Abstract
New antibacterial agents with novel target and mechanism of action are urgently needed to combat problematic bacterial infections and mounting antibiotic resistances. Topoisomerase IA represents an attractive and underexplored antibacterial target, as such, there is a growing interest in developing selective and potent topoisomerase I inhibitors for antibacterial therapy. Based on our initial biological screening, fluoroquinophenoxazine 1 was discovered as a low micromolar inhibitor against E. coli topoisomerase IA. In the literature, fluoroquinophenoxazine analogs have been investigated as antibacterial and anticancer agents, however, their topoisomerase I inhibition was relatively underexplored and there is little structure-activity relationship (SAR) available. The good topoisomerase I inhibitory activity of 1 and the lack of SAR prompted us to design and synthesize a series of fluoroquinophenoxazine analogs to systematically evaluate the SAR and to probe the structural elements of the fluoroquinophenoxazine core toward topoisomerase I enzyme target recognition. In this study, a series of fluoroquinophenoxazine analogs was designed, synthesized, and evaluated as topoisomerase I inhibitors and antibacterial agents. Target-based assays revealed that the fluoroquinophenoxazine derivatives with 9-NH2 and/or 6-substituted amine functionalities generally exhibited good to excellent inhibitory activities against topoisomerase I with IC50s ranging from 0.24–3.9 µM. Notably, 11a bearing the 6-methylpiperazinyl and 9-amino motifs was identified as one of the most potent topoisomerase I inhibitors (IC50 = 0.48 µM), and showed broad spectrum antibacterial activity (MICs = 0.78–7.6 µM) against all the bacteria strains tested. Compound 11g with the 6-bipiperidinyl lipophilic side chain exhibited the most potent antituberculosis activity (MIC = 2.5 µM, SI = 9.8). In addition, CoMFA analysis was performed to investigate the 3D-QSAR of this class of fluoroquinophenoxazine derivatives. The constructed CoMFA model produced reasonable statistics (q2 = 0.688 and r2 = 0.806). The predictive power of the developed model was obtained using a test set of 7 compounds, giving a predictive correlation coefficient r2pred of 0.767. Collectively, these promising data demonstrated that fluoroquinophenoxazine derivatives have the potential to be developed as a new chemotype of potent topoisomerase IA inhibitors with antibacterial therapeutic potential.
Keywords: Antibacterial, CoMFA analysis, 3D-QSAR, fluoroquinophenoxazine, topoisomerase IA, SAR
Graphical Abstract
1. Introduction
DNA topoisomerases maintain the helical and superhelical structure of DNA and are essential enzymes required for cellular processes and functions including DNA replication, transcription, and repair [1, 2]. Therefore, poison inhibitors of topoisomerase enzymes can lead to the accumulation of the intermediate topoisomerase-DNA cleavage complex and subsequently result in bacterial or cancer cell death [2, 3]. Clinically, topoisomerase enzymes represent attractive and successful targets for anticancer and antibacterial chemotherapy [4, 5]. In contrast, topoisomerase I is underexplored as antibacterial target, thus potent and selective topoisomerase I inhibitors can serve as a promising class of chemotherapeutic agents toward the treatment of problematic bacterial infections [6, 7].
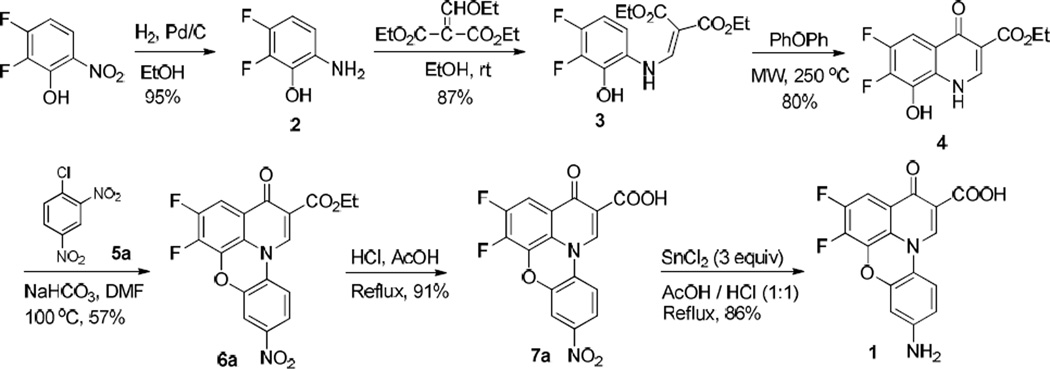
In our ongoing efforts to discover new topoisomerase I inhibitors as antibacterial agents and to probe the topoisomerase I recognition [8, 9], we identified during high-throughput screening (HTS) assay development a compound NSC648059 (1, Fig. 1) with low micromolar inhibitory activity (IC50 = 0.8–2.0 µM) against Escherichia coli topoisomerase I. Structurally, 1 is a fluoroquinophenzoxazine derivative with a unique planar tetracyclic ring system, belonging to a member of an extended chemical class of fluoroquinolone antibiotics. In the clinic, fluoroquinolones including norfloxacin and ciprofloxacin (Fig. 1) represent some of the most successful antibiotic classes, whose mechanisms of action are to inhibit bacterial DNA gyrase and topoisomerase IV as well as relaxation of supercoiled DNA and thus to promote breakage of double-stranded DNA [10]. Specifically, fluoroquinophenoxazines such as A-62176 and A-85226 (Fig. 1) have been reported as antibacterial [11, 12] and anticancer [13–16] agents. For example, A-62176 exhibited good activity against several cancer cell lines with IC50 values ranging from 0.87–4.34 µM [13]. However, their bacterial topoisomerase I inhibition was relatively underexplored and there is little structure-activity relationship (SAR) of fluoroquinophenoxazine derivatives available in the literature [13]. Herein, on the basis of this HTS hit 1, we describe the design, synthesis, and evaluation of a series of fluoroquinophenoxazine structural analogs, probing their SAR toward bacterial topoisomerase I and other topoisomerases as well as their whole cell antibacterial activity. Additionally, we built and developed a quantitative structure-activity relationship (QSAR) and comparative molecular field analysis (CoMFA) model in an effort to guide further design and synthesis of this class of topoisomerase IA inhibitors.
