Abstract
Metabolic activation of the carcinogenic tobacco-specific N-nitrosamines leads to the formation of 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB)-releasing DNA adducts. We recently developed a liquid chromatography (LC)-tandem mass spectrometry (MS/MS) method for the analysis of HPB-releasing DNA adducts in human oral cells. However, given the limited amounts of DNA that can be extracted from oral cells, higher sensitivity and selectivity are required for the reliable analysis of these adducts in future studies. We have developed a new sensitive LC-nanoelectrospray ionization-high resolution MS/MS method for the analysis of HPB-releasing DNA adducts in oral cells. A new procedure was also developed for guanine analysis by LC-MS/MS. The detection limit of the developed assay is 5 amol, and the limit of quantitation is 0.35 fmol HPB on-column, starting with 50 pg DNA. The method was tested by analyzing oral samples from 65 smokers, including 30 head and neck squamous cell carcinoma (HNSCC) patients and 35 cancer-free controls. In all smokers, the levels of HPB-releasing DNA adducts averaged 6.22±16.18 pmol/mg DNA, with significant inter-individual variation being consistent with previous reports. The median HPB-releasing DNA adduct level was 6.6 time greater for those with HNSCC than for smokers without HNSCC (p=0.002). The developed highly sensitive and selective method is valuable tool for future measurement of HPB-releasing DNA adducts in tobacco users, which can potentially provide critical insights for the identification of individuals at risk for cancer.
Keywords: HPB, DNA adduct, oral cells, oral cancer, nanoelectrospray, high resolution mass spectrometry
INTRODUCTION
Exposure to chemical carcinogens present in tobacco and cigarette smoke is largely responsible for the high cancer morbidity and mortality rates among tobacco users worldwide.1-3 DNA adduct formation is central to the process of chemical carcinogenesis.4, 5 In exposed individuals, the levels of DNA adducts derived from a specific carcinogen are likely to depend on the level of exposure, the relative efficiency of the carcinogen metabolic activation and detoxification pathways, as well as the efficiency of DNA repair mechanisms.3 Therefore, measurement of tobacco carcinogen-derived DNA adducts can potentially provide critical insights into individual susceptibility to tobacco-induced cancers in tobacco users.
Tobacco-specific N-nitrosamines (TSNA) are among the most important carcinogens in tobacco and cigarette smoke. Two compounds from this group – 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N′-nitrosonornicotine (NNN) – are organ-specific carcinogens inducing cancers of the lung, pancreas, oral and nasal cavity, and esophagus, and are classified as carcinogenic to humans by the International Agency for Research on Cancer.6, 7 Since NNK and NNN are formed from tobacco alkaloids, human uptake of these carcinogens occurs primarily through exposure to tobacco or cigarette smoke, and to a lesser degree, from some tobacco alkaloid-containing products such as nicotine replacement therapy products or electronic cigarettes.6, 8-10 Measurement of urinary biomarkers of NNK and NNN exposure generated critical insights into human uptake and metabolism of these carcinogens.8, 11, 12 Notably, recent studies demonstrated the association between NNK and NNN exposure and the risk of lung and esophageal cancer, respectively, in smokers from the prospective Shanghai cohort study.11, 13
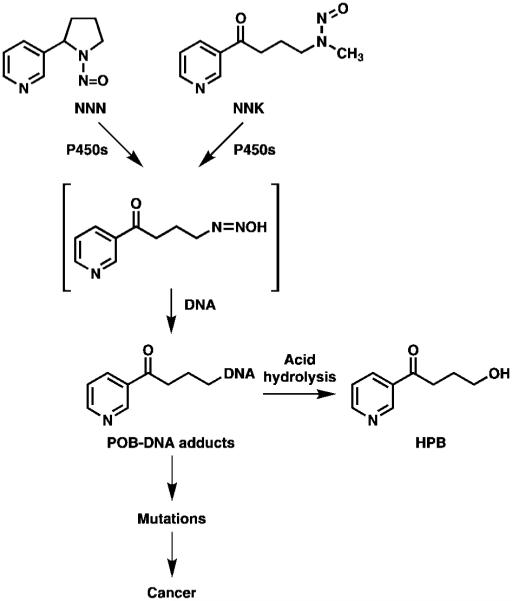
Metabolic activation of NNK and NNN leads to the formation of pyridyloxobutyl (POB) DNA adducts.4, 6 Under strong acid hydrolysis conditions, the majority of POB-DNA adducts decompose to release 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB). 14-16 In vitro and laboratory animal studies have demonstrated the importance of HPB-releasing adducts in NNK- and NNN-induced carcinogenesis.6, 17 A limited number of studies measured HPB-releasing DNA adducts in human tissues, supporting the potential utility of this biomarker as a direct measure of tobacco-induced DNA damage and cancer risk.18-21 The recently proposed use of oral cells as a surrogate source of DNA for HPB-releasing DNA adduct measurement in smokers presents a promising approach to overcome the invasiveness of tissue collection from the most organs targeted by NNK and NNN.16 In addition, given that NNN is a potent oral carcinogen,17 measurement of HPB-releasing DNA adducts in oral cells could potentially provide the most direct quantitative and mechanistic link between tobacco use and oral cancer in humans.
