the modified CHIRP pulse program. The authors would like to acknowledge the NIH/NCI Cancer Center Support Grant P30 CA008748 (PI: Thompson) and NIH/NIBIB R00 EB014328 (PI: Keshari), the Center for Molecular Imaging and Nanotechnology (CMINT) at MSK, and the American Italian Cancer Foundation.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Additional information as noted in text (PDF).
The authors declare no competing financial interest.
REFERENCES
- 1.Nagana Gowda GA, Raftery D. J. Magn. Reson. 2015;260:144–160. doi: 10.1016/j.jmr.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emwas AH, Luchinat C, Turano P, Tenori L, Roy R, Salek RM, Ryan D, Merzaban JS, Kaddurah-Daouk R, Zeri AC, Nagana Gowda GA, Raftery D, Wang Y, Brennan L, Wishart DS. Metabolomics. 2015;11:872–894. doi: 10.1007/s11306-014-0746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holt RM, Newman MJ, Pullen FS, Richards DS, Swanson AG. J. Mass Spectrom. 1997;32:64–70. doi: 10.1002/(SICI)1096-9888(199701)32:1<64::AID-JMS450>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Bajad S, Coumar M, Khajuria R, Suri OP, Bedi KL. Eur. J. Pharm. Sci. 2003;19:413–421. doi: 10.1016/s0928-0987(03)00143-x. [DOI] [PubMed] [Google Scholar]
- 5.Karimpour M, Surowiec I, Wu J, Gouveia-Figueira S, Pinto R, Trygg J, Zivkovic AM, Nording ML. Anal. Chim. Acta. 2016;908:121–131. doi: 10.1016/j.aca.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Fan TW, Lane AN. Prog. Nucl. Magn. Reson. Spectrosc. 2016;92–93:18–53. doi: 10.1016/j.pnmrs.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lane AN, Fan TW, Higashi RM. Methods Cell Biol. 2008;84:541–588. doi: 10.1016/S0091-679X(07)84018-0. [DOI] [PubMed] [Google Scholar]
- 8.Malloy CR, Sherry AD, Jeffrey FM. Am. J. Physiol. 1990;259:H987–H995. doi: 10.1152/ajpheart.1990.259.3.H987. [DOI] [PubMed] [Google Scholar]
- 9.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, Pascual JM, Madden CJ, Mickey BE, Malloy CR, Bachoo RM, Maher EA. Nat. Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, Cai L, Good L, Tu BP, Hatanpaa KJ, Mickey BE, Mates JM, Pascual JM, Maher EA, Malloy CR, Deberardinis RJ, Bachoo RM. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bingol K, Bruschweiler R. Anal. Chem. 2014;86:47–57. doi: 10.1021/ac403520j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An YJ, Xu WJ, Jin X, Wen H, Kim H, Lee J, Park S. ACS Chem. Biol. 2012;7:2012–2018. doi: 10.1021/cb3004226. [DOI] [PubMed] [Google Scholar]
- 13.Carvalho RA, Zhao P, Wiegers CB, Jeffrey FM, Malloy CR, Sherry AD. Am. J. Physiol. 2001;281:H1413–H1421. doi: 10.1152/ajpheart.2001.281.3.H1413. [DOI] [PubMed] [Google Scholar]
- 14.Benjamin DI, Cravatt BF, Nomura DK. Cell Metab. 2012;16:565–577. doi: 10.1016/j.cmet.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tredwell GD, Behrends V, Geier FM, Liebeke M, Bundy JG. Anal. Chem. 2011;83:8683–8687. doi: 10.1021/ac202123k. [DOI] [PubMed] [Google Scholar]
- 16.Maharjan RP, Ferenci T. Anal. Biochem. 2003;313:145–154. doi: 10.1016/s0003-2697(02)00536-5. [DOI] [PubMed] [Google Scholar]
- 17.Keshari KR, Sriram R, Koelsch BL, Van Criekinge M, Wilson DM, Kurhanewicz J, Wang ZJ. Cancer Res. 2013;73:529–538. doi: 10.1158/0008-5472.CAN-12-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marin-Valencia I, Cho SK, Rakheja D, Hatanpaa KJ, Kapur P, Mashimo T, Jindal A, Vemireddy V, Good LB, Raisanen J, Sun X, Mickey B, Choi C, Takahashi M, Togao O, Pascual JM, Deberardinis RJ, Maher EA, Malloy CR, Bachoo RM. NMR Biomed. 2012;25:1177–1186. doi: 10.1002/nbm.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tannus A, Garwood M. NMR Biomed. 1997;10:423–434. doi: 10.1002/(sici)1099-1492(199712)10:8<423::aid-nbm488>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 20.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Anal. Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 21.Wise DR, Thompson CB. Trends Biochem. Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang CV. Cell Cycle. 2010;9:3884–3886. doi: 10.4161/cc.9.19.13302. [DOI] [PubMed] [Google Scholar]
- 23.Pavlova NN, Thompson CB. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP, Yuan C, Bao Y, Townsend MK, Tworoger SS, Davidson SM, Papagiannakopoulos T, Yang A, Dayton TL, Ogino S, Stampfer MJ, Giovannucci EL, Qian ZR, Rubinson DA, Ma J, Sesso HD, Gaziano JM, Cochrane BB, Liu S, Wactawski-Wende J, Manson JE, Pollak MN, Kimmelman AC, Souza A, Pierce K, Wang TJ, Gerszten RE, Fuchs CS, Vander Heiden MG, Wolpin BM. Nat. Med. 2014;20:1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganti S, Taylor SL, Abu Aboud O, Yang J, Evans C, Osier MV, Alexander DC, Kim K, Weiss RH. Cancer Res. 2012;72:3471–3479. doi: 10.1158/0008-5472.CAN-11-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Milito A, Fais S. Future Oncol. 2005;1:779–786. doi: 10.2217/14796694.1.6.779. [DOI] [PubMed] [Google Scholar]
- 27.Benjamin D, Colombi M, Moroni C, Hall MN. Nat. Rev. Drug Discovery. 2011;10:868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 28.Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X, Wang Y, Jing Y, Yang H, Chen R, Chang L, Zhang Y, Goto J, Onda H, Chen T, Wang MR, Lu Y, You H, Kwiatkowski D, Zhang H. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edinger AL, Linardic CM, Chiang GG, Thompson CB, Abraham RT. Cancer Res. 2003;63:8451–8460. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.