Fig. 1.
Chemical structures of clinical fluoroquinolone antibiotics and fluoroquinophenzoxazine derivatives including our topo I inhibitor 1.
2. Results and discussion
2.1. Chemistry
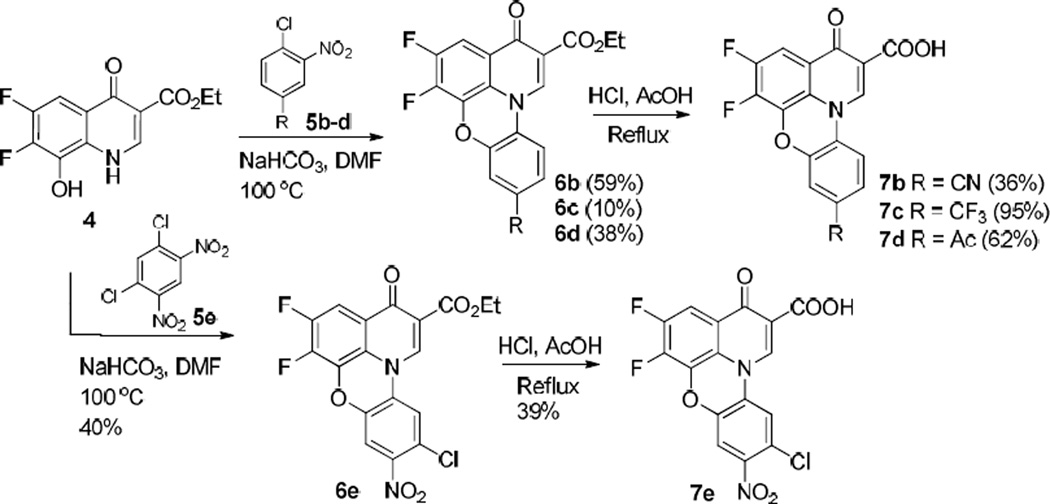
To validate the initial assay hit, we first resynthesized 9-amino-5, 6-difluoro-3-oxo-3H–pyrido[3,2,1-kl] phenoxazine-2-carboxylic acid (1) and retested its biochemical activity against E. coli topoisomerase I. Resynthesized fluoroquinophenoxazine hit 1 showed reproducible topoisomerase I inhibitory activity with an IC50 of 1.95 µM. The synthesis of 1 is shown in Scheme 1. Briefly, commercially available 2,3-difluoro-6-nitrophenol was hydrogenated using Pd/C (20 mol %) as the catalyst to the corresponding amino product 2, which can be used in next step without further purification [17]. Subsequent reaction of the aniline derivative 2 with diethyl 2-(ethoxymethylene)malonate under ambient temperature gave 3 in 87% yield. For next nucleophilic displacement cyclization, an improved protocol was developed for this intramolecular cyclization reaction under microwave irradiation at 250 °C instead of conventional heating [11]. Following simple filtration, 4 was obtained in 80% yield. Subsequently, 4 reacted with 1-chloro-2,4-dinitrobenzene (5a) in DMF at 100 °C to give the desired tetracyclic product 6a in 57% yield. Upon treatment of 6a in acidic condition (AcOH/HCl) under reflux for 4 h, the free carboxylic acid derivative 7a was obtained by simple filtration in 91% yield. Finally, hydrogenation of 7a under H2 (1.0 bar) using FeSO4·6H2O as the catalyst failed to yield the amine product 1 after 14 h. However, when SnCl2 was used as catalyst and AcOH as the solvent, the nitro group was smoothly reduced into the amino group under reflux for 3 h and the final product 1 was obtained in 86% yield.
Scheme 1.
Synthesis of 9-amino-5, 6-difluoro-3-oxo-3H–pyrido[3,2,1-kl] phenoxazine-2-carboxylic acid (1).
Under the optimized conditions, we next expanded the structural diversity of 1 to evaluate the effect of various substituents on the quinophenoxazine skeleton and to explore their SAR. First, we used various substrates 5b–e in Scheme 2 in an effort to generate a focused set of quinophenoxazine derivatives 7b–e. It is worthwhile noting that a dramatic difference was observed in terms of the reactivity of the substrate 5 with different electronic and/or physiochemical properties. For example, 5b bearing an electron-withdrawing nitrile group facilitated the completion of cyclization in 2 h and 6b was obtained in 59% yield. On the other hand, 5c with the lipophilic trifluoromethyl group could finish the reaction by extending the reaction time, affording the desired product 6c in a lower yield (10%). In addition, when the substrate 5d with an acetyl group was tried, 6d was obtained in 38% yield. However, for the substrate with the corresponding fluorine substitution, no reaction occurred even when the reaction temperature was raised to 120 °C. Furthermore, more harsh conditions were tried in an attempt to facilitate the reaction by heating the reaction to 160 °C in a sealed pressure tube or heating the reaction to 200 °C under microwave irradiation, but with not much success. In the case of 5e with additional nitro and chlorine substituents, the reaction proceeded smoothly and 6e was obtained in 40% yield. Final ester hydrolysis was performed in acetic acid/hydrochloric acid under reflux, affording free carboxylic acid products 7b–e in 36–95% yields.