We recently developed a liquid chromatography–tandem mass spectrometry (LC-MS/MS) method for the analysis of HPB-releasing DNA adducts in human oral cells.16 However, application of this method in studies in which only limited oral cells are available for DNA extraction requires further modification to increase its sensitivity and selectivity. Collection of oral brushings from oral cancer patients or from children exposed to secondhand smoke are some examples of scenarios in which only small amounts of DNA may be extracted from the collected samples. Our purpose in this study was to modify the previously developed method by optimizing the sample preparation procedure and switching to LC-nanoelectrospray ionization-high resolution tandem mass spectrometry (LC-NSI-HRMS/MS). To demonstrate the utility of this optimized method, we applied it to the analysis of oral brushings collected from smokers with head and neck squamous cell carcinoma (HNSCC). In addition, since in our previous study smokers with HNSCC showed increased exposures to NNN than those without HNSCC,22 oral samples from cancer-free smokers were analyzed here for comparison. The ultimate goal of the development of this methodology is its application in future studies to understand the relationship between tobacco exposure and HPB-releasing DNA adduct formation in tobacco users, and to evaluate the utility of these adducts as biomarkers of cancer risk in tobacco users.
EXPERIMENTAL PROCEDURES
Chemicals and enzymes
HPB, [pyridine-D4]HPB, [3,3,4,4-D]HPB, and [13C215N]guanine were purchased from Toronto Research Chemicals (North York, Ontario, Canada). Reagents and enzymes for DNA isolation were obtained from QIAGEN Sciences (Germantown, MD). Calf thymus DNA was purchased from Worthington Biochemical Corporation (Lakewood, NJ). All other chemicals and solvents were purchased from Sigma-Aldrich Chemical Co. (Milwaukee, WI).
Subjects and oral cell collection
Oral brushings were collected from 30 smokers with head and neck squamous cell carcinoma (HNSCC) and 35 smokers without HNSCC recruited through the Department of Otolaryngology-Head and Neck Surgery at the University of Minnesota. Study participants were included if they were daily smokers of at least 5 cigarettes per day for at least 5 years. HNSCC patients were identified upon presentation to the Otolaryngology-Head and Neck Surgery clinic with a new diagnosis of squamous cell carcinoma of the upper aerodigestive tract. This included tumors of the oral cavity, oropharynx, larynx and hypopharynx. Cancer-free controls were recruited in the same outpatient clinic. These subjects were visiting the clinic for clinical evaluation of problems other than cancer (i.e. sinusitis, hearing loss). The study was approved by the University of Minnesota Research Subjects’ Protection Programs Institutional Review Board: Human Subjects Committee (IRB Study # 0903M62203). Oral cells were collected by brushing the oral mucosa inside one cheek with a clean toothbrush and swirling the brush in a sterile polypropylene centrifuge tube with a commercial mouthwash to transfer the collected buccal cells from the brush into the liquid. After the collection, the samples were centrifuged at 2,700 rpm to pellet cells; the pellets were washed with Tris-EDTA buffer (pH 7.4) and stored at −20 °C until DNA isolation and analysis.
DNA isolation from oral cells
DNA isolation from the collected samples was performed using the commercial protocol for DNA purification from oral cells (Qiagen, Valencia, CA). Briefly, oral cells were thawed and centrifuged at 3,000 × g for 15 min at 4°C. Supernatant was discarded and pellets were re-suspended in 200 μL 100 mM phosphate buffer (pH 6.8). Cells were lysed using Qiagen AL buffer, treated with proteinase K and RNase A, and DNA was purified using QiaAmp DNA Mini spin columns. The amount of isolated DNA was calculated based on the amount of guanine,23 which was analyzed as part of our developed method as described below.
DNA hydrolysis and sample purification
The isolated DNA samples were mixed with 0.12 pmol of [pyridine-D4]HPB and 1000 pg of [13C215N]guanine, which served as internal standards for HPB and guanine, respectively. A water blank (0.5 mL) and a sample of calf thymus DNA (5μg dissolved in 0.5 mL deionized water) were added to each sample set as negative controls. Hydrochloric acid was added to each sample to bring the final concentration of the acid to 0.8 N, and the samples were incubated at 80 °C for 3 h to release HPB. 14, 16, 20 The hydrolysates were adjusted to neutral pH with NaOH, and loaded on HyperSep Hypercarb cartridges (25 mg, Thermo Scientific, Rockwood, TN) activated with 2 mL MeOH and 2 mL H2O. The cartridges were then washed with 2 mL H2O. HPB was eluted with 1 mL of 65% MeOH, and guanine was eluted with 1 mL MeOH (containing 1% TFA) sequentially. The two fractions were collected in separate collection tubes, concentrated to dryness in a centrifugal evaporator, and re-dissolved in H2O prior to mass spectrometry analysis.