Scheme 2.
Synthesis of fluoroquinophenoxazine derivatives 7b–e.
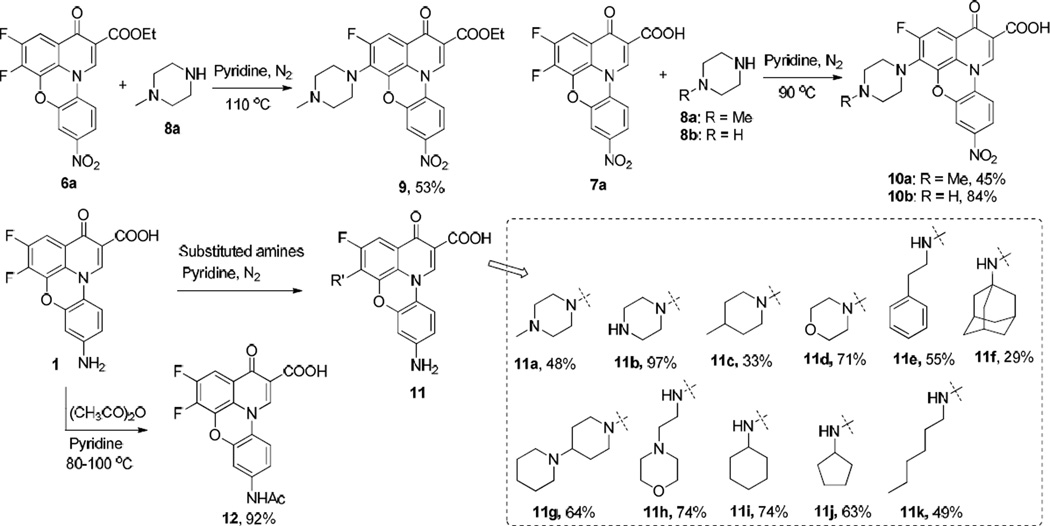
In the course of antibacterial evaluation, we observed compound 1 lost 2–4 fold whole cell antibacterial activity after 4 weeks of storage, indicating that 1 may have stability and/or solubility issue. From our 1H NMR experiments, we also noted that 1 could be easily precipitated out in d6-DMSO solvent and no degradation-related evidence was observed following several weeks of monitoring 1 in both d6-DMSO NMR and HPLC experiments (data not shown). Thus, to enhance the overall solubility profile of this class of quinophenoxazine derivatives, we introduced a variety of solubilizing and polar groups into the fluoroquinophenoxazine scaffold by displacing the 6-fluorine atom of 6a, 7a, or 1 with different amine functionalities such as piperazine, 1-methylpiperazine, and morpholine (Scheme 3) [18]. Specifically, 6a reacted with 1-methylpiperazine in pyridine at 110 °C to give 9 in 53% yield. Accordingly, 7a reacted with 1-methylpiperazine and piperazine in pyridine under nitrogen atmosphere at 90 °C, and both reactions proceeded smoothly to afford 10a and 10b in 45% and 84% yields, respectively [12]. In the cases of 1 and some other amine substrates, notably, reaction temperature appeared to play a critical role in this nucleophilic displacement reaction. For example, when morpholine was used as a nucleophile, the reaction became complicated under 90 °C, which is suitable for 1-methylpiperazine and piperazine. In contrast, when temperature was reduced to 70 °C, the reaction could be completed in 16 h to produce 11d in 71% yield. Other functional amines were also subjected to this substitution reaction, and most of the reactions could lead to the desired products 11c–k under 90 °C in moderate yields except for 1-adamantylamine. We found that the reaction of 1 with 1-adamantylamine could finish under reflux after 4 days in 29% yield, presumably due to steric effect. In addition, to investigate the effect of the free amine functionality of 1 on topoisomerase inhibition and antibacterial activity, we next tried to protect the free amino group with acetyl functionality. The N-acetyl derivative 12 was synthesized from 1 and acetic anhydride in pyridine at 80–100 °C and the solid product was collected by simple filtration in high yield (92%).
Scheme 3.
Synthesis of diverse amino-substituted fluoroquinophenoxazines 9–11 and acetyl-protected quinophenoxazine 12.
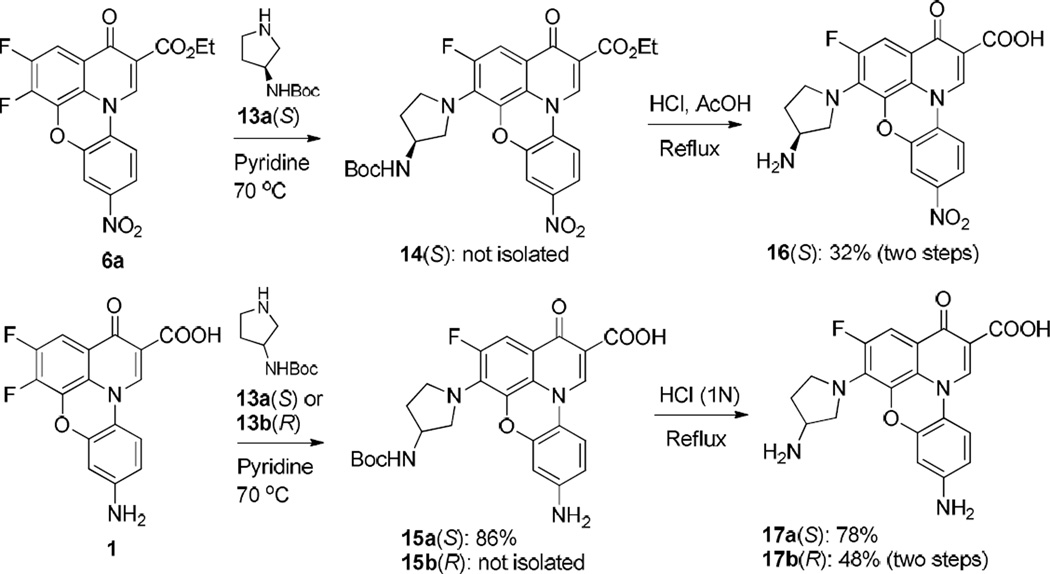
Finally, to evaluate the potential stereospecific effect at the 6 position of fluoroquinophenoxazine derivatives on biological activity, we designed and synthesized several chiral fluoroquinophenoxazine amine derivatives from 6a or 1 and chiral amine building blocks [12, 13]. The nucleophilic substitution reaction of 1 and (S)-3-(Boc-amino)pyrrolidine (13a) in pyridine was completed in 20 h, affording 15a in 86% yield. Subsequent N-Boc deprotection of 15a produced 17a in 78% yield upon the treatment with diluted hydrochloric acid. With regard to the reaction of 1 and the corresponding (R)-3-(Boc-amino)pyrrolidine (13b), the substituted compound 15b could not be obtained by filtration upon the completion of reaction. Therefore, the crude product 15b was used for the following deprotection reaction and the corresponding fluorophenoxazine derivative 17b (R) was obtained in 48% yield over two steps. Accordingly, compound 16 was synthesized from 6a as the starting material in 32% yield over two steps (Scheme 4).
Scheme 4.
Synthesis of fluoroquinophenoxazine chiral amine derivatives 16 and 17.
2.2. Biological testing
All the synthesized target molecules were tested for the ability to inhibit the relaxation activity of E. coli topoisomerase I in target-based assay, as well as against a panel of bacterial strains including the wild-type E. coli MG1655 K12 strain, E. coli strain BAS3023 with imp mutation conferring membrane permeability to small molecules [19, 20], the wild-type Gram-positive B. subtilis (ATCC 6633) strain, and M tuberculosis (H37Rv). The results are summarized in Table 1.
Table 1.
E. coli topoisomerase I inhibition and whole cell antibacterial activities (µM) of fluoroquinophenoxazine derivativesa
| Topoisomerase inhibitory activity (IC50, µM) | Whole cell based antibacterial activity (MIC, µM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compd |
E. coli topo I (type IA) |
E. coli DNA gyrase (type IIA) |
Human topo I (type IB) |
Human topo IIα (type IIA) |
E. coli Imp4213 (BAS3023) |
E. coli (MG1655) WT |
B. subtilis (ATCC 6633) WT |
M tuberculosis (H37Rv) |
Vero cell IC50 |
SIb |
| 1 | 1.95 | >125 | 31.3 | >500 | 0.78–1.56 | >200 | 6.25 | 11.2 | 75.7 | 6.8 |
| 6a | 15.6 | 50 | >200 | 25 | ||||||
| 6b | 31.25 | >200 | >200 | 200 | ||||||
| 6c | >125 | >200 | >200 | >200 | ||||||
| 7a | 15.6 | 0.78 | 200 | 12.5 | ||||||
| 7b | 31.25 | 200 | >200 | >200 | ||||||
| 7c | >125 | >200 | >200 | 25 | ||||||
| 7d | 15.6 | >200 | >200 | >200 | ||||||
| 7e | 31.25 | 62.5 | 31.25 | 125 | 0.39 | 50 | 3.12 | 29 | >127 | >4.4 |
| 9 | 1.95 | 25 | >200 | 50 | ||||||
| 10a | 1.95 | 62.5–125 | 31.25 | 250–500 | 1.56 | 100 | 0.78 | 19 | 95 | 5.0 |
| 10b | 3.9 | >200 | >200 | >200 | ||||||
| 11a | 0.48 | 15.6–31.25 | 15.6 | 3.9–7.8 | 0.78 | 6.25 | 0.78 | 7.6 | 29 | 3.8 |
| 11b | 0.24 | 7.8–15.6 | 7.8 | 1.95–3.9 | 0.39 | >200 | 25 | 29.5 | >126 | >4.3 |
| 11c | 3.9 | 3.12 | >200 | 0.78 | ||||||
| 11d | 0.97 | 7.8 | 15.6 | 15.6 | 0.19–0.39 | >200 | 0.19 | 3.5 | 24.7 | 7.1 |
| 11e | 3.9 | 50 | >200 | 25–50 | ||||||
| 11f | 3.9–7.8 | 3.12 | >200 | 12.5 | 38.4 | 30.7 | 0.8 | |||
| 11g | 0.48 | 15.6 | 3.9 | 1.95–3.9 | 0.39–0.78 | >200 | 1.56 | 2.5 | 24.4 | 9.8 |
| 11h | 0.97 | 15.6–31.25 | 7.8–15.6 | 3.9–7.8 | 0.78 | >200 | 1.56 | 21.6 | 43.0 | 2.0 |
| 11i | 3.9 | 25 | >200 | 12.5 | ||||||
| 11j | 3.9 | 25 | >200 | 12.5 | ||||||
| 11k | 15.6 | >200 | >200 | >200 | ||||||
| 12 | 7.8 | >200 | >200 | >200 | ||||||
| 15a | 3.9 | 12.5 | >200 | 12.5 | ||||||
| 16 | 0.48 | 3.9 | 7.8 | 1.95–3.9 | 1.56 | >200 | 0.78 | >50 | >50 | |
| 17a | 0.48–0.97 | 3.9 | 3.9 | 0.97–1.95 | 1.56 | >200 | 6.25 | >63.1 | >63.1 | |
| 17b | 0.48–0.97 | 3.9 | 3.9 | 3.9 | 6.25 | >200 | 12.5 | >63.1 | >63.1 | |
Blank cells indicate Not Determined.