Analysis of released HPB by LC-NSI-HRMS/MS
The analysis of HPB in the prepared samples was carried out on an LTQ Orbitrap Velos instrument (Thermo Scientific, Waltham, MA) interfaced with a Nano2D-LC HPLC (Eksigent, Dublin, CA) system using nanoelectrospray ionization. The analysis was performed using a capillary column (75 μm ID, 10 cm length, 15 μm orifice) prepared by hand packing a commercially available fused-silica emitter (New Objective, Woburn MA) with Luna C18 bonded separation media (Phenomenex, Torrance, CA). The mobile phase consisted of 5 mM NH4OAc and CH3CN, using a linear gradient at a flow rate of 300 nL/min with increasing CH3CN from 2 to 30% over 15 min, followed by ramping to 98% CH3CN within 1 min, and holding at this composition for 2 min. The gradient was then returned to 2% CH3CN in 1 min and the system was re-equilibrated at this mobile phase composition for 5 min at 1000 nL/min before next injection. For injections, a 5 μL injection loop was used and the sample (4 μL) was loaded onto the capillary column with a 1000 nL/min flow at the initial conditions (2% CH3CN) for 5.5 min. The nanoelectrospray source voltage was set at 2.2 kV. The capillary temperature was 350°C, and the S-Lens RF Level was set at 40%. The analysis was performed using accurate mass extracted ion chromatograms of m/z 106.0287 [C6H4NO]+ (parent ion m/z 166.1) for HPB and corresponding fragment (m/z 110.0538, parent ion m/z 170.1) for [pyridine-D4]HPB with a mass tolerance of 5 ppm. The scan events were performed using higher-energy collisional dissociation (HCD) fragmentation with a normalized collision energy of 60 units, isolation widths of 3 Da for both the analyte and internal standard, and product ion spectra acquisition at a resolution of 50,000.
Analysis of guanine by LC-MS/MS
Analysis of guanine was performed on a TSQ Vantage triple quadrupole mass spectrometer (Thermo Scientific, Waltham, MA) interfaced with an Agilent 1100 capillary HPLC system (Agilent, Palo Alto, CA). Analysis was performed on a Luna C18(2) column (5 μm, 250 × 0.5 mm, Phenomenex) at a flow rate of 10 μL/min with the temperature maintained at 30°C. Sample injection volume was 8 μL. The mobile phase consisted of H2O (containing 0.01% acetic acid) and CH3CN with a linear gradient from 3 to 18% CH3CN over a period of 15 min, followed by ramping to 70% CH3CN within 1 min and holding at this composition for 4 min. The gradient was then returned to 3% CH3CN in 2 min followed by 15 min re-equilibration. The ESI source was operated in positive ion mode, monitoring m/z 152 [M+H]+→135 [C5H3N4O]+, and m/z 152→110 [C4H4N3O]+ for guanine, and corresponding transitions m/z 155→138 and m/z 155→113 for [13C215N]guanine. The collision gas was Ar at 1 mTorr with collision energy of 19 eV. The quadrupoles were operated at a resolution of 0.2 (Q1) and 0.7 (Q3) Da.
Quantitation of HPB and guanine
The quantitation of HPB and guanine was based on the peak area ratio of the analytes to their corresponding isotope-labeled internal standards, the constructed calibration curves, and the amount of internal standards added. Calibration curves were constructed before each analysis using a series of standard solutions of analytes and internal standards. The calibration standard solutions of HPB contained a constant amount of [pyridine-D4]HPB (4.7 fmol on-column) and varying amounts of HPB (0.005, 0.012, 0.024, 0.05, and 0.12 fmol on-column). The calibration standard solutions of guanine contained a constant amount of [13C215N]guanine (80 ng on-column) and varying amounts of guanine (1.6, 4, 8, 40, 80, 400, and 800 ng on-column).
Method characterization
To eliminate any potential interference of background HPB contamination, we used [pyridine-D4]HPB to characterize the accuracy and precision of the developed method. To determine the amounts of [pyridine-D4]HPB in the samples processed in these experiments, we used [3,3,4,4-D4]HPB as internal standard. The two deuterium-labeled standards have the same parent ion m/z 170.1, but form different fragments and therefore can be distinguished by extracting ion chromatograms of m/z 110.0538 for [pyridine-D4]HPB and m/z 106.0287 for [3,3,4,4-D4]HPB. Accuracy and precision were determined by adding different amounts of [pyridine-D4]HPB (0.35, 0.7, 3.5, 7, and 35 fmol) and 3.5 fmol of [3,3,4,4-D4]HPB to 50 pg calf thymus DNA in 0.5 mL H2O, followed by hydrolysis and purification as described above. Samples at each level of added [pyridine-D4]HPB were prepared and analyzed in triplicate.24 Accuracy was determined by comparing added and measured amounts of [pyridine-D4]HPB at each level. Precision was determined as intra-day and inter-day coefficients of variation (% CV) for the triplicate samples analyzed on three separate days.
The limit of detection (LOD) was determined using standard solutions of [pyridine-D4]HPB. The limit of quantitation (LOQ) was established in calf thymus DNA samples by adding [pyridine-D4]HPB (0.07, 0.35, 0.7, and 3.5 fmol) and [3,3,4,4-D4]HPB (3.5 fmol) to calf thymus DNA samples, followed by hydrolysis and purification, and analyzing each sample in triplicate. The LOQ was defined by identification of the lowest [pyridine-D4]HPB level that produced a coefficient of variation (CV) lower than 15%.25
Recovery was determined by comparing the results of samples to which [3,3,4,4-D4]HPB (240 pmol) was added to 5 μg of calf thymus DNA at the beginning and at the end of sample preparation procedure.
Statistical analyses
The HPB values below the LOQ were estimated based on the lowest quantifiable HPB level among the analyzed oral samples (see footnote ‘b’ to Table 1). DNA yields and the HPB-releasing DNA adduct levels were summarized by the mean, standard deviation (SD), median and inter-quartile range (IQR) for HNSCC patients and cancer-free smokers separately and combined. Due to an extremely skewed distribution for the HPB level, the analysis was performed in the natural logarithmic scale using a two-sided, two-sample t-test with unequal variances. To verify these results, the two-sample non-parametric Wilcoxon rank sum test was applied to the original arithmetic values without imputing a value below the LOQ, but instead assigning the same lowest rank for these cases. The Fisher’s exact test compared the percent of cases with HPB-releasing DNA adducts below the LOQ between cancer and cancer-free smokers.