Selectivity index = cytotoxic IC50 against Vero cells/MIC against M. tuberculosis.
2.2.1. E. coli topoisomerase I inhibition
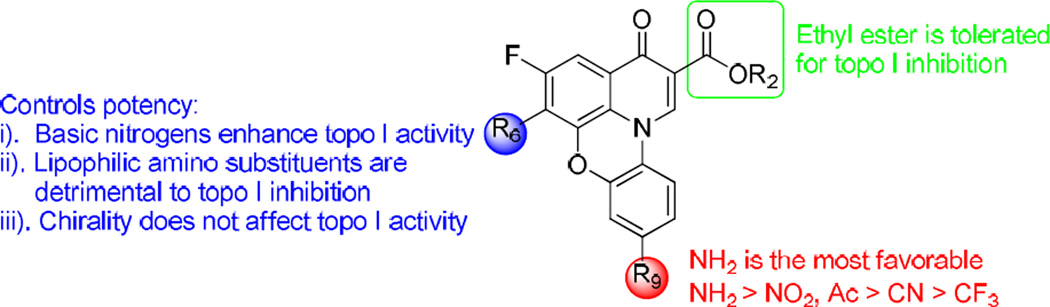
Biochemical evaluation for inhibition of the relaxation activity of E. coli topoisomerase I revealed that the majority of our synthesized compounds possessed good activity against E. coli topoisomerase I. On the basis of these topoisomerase I inhibition data (Table 1), an illuminating SAR has been obtained and is summarized in Fig. 2. i). The 9 position substituent plays a very important role in topoisomerase I inhibitory activity. Among the 5,6-difluoroquinophenoxazine derivatives, the hit compound 1 with the electron-donating 9-NH2 group showed the most potent activity (IC50 = 195 µM) against E. coli topoisomerase I. Both free carboxylic acid 7c and its ethyl ester derivative 6c with the 9-CF3 functionality were inactive against topoisomerase I when tested at 125 µM. In addition, compared to 1 (9-NH2, 1.95 µM), all the other 5,6-difluoro derivatives with 9-substituted electron-withdrawing groups (7a with 9-NO2, 15.6 µM; 7b with 9-CN, 31.25 µM; 7d with 9-Ac, 15.6 µM; 7e with 9-NO2 and 10-Cl, 31.25 µM) were 8–16 fold less active with IC50 values ranging from 15.6–31.25 µM. ii). In general, the basic amine functionality at the 6 position significantly enhanced topoisomerase I inhibitory activity. For example, the 6-substituted amine derivatives 11a with 6-methylpiperazinyl, 11b with piperazinyl, 11d with morpholino, 11g with bipiperidinyl, 11h with morpholinoethyl, as well as the 6-substituted aminopyrrolidinyl derivatives 16, 17a, and 17b demonstrated the most potent topoisomerase I inhibitory activity with IC50 values of 0.24–0.97 µM; and within this group, 11d and 11h with the morpholino group had an IC50 value of 0.97 µM. In contrast, all the other 6-substituted amine derivatives with a more lipophilic side chain, including 11c with methylpiperidinyl, 11e with phenethyl, 11f with adamantanyl, 11i with cyclohexyl, 11j with cyclopentyl, 11k with n-hexyl, and 15a with t-Boc-aminopyrrolidinyl, showed weaker topoisomerase I inhibition with IC50 values ranging from 3.9 to 15.6 µM. Notably, both 6-substituted aminopyrrolidinyl S- and R- stereoisomers 17a and 17b exhibited the same topoisomerase I inhibitory activity (IC50 = 0.48–0.97 µM), suggesting the stereochemistry at the 6 position is not required for topoisomerase I inhibition. iii) Esterification of the carboxylic acid group had little effect on the E. coli topoisomerase I inhibitory activity by comparing 6a and 7a (IC50 = 15.6 µM), 6b and 7b (IC50 = 31.25 µM), as well as 6c and 7c (IC50 >125 µM), indicating the ethyl ester functionality is tolerated for topoisomerase I enzyme inhibition. Representative inhibition results of 11a and 11b against E. coli topoisomerase relaxation activity are shown in Fig. 3.