Table 1.
Levels of HPB-releasing DNA adducts in oral cells from smokers with HNSCC and smokers without HNSCC.
| Smokers without HNSCC | Smokers with HNSCC | ||||
|---|---|---|---|---|---|
|
|
|
||||
| subject # | DNA yield (μg) |
HPB (pmol/mg DNA) |
subject # | DNA yield (μg) |
HPB (pmol/mg DNA) |
|
|
|
||||
| 1 | 1.24 | LOQa | 1 | 0.96 | 0.59 |
| 2 | 0.21 | LOQ | 2 | 1.43 | 0.29 |
| 3 | 0.87 | 0.80 | 3 | 0.47 | 1.61 |
| 4 | 0.17 | 2.81 | 4 | 0.089 | 4.53 |
| 5 | 1.71 | 0.23 | 5 | 1.15 | 0.13 |
| 6 | 0.66 | 0.27 | 6 | 0.28 | 0.32 |
| 7 | 0.14 | 0.66 | 7 | 0.35 | 0.27 |
| 8 | 0.092 | 4.53 | 8 | 0.092 | 1.26 |
| 9 | 0.19 | LOQ | 9 | 0.14 | 2.56 |
| 10 | 0.27 | 0.68 | 10 | 2.46 | 0.13 |
| 11 | 0.48 | 0.56 | 11 | 0.063 | 29.8 |
| 12 | 2.25 | 0.028 | 12 | 0.057 | 31.8 |
| 13 | 0.068 | 2.68 | 13 | 8.71 | 0.10 |
| 14 | 0.11 | 0.37 | 14 | 0.035 | 92.2 |
| 15 | 0.021 | LOQ | 15 | 1.32 | 0.93 |
| 16 | 1.17 | 0.79 | 16 | 0.40 | 0.68 |
| 17 | 0.18 | LOQ | 17 | 0.087 | 22.9 |
| 18 | 0.23 | LOQ | 18 | 0.022 | 8.47 |
| 19 | 0.026 | LOQ | 19 | 0.23 | 2.33 |
| 20 | 0.006 | 47.0 | 20 | 0.48 | 1.47 |
| 21 | 0.78 | 0.68 | 21 | 0.49 | LOQ |
| 22 | 0.002 | LOQ | 22 | 0.038 | 3.90 |
| 23 | 0.60 | LOQ | 23 | 0.14 | 11.3 |
| 24 | 0.052 | LOQ | 24 | 0.33 | 17.9 |
| 25 | 0.050 | LOQ | 25 | 0.36 | 2.01 |
| 26 | 0.026 | 19.0 | 26 | 0.75 | 5.12 |
| 27 | 0.091 | 2.01 | 27 | 0.25 | 0.80 |
| 28 | 0.042 | LOQ | 28 | 0.052 | 1.55 |
| 29 | 0.039 | LOQ | 29 | 0.33 | 0.63 |
| 30 | 0.024 | 71.5 | 30 | 0.69 | 0.067 |
| 31 | 0.11 | LOQ | Averageb | 0.74 | 8.19 |
| 32 | 0.18 | 1.61 | SD | 1.57 | 17.8 |
| 33 | 0.10 | 0.11 | Median | 0.33 | 1.51 |
| 34 | 0.028 | 2.21 | IQR | 0.089, 0.69 | 0.32, 5.12 |
| 35 | 0.075 | LOQ | |||
| Averageb | 0.35 | 4.53 | |||
| SD | 0.52 | 14.36 | |||
| Median | 0.11 | 0.23 | |||
| IQRc | 0.042, 0.48 | 0.001, 1.61 | |||
LOQ, below the limit of quantitation (2 amol on-column)
To calculate average values, 1/2 of the lowest level of HPB in the samples that HPB was quantifiable (0.028 pmol/mg DNA in subject 12 of smokers without HNSCC) was used for samples in which HPB levels were below LOQ. The value (0.028 pmol) was divided by its DNA yield (2.2 μg), then multiplied by the DNA yield of samples with LOQ, and half of that value was used as the HPB levels in these samples.
IQR, inter-quartile range.
RESULTS
Development of the analytical procedure
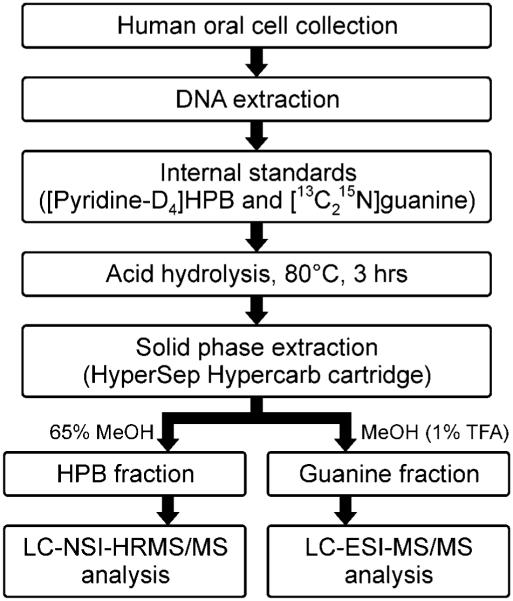
The developed protocol is outlined in Figure 1. Changes in sample purification procedure introduced in this modified method were driven in part by the need to optimize DNA quantitation in samples obtained from oral brushings. In our previously reported method, an aliquot (40 μL) of unpurified DNA hydrolysate from each sample was collected for analysis of guanine by HPLC-UV,16 which is used to calculate DNA yield.23 However, due to the small amounts of DNA that can be obtained from oral cells collected with a cytobrush, we were not able to measure guanine reliably by HPLC-UV in most samples. The development of a more sensitive and selective mass spectrometry-based method required introduction of a solid phase extraction clean-up step for hydrolysate purification to remove interfering salts and thus prevent ion suppression during guanine analysis. Initially, we explored the use of Strata X (Phenomenex) – the cartridge used in our originally developed method – to wash out interfering salts in H2O and subsequently elute both guanine and HPB by applying stronger solvents. However, guanine was not retained on the cartridge due to its high polarity. Therefore, we switched to Hypercarb solid phase extraction cartridge (Thermo) which has high affinity towards polar analytes. Both guanine and HPB present in DNA hydrolysates were successfully retained on the cartridge; the analytes were eluted from the cartridge in two wash-out steps (Figure 1), generating two separate fractions for mass spectrometry analyses.
Figure 1.
Analytical scheme for the analysis of HPB-releasing DNA adducts and guanine in human oral cell DNA.
Analysis of HPB released from DNA
Using HCD fragmentation of the LC-NSI-HRMS/MS system, the product ion spectra of HPB and [pyridine-D4]HPB generated several major fragment ions (Figure S1). Because the highest signal intensities in the spectra of both HPB and [pyridine-D4]HPB were for m/z 106.0287 and m/z 110.0538, respectively, these fragments were selected for quantitative analysis. Two additional fragments, m/z 148.0757 and m/z 122.0600, were used to confirm the identity of HPB; the corresponding deuterium-labeled fragment ions were used to confirm the identity of [pyridine-D4]HPB.
Analysis of guanine
Based on the fragmentation patterns determined in the product ion scan mode, two major ions were monitored for guanine and its internal standard in selected reaction monitoring (SRM) mode: m/z 135 and m/z 110 for guanine, and m/z 138 and m/z 113 for [13C215N]guanine. Because of their higher signal intensities, the transitions m/z 152→135 and m/z 155→138 were selected for quantitation of guanine and [13C215N]guanine, respectively. The peak area ratio between m/z 152→135 and m/z 152→110 was used to confirm the identity of guanine, and corresponding ratio was used to confirm the identity of the internal standard (Figure S2). With this developed LC-ESI-MS/MS method, guanine exhibited excellent linearity within the determined concentration range (Figure S3), and it was accurately quantified in all oral cell DNA samples. Typical SRM chromatograms obtained upon analysis of guanine and [13C215N]guanine in a standard mix and in oral cells from a smoker are presented in Figure 3.
Figure 3.
Typical SRM chromatograms obtained upon analysis of guanine and [13C215N]guanine in (A) standard mix and (B) oral cell sample from a smoker.
Method characteristics
The average accuracy of measured levels of [pyridine-D4]HPB (expressed as % of added amount) at 0.35, 0.7, 3.5, 7, and 35 fmol was 106%, 116%, 108%, 107%, and 109%, respectively, exhibiting excellent linearity (R2 = 1, Figure 4A). The interday and intraday CVs were 11% and 13%, respectively. The recovery of the assay was 65 ± 4% (n=5). An LOD of 5 amol [pyridine-D4]HPB (on-column) was achieved for a standard HPB solution. In CT-DNA samples mixed with HPB, an LOQ of 0.35 fmol (on-column) was achieved based on a CV of 13%. During the analysis of oral samples from smokers, the instrument response and the HPB/[pyridine-D4]HPB ratio for the calibration standard mixes were linear in the 0.005–0.12 fmol (on-column) range of HPB (R2 = 0.998, Figure 4B).
Figure 4.
(A) Relationship between added and measured [pyridine-D4]HPB in the accuracy experiment. Various amounts of [pyridine-D4]HPB (0.35, 0.7, 3.5, 7, and 35 fmol) were added to calf thymus DNA (50 pg), and analyzed by the method described in the text; (B) Linearity of HPB/[pyridine-D4]HPB at constant [pyridine-D4]HPB amount (4.8 fmol on-column) and HPB ranging from 0.005 to 0.12 fmol (on-column).
Quantitation of HPB-releasing DNA adducts in human oral cells
The developed method was applied to the analysis of oral cell DNA samples from 65 smokers, including 30 smokers with HNSCC and 35 smokers without HNSCC. Typical chromatograms obtained upon analysis of HPB in oral cells from a smoker with HNSCC and a smoker without HNSCC are presented in Figure 5.
Figure 5.
Typical accurate mass extracted ion chromatograms obtained upon analysis of HPB-releasing DNA adducts in human oral cells from a (A) smoker with HNSCC and (B) smoker without HNSCC.