Fig. 2.
SAR toward E. coli topoisomerase I inhibition.
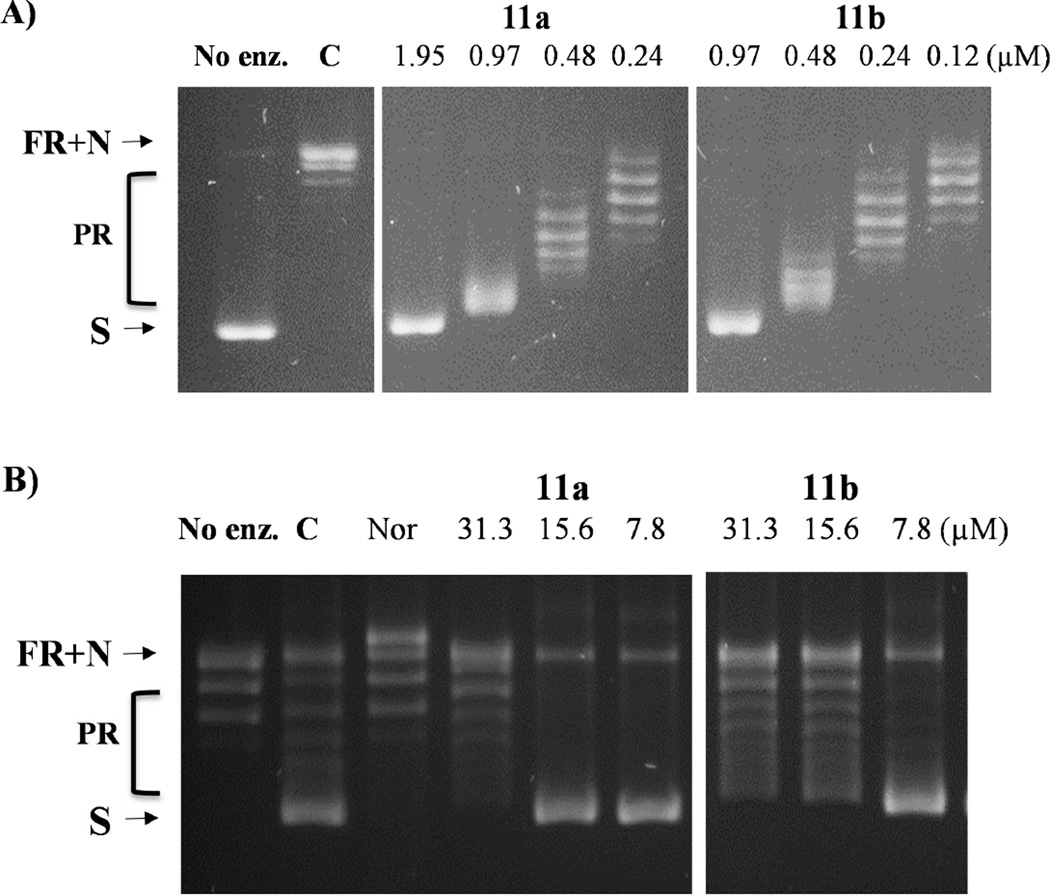
Fig. 3.
Inhibition of E. coli topoisomerase relaxation activity by representative 11a and 11b. A) E. coli topoisomerase I inhibition assays with supercoiled plasmid DNA. B) E. coli DNA gyrase inhibition assays with relaxed plasmid DNA. C: DMSO control; Nor: Norfloxacin (125 µM) control; S: Supercoiled plasmid DNA; N: Nicked DNA; FR: Fully relaxed DNA; PR: Partially relaxed DNA.
2.2.2. Selectivity and specificity against other DNA topoisomerase enzymes
In addition, to determine the selectivity and specificity profiles of this class of fluoroquinophenoxazine derivatives, selected compounds were also investigated for the ability to inhibit other DNA topoisomerases including E. coli DNA gyrase as well as human topoisomerase I and IIα enzymes. Overall, these compounds were more selective toward E. coli topoisomerase I than other enzymes tested. Given that this series of compounds has close structural similarity to quinolone antibiotic class, as such, inhibition against E. coli gyrase can be also observed. Specifically, most of the compounds showed 4–64 fold selectivity toward topoisomerase I over DNA gyrase except that the moderately active compound 7e with the 9-NO2 and 10-chloro substituents showed less specificity with 2 fold selectivity toward topoisomerase I. With respect to human topoisomerases I and IIα inhibition, these compounds also showed inhibitory activity against both human topoisomerase I (IC50 = 3.9–31.25 µM) and topoisomerase IIα (IC50 = 0.97–250 µM), with approximate 4–32 fold selectivity. Taken together, compounds 11a (IC50 = 0.48 µM) and 11b (IC50 = 0.24 µM) (Fig. 3) bearing both 9-NH2 and 6-piperazinyl motifs exhibited the most potent topoisomerase I inhibitory activity with the more favorable selectivity profile (8–64 fold) toward E. coli topoisomerase I against all the other enzymes tested.