The results of HPB levels from all the subjects are listed in Table 1. The yield of DNA isolated from oral cells in this study averaged 0.53 ± 1.16 μg (median 0.18, IQR 0.063, 0.49). The levels of HPB in the analyzed samples ranged from non-quantifiable to 92.2 pmol/mg DNA, averaging (±SD) 6.22 ± 16.18 pmol/mg DNA in all subjects (median 0.68, IQR 0.028, 2.56). Only one out of 30 (3.3%) HNSCC patients had HPB-releasing DNA adduct levels below the LOQ, compared to 15 out of 35 (42.9%) cancer-free smokers (p<0.001). In HNSCC patients, HPB-releasing DNA adducts averaged 8.19 ± 18.12 pmol/mg DNA (median 1.51, IQR 0.32, 5.12). In cancer-free smokers, the adduct level averaged 4.53 ± 14.36 pmol/mg DNA (median 0.23, IQR 0.001, 1.61). Comparing the means in the log scale resulted in a significantly higher HPB-releasing DNA adducts in HNSCC patients than cancer-free smokers (p<0.001). Comparing the medians plus percentiles between these groups using the non-parametric method resulted in a similar finding, with the median HPB-releasing DNA adducts for the cancer cases ~6.6 times greater than non-cancer smokers (p=0.002).
DISCUSSION
Analysis of HPB-releasing DNA adducts in oral cells can potentially be used as a non-invasive measure of DNA damage caused by the tobacco carcinogens NNK and NNN in tobacco users. We previously developed a method for the analysis of HPB-releasing DNA adducts in human oral cells. However, the amount of DNA that can be isolated from oral cells is usually low, and application of the previously developed method in our new studies often resulted in undetectable levels of HPB or inability to reliably quantify DNA in the available oral samples. Therefore, we aimed to modify our previously developed method in order to achieve lower limit of quantitation and better selectivity for HPB-releasing DNA adducts, as well as higher accuracy of DNA quantitation in human oral cells.
The previous method for quantitation of HPB in oral cells was based on the use of a triple quadrupole mass spectrometer and achieved an LOQ of 2.4 fmol HPB on-column.16 In that study, a high resolution mass spectrometer (HRMS) with accurate mass capability (Orbitrap) was used to confirm the identity of HPB in several oral cell DNA samples.16 That experiment revealed that when the same processed oral cell sample was analyzed on the two instruments, a dramatic improvement in selectivity was observed in the case of HRMS, producing clear peaks and no baseline noise or co-eluting interferences. Based on this observation, and our previous successful experience with the quantitation of another DNA adduct, 3-(2-deoxy-β-D-erythro-pentafuranosyl)pyrimido[1,2-α]purin-10(3H)-one deoxyguanosine (M1dG),26 we decided to switch to the use of LC-NSI-HRMS/MS for the development of the new optimized method. An LOQ of 0.35 fmol on-column was achieved using the current method, improving the sensitivity by approximately 7-fold compared to previous method. Consistent with the observation from our first study, the baseline noise was completely eliminated with the use of LC-NSI-HRMS/MS, greatly improving selectivity of the method (Figure 5). In addition to the improvement in the measurement of HPB-releasing DNA adducts, our procedure allows for simultaneous extraction of guanine during solid-phase purification of DNA hydrolysates, and the quantitation of guanine by the newly developed mass spectrometry-based method. Among the analyzed samples, the lowest calculated DNA yield based on guanine analysis was 0.002 μg (Table 1), demonstrating excellent sensitivity of the developed method and its value for studies in which only limited amounts of DNA are available.
Application of the modified method to the analysis of oral cells from smokers showed large inter-individual variation in the levels of HPB-releasing DNA adducts, which encourages further research on the role of these adducts in cancer risk in tobacco users. This variation and the range of adduct levels – from non-quantifiable to 92.2 pmol/mg DNA – are consistent with our previous report.16 In that report, one out of 30 oral cell brushings from smokers contained HPB in the amount of 246 pmol/mg DNA; the remaining 29 samples ranged from non-quantifiable to 109 pmol/mg DNA, which is similar to the current study. The average level of HPB-releasing DNA adducts in the current study however, is 6.22 ± 16.18 pmol/mg DNA in all 65 subjects, which is lower than 45 ± 57 pmol/mg DNA in 30 smokers in the previous report. This difference could be due to the differences in personal characteristics (for instance smoking level and history) across the two groups of smokers: the previous study used samples collected through the NCI-supported TobPRAC biorepository, while the subjects for the current study were recruited in the outpatient clinic at the University of Minnesota, and the time of subject recruitment and sample collection is at least 5 years apart for the two studies. Independent of the reason, the lower average levels of HPB-releasing DNA adducts, as well as the slightly lower DNA yields, in this study as compared to the previous report further stress the importance of our new highly sensitive methodology for future investigations.
In this study, we included smokers with and without HNSCC to provide preliminary insights into the potential differences in HPB-releasing DNA adduct levels between oral cancer patients and cancer-free smokers. The comparison between the two groups of smokers shows that HPB-releasing DNA adducts were detected in 97% of HNSCC patients and only 57% of cancer-free smokers, with the median levels being more than 6-fold higher in the oral cells of HNSCC patients than in controls (Table 1). This difference is highly statistically significant; however, these results should be validated with further investigations. Tobacco is a major etiologic factor in the development of oral cancer, with approximately 75% of all oral cancer patients being tobacco users,27-29 and NNN is a potent oral carcinogen. Furthermore, our previous study has shown that urinary total NNN was higher in smokers with HNSCC than in cancer-free smokers.22 In the present study, all enrolled subjects completed an in-depth questionnaire on lifetime tobacco use and provided additional biological samples, including urine. While in-depth analysis of smoking histories and constituent exposures is beyond the scope of this report, additional research is underway to compare these measures between the two groups of smokers and investigate the potential utility of oral cell HPB-releasing DNA adducts in identifying tobacco users at risk for oral cancer. Given the high inter-individual variability of HPB-releasing adduct levels with some being below the LOQ, future studies comparing different tobacco and disease groups could utilize the Tobin model,30 which allows for the uncertainty about the unobserved values below the LOQ and permits the addition of multiple covariates related to subject characteristics to explain more of the inter-individual variations in the HPB-releasing DNA adducts.