2.2.3. Cell-based antibacterial activity
In addition to target-based topoisomerase enzyme inhibition, whole cell antibacterial activities of the synthesized compounds were also assessed against a panel of bacterial strains. The results are also shown in Table 1. From these data, the majority of these fluoroquinophenoxazine derivatives exhibited good to excellent antibacterial activity against the membrane permeable E. coli strain BAS3023 and Gram-positive B. subtilis strain, and were inactive against the wide type E. coli strain. Additionally, the antibacterial activity of most fluoroquinophenoxazine derivatives (e.g., 1, 7a, 7c, 9, 10a, 11a–d, 11f–i, 15a, 16, and 17a–b) generally correlated with E. coli topoisomerase I inhibitory activity, suggesting that the antibacterial basis of these compounds may be in part due to the inhibition of topoisomerase I. The only type IA topoisomerase present in M. tuberculosis has recently been validated as an antitubercular target [21]. The topoisomerase I activity has been shown to be essential for viability and infection in a murine model of tuberculosis [21, 22]. To further determine the antituberculosis profile for this chemical class of fluoroquinophenoxazine derivatives, twelve compounds were selected and evaluated against M. tuberculosis. Among them, 11g with the 6-bipiperidinyl lipophilic side chain and 11d with the 6-morpholino heterocyclic ring system showed the most potent antituberculosis activity with minimum inhibitory concentration (MIC) values of 2.5 and 3.5 µM, respectively. In addition, compared to 11d, its corresponding 6-piperazinyl structural analogs 11a with tertiary amine and 11b with secondary amine functionality was about 2- and 8-fold less active with the MIC values of 7.6 and 29.5 µM, respectively. In contrast, both 6-substituted aminopyrrolidinyl derivatives 17a (S) and 17b (R) with primary amine functionality were not active (MIC > 63.1 µM) against M. tuberculosis. These data strongly suggest that the decreased or lost whole cell antituberculosis activity of these compounds are most likely due to their decreased lipophilicity and subsequent cell membrane penetration. Unfortunately, cytotoxicity evaluation of our tested compounds against healthy normal Vero cells showed that they generally had narrow selectivity index, with 11g (SI = 9.8) being the most promising compound. It is also worthwhile noting that, one of the most potent topoisomerase I inhibitors, 11a (IC50 = 0.48 µM) bearing the 6-methylpiperazinyl and 9-amino motifs, showed broad spectrum antibacterial activity against all the test bacteria strains with MICs ranging from 0.78 to 7.6 µM (SI = 3.8–37).
2.3. CoMFA analysis
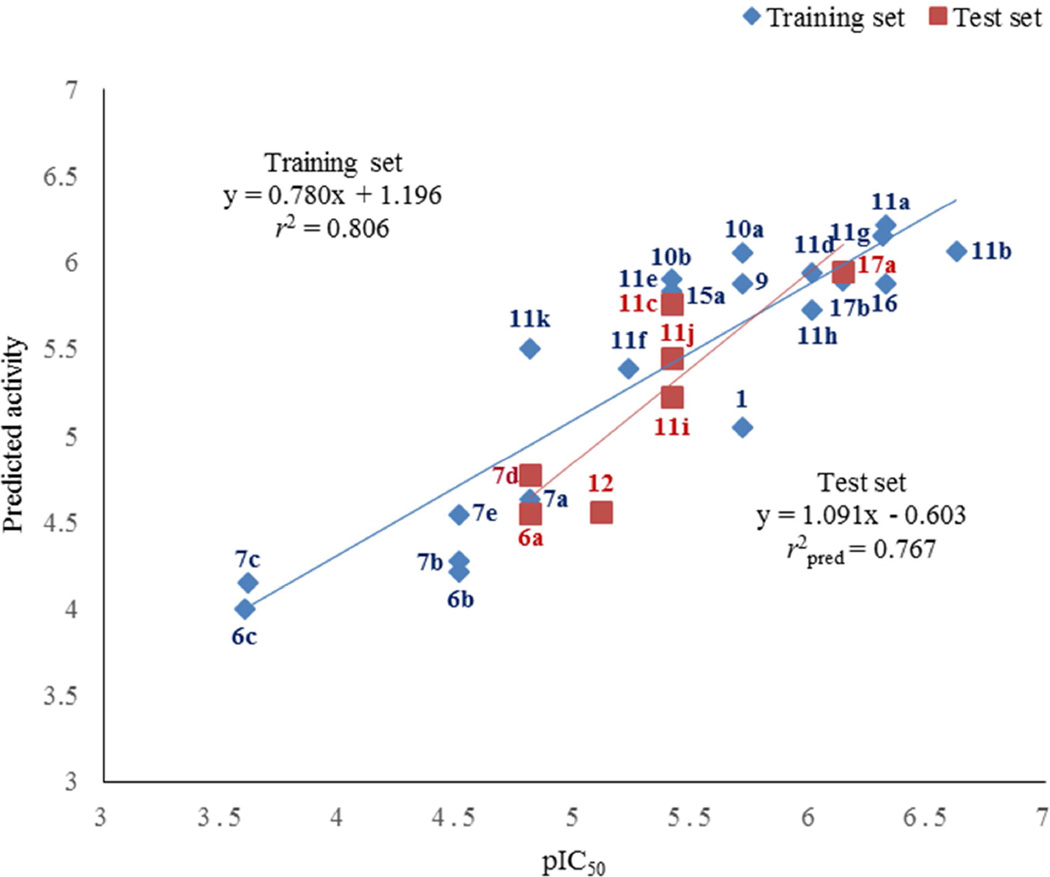
To further understand the structural basis for topoisomerase I inhibitory activity of this set of fluoroquinophenoxazine derivatives, we subsequently performed three dimensional QSAR (3D-QSAR) study using CoMFA analysis [23]. Because of the relatively rigid core structural feature of this class of fluoroquinophenoxazine molecules, we directly applied core structure based alignment to build reliable 3D-QSAR models. The CoMFA study was carried out using a total of 21 compounds (entries 1–21, Table 2). Statistical parameters of the CoMFA model showed a reasonable cross-validated correlation coefficient q2 of 0.688, indicating a good internal prediction of the model. The CoMFA model also exhibited a conventional correlation coefficient r2 of 0.806. To evaluate the predictive ability of our developed model, a test set of 7 compounds (entries 22–28, Table 2) which was not included in model generation was subsequently used. The predicative correlation coefficient r2pred of 0.767 indicates good external predicative ability of the CoMFA model. The experimental and predicted values as well as their residuals from the training and test set molecules are listed in Table 2. The correlation between the predicted and experimental values of all compounds was plotted and the resulting chart is shown in Fig. 4.
Table 2.
Experimental (pIC50) and CoMFA predicted activity (PA) values and residuals for the training and test set compoundsa
| Entry | Compd. | IC50 (µM) | pIC50 | CoMFA PAb | Δc |
|---|---|---|---|---|---|
| 1 | 1 | 1.95 | 5.71 | 5.05 | 0.66 |
| 2 | 6b | 31.25 | 4.51 | 4.21 | 0.30 |
| 3 | 6c | 250 | 3.60 | 4.00 | −0.40 |
| 4 | 7a | 15.6 | 4.81 | 4.63 | 0.18 |
| 5 | 7b | 31.25 | 4.51 | 4.27 | 0.24 |
| 6 | 7c | 250 | 3.61 | 4.15 | −0.54 |
| 7 | 7e | 31.25 | 4.51 | 4.54 | −0.03 |
| 8 | 9 | 1.95 | 5.71 | 5.88 | −0.17 |
| 9 | 10a | 1.95 | 5.71 | 6.05 | −0.34 |
| 10 | 10b | 3.9 | 5.41 | 5.90 | −0.49 |
| 11 | 11a | 0.48 | 6.32 | 6.21 | 0.11 |
| 12 | 11b | 0.24 | 6.62 | 6.06 | 0.56 |
| 13 | 11d | 0.97 | 6.01 | 5.94 | 0.07 |
| 14 | 11e | 3.9 | 5.41 | 5.83 | −0.42 |
| 15 | 11f | 5.85 | 5.23 | 5.39 | −0.16 |
| 16 | 11g | 0.48 | 6.31 | 6.15 | 0.16 |
| 17 | 11h | 0.97 | 6.01 | 5.72 | 0.29 |
| 18 | 11k | 15.6 | 4.81 | 5.50 | −0.69 |
| 19 | 15a | 3.9 | 5.41 | 5.77 | −0.36 |
| 20 | 16 | 0.48 | 6.32 | 5.88 | 0.44 |
| 21 | 17b | 0.73 | 6.14 | 5.89 | 0.25 |
| 22 | 6a | 15.6 | 4.81 | 4.55 | 0.26 |
| 23 | 7d | 15.6 | 4.81 | 4.77 | 0.04 |
| 24 | 11c | 3.9 | 5.41 | 5.76 | −0.35 |
| 25 | 11i | 3.9 | 5.41 | 5.23 | 0.18 |
| 26 | 11j | 3.9 | 5.41 | 5.45 | −0.04 |
| 27 | 12 | 7.8 | 5.11 | 4.56 | 0.55 |
| 28 | 17a | 0.73 | 6.14 | 5.95 | 0.19 |
Entries 1–21 for training set; entries 22–28 for test set.
Predicted activity.
Residual of experimental and predicted activity values.
Fig. 4.
The correlation chart of experimental versus predicted values for the training and test set compounds.
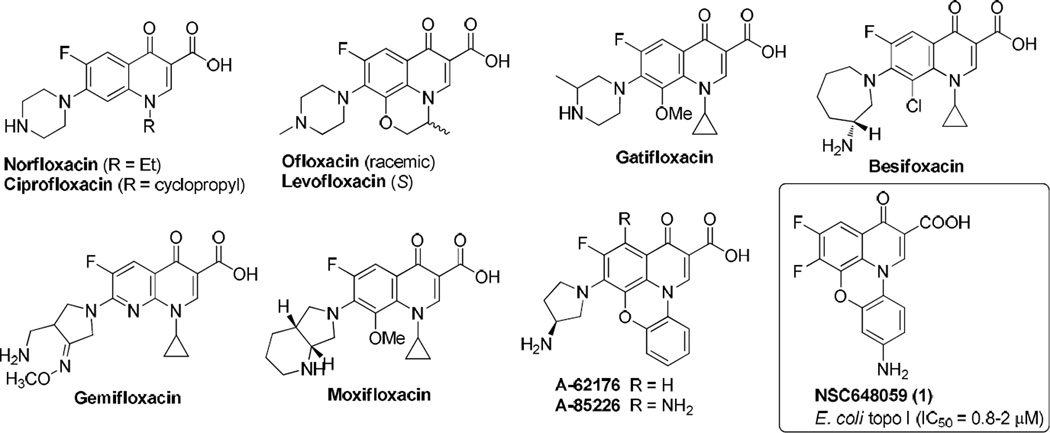