In summary, we have developed an improved highly sensitive LC-NSI-HRMS/MS method for the analysis of HPB-releasing DNA adducts in small amounts of oral cell DNA, and successfully applied this method to the analysis of oral samples from 65 smokers. This methodology can be applied in future studies on the role of HPB-releasing DNA adduct formation in tobacco-induced cancers in humans.
Supplementary Material
Figure 2.
Formation of HPB-releasing DNA adducts from NNK and NNN.
Acknowledgments
We thank Xun Ming and Peter Villalta for their help with the mass spectrometry analysis, Rebecca Dove for assistance with subject enrollment and specimen collection, and Bob Carlson for editorial assistance.
Funding Sources
This study was supported by grants P01-CA138338 and R01-CA179246 from the National Cancer Institute, grant K23-DE023572 from the National Institute of Dental and Craniofacial Research, and by startup funds to IS from the Masonic Cancer Center. Mass spectrometry analysis was carried out in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center, supported in part by grant CA-77598 from the National Cancer Institute.
Abbreviations
- CV
coefficient of variation
- ESI
electrospray ionization
- HCD
higher-energy collisional dissociation
- HNSCC
head and neck squamous cell carcinoma
- HPB
4-hydroxy-1-(3-pyridyl)-1-butanone
- IQR
inter-quartile range
- LC-NSI-HRMS/MS
liquid chromatography-nanoelectrospray ionization-high resolution tandem mass spectrometry
- LOD
limit of detection
- LOQ
limit of quantitation
- NNAL
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNN
N’-nitrosonornicotine
- POB DNA adducts
pyridyloxobutyl DNA adducts
- SD
standard deviation
- SRM
selected reaction monitoring
- TSNA
tobacco-specific nitrosamines
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information
Product ion spectra of HPB and [pyridine-D4]HPB; Typical SRM chromatograms of guanine; Linearity of guanine. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Jha P, MacLennan M, Chaloupka FJ, Yurekli A, Ramasundarahettige C, Palipudi K, Zatonksi W, Asma S, Gupta PC. Cancer: Disease Control Priorities. Third. The International Bank for Reconstruction and Development/The World Bank; Washington (DC): 2015. Global hazards of tobacco and the benefits of smoking cessation and tobacco taxes. Chapter 10. [PubMed] [Google Scholar]
- (2).Pan A, Wang Y, Talaei M, Hu FB. Relation of smoking with total mortality and cardiovascular events among patients with diabetes mellitus: A meta-analysis and systematic review. Circulation. 2015;132:1795–1804. doi: 10.1161/CIRCULATIONAHA.115.017926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hecht SS, Szabo E. Fifty years of tobacco carcinogenesis research: from mechanisms to early detection and prevention of lung cancer. Cancer Prev. Res. (Phila) 2014;7:1–8. doi: 10.1158/1940-6207.CAPR-13-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hecht SS. Lung carcinogenesis by tobacco smoke. Int. J. Cancer. 2012;131:2724–2732. doi: 10.1002/ijc.27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Poirier MC. Chemical-induced DNA damage and human cancer risk. Discov. Med. 2012;14:283–288. [PMC free article] [PubMed] [Google Scholar]
- (6).Hecht SS, Stepanov I, Carmella SG. Exposure and metabolic activation biomarkers of carcinogenic tobacco-specific nitrosamines. Acc. Chem. Res. 2016;49:106–114. doi: 10.1021/acs.accounts.5b00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).International Agency for Research on Cancer . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 89. Lyon, FR: 2007. Smokeless Tobacco and Tobacco-Specific Nitrosamines; pp. 421–583. [PMC free article] [PubMed] [Google Scholar]
- (8).Yuan JM, Butler LM, Stepanov I, Hecht SS. Urinary tobacco smoke-constituent biomarkers for assessing risk of lung cancer. Cancer Res. 2014;74:401–411. doi: 10.1158/0008-5472.CAN-13-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Goniewicz ML, Knysak J, Gawron M, Kosmider L, Sobczak A, Kurek J, Prokopowicz A, Jablonska-Czapla M, Rosik-Dulewska C, Havel C, Jacob P, III, Benowitz N. Levels of selected carcinogens and toxicants in vapour from electronic cigarettes. Tob. Control. 2014;23:133–139. doi: 10.1136/tobaccocontrol-2012-050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Stepanov I, Carmella SG, Briggs A, Hertsgaard L, Lindgren B, Hatsukami D, Hecht SS. Presence of the carcinogen N'-nitrosonornicotine in the urine of some users of oral nicotine replacement therapy products. Cancer Res. 2009;69:8236–8240. doi: 10.1158/0008-5472.CAN-09-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yuan JM, Koh WP, Murphy SE, Fan Y, Wang R, Carmella SG, Han S, Wickham K, Gao YT, Yu MC, Hecht SS. Urinary levels of tobacco-specific nitrosamine metabolites in relation to lung cancer development in two prospective cohorts of cigarette smokers. Cancer Res. 2009;69:2990–2905. doi: 10.1158/0008-5472.CAN-08-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Stepanov I, Hecht SS. Tobacco-specific nitrosamines and their pyridine-N-glucuronides in the urine of smokers and smokeless tobacco users. Cancer Epidemiol. Biomarkers Prev. 2005;14:885–891. doi: 10.1158/1055-9965.EPI-04-0753. [DOI] [PubMed] [Google Scholar]
- (13).Hecht SS, Murphy SE, Stepanov I, Nelson HH, Yuan JM. Tobacco smoke biomarkers and cancer risk among male smokers in the Shanghai cohort study. Cancer Lett. 2013;334:34–38. doi: 10.1016/j.canlet.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hecht SS, Spratt TE, Trushin N. Evidence for 4-(3-pyridyl)-4-oxobutylation of DNA in F344 rats treated with the tobacco-specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and N'-nitrosonornicotine. Carcinogenesis. 1988;9:161–165. doi: 10.1093/carcin/9.1.161. [DOI] [PubMed] [Google Scholar]
- (15).Wang M, Cheng G, Sturla SJ, Shi Y, McIntee EJ, Villalta PW, Upadhyaya P, Hecht SS. Identification of adducts formed by pyridyloxobutylation of deoxyguanosine and DNA by 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone, a chemically activated form of tobacco specific carcinogens. Chem. Res. Toxicol. 2003;16:616–626. doi: 10.1021/tx034003b. [DOI] [PubMed] [Google Scholar]
- (16).Stepanov I, Muzic J, Le CT, Sebero E, Villalta P, Ma B, Jensen J, Hatsukami D, Hecht SS. Analysis of 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB)-releasing DNA adducts in human exfoliated oral mucosa cells by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 2013;26:37–45. doi: 10.1021/tx300282k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- (18).Heppel CW, Heling AK, Richter E. Ultrasensitive method for the determination of 4-hydroxy-1-(3-pyridyl)-1-butanone-releasing DNA adducts by gas chromatography-high resolution mass spectrometry in mucosal biopsies of the lower esophagus. Anal. Bioanal. Chem. 2009;393:1525–1530. doi: 10.1007/s00216-008-2566-y. [DOI] [PubMed] [Google Scholar]
- (19).Holzle D, Schlobe D, Tricker AR, Richter E. Mass spectrometric analysis of 4-hydroxy-1-(3-pyridyl)-1-butanone-releasing DNA adducts in human lung. Toxicology. 2007;232:277–85. doi: 10.1016/j.tox.2007.01.016. [DOI] [PubMed] [Google Scholar]
- (20).Foiles PG, Akerkar SA, Carmella SG, Kagan M, Stoner GD, Resau JH, Hecht SS. Mass spectrometric analysis of tobacco-specific nitrosamine-DNA adducts in smokers and nonsmokers. Chem. Res. Toxicol. 1991;4:364–368. doi: 10.1021/tx00021a017. [DOI] [PubMed] [Google Scholar]
- (21).Schlobe D, Holzle D, Hatz D, von Meyer L, Tricker AR, Richter E. 4-Hydroxy-1-(3-pyridyl)-1-butanone-releasing DNA adducts in lung, lower esophagus and cardia of sudden death victims. Toxicology. 2008;245:154–161. doi: 10.1016/j.tox.2007.12.021. [DOI] [PubMed] [Google Scholar]
- (22).Khariwala SS, Carmella SG, Stepanov I, Fernandes P, Lassig AA, Yueh B, Hatsukami D, Hecht SS. Elevated levels of 1-hydroxypyrene and N'-nitrosonornicotine in smokers with head and neck cancer: A matched control study. Head & Neck. 2013;35:1096–1100. doi: 10.1002/hed.23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 2007;20:235–245. doi: 10.1021/tx060207r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).U.S. FDA . Guidance for industry: Bioanalytical method validation. U.S. Department of Health and Human Services; 2001. http://www.fda.gov/downloads/Drugs/Guidances/ucm070107.pdf. [Google Scholar]
- (25).Dolan JW. Calibration curves, part II: What are the limits? LCGC North America. 2009;27:306–312. [Google Scholar]
- (26).Ma B, Villalta PW, Balbo S, Stepanov I. Analysis of a malondialdehyde-deoxyguanosine adduct in human leukocyte DNA by liquid chromatography nanoelectrospray-high-resolution tandem mass spectrometry. Chem. Res. Toxicol. 2014;27:1829–1836. doi: 10.1021/tx5002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).International Agency for Research on Cancer Global data on incidence of oral cancer. 2005 http://screening.iarc.fr/doc/oral_cancer_brochure.pdf. [Google Scholar]
- (28).Sankaranarayanan R, Ramadas K, Amarasinghe H, Subramanian S, Johnson N. Cancer: Disease Control Priorities. Third. The International Bank for Reconstruction and Development/The World Bank; Washington (DC): 2015. Oral Cancer: Prevention, Early Detection, and Treatment. Chapter 5. [PubMed] [Google Scholar]
- (29).Oral Cancer Foundation Oral cancer: What you need to know. 2006 http://www.oralcancerfoundation.org/presskit/pdf/what_you_need_to_know_2006.pdf.
- (30).Tobin J. Estimation of Relationships for Limited Dependent-Variables. Econometrica. 1958;26:24–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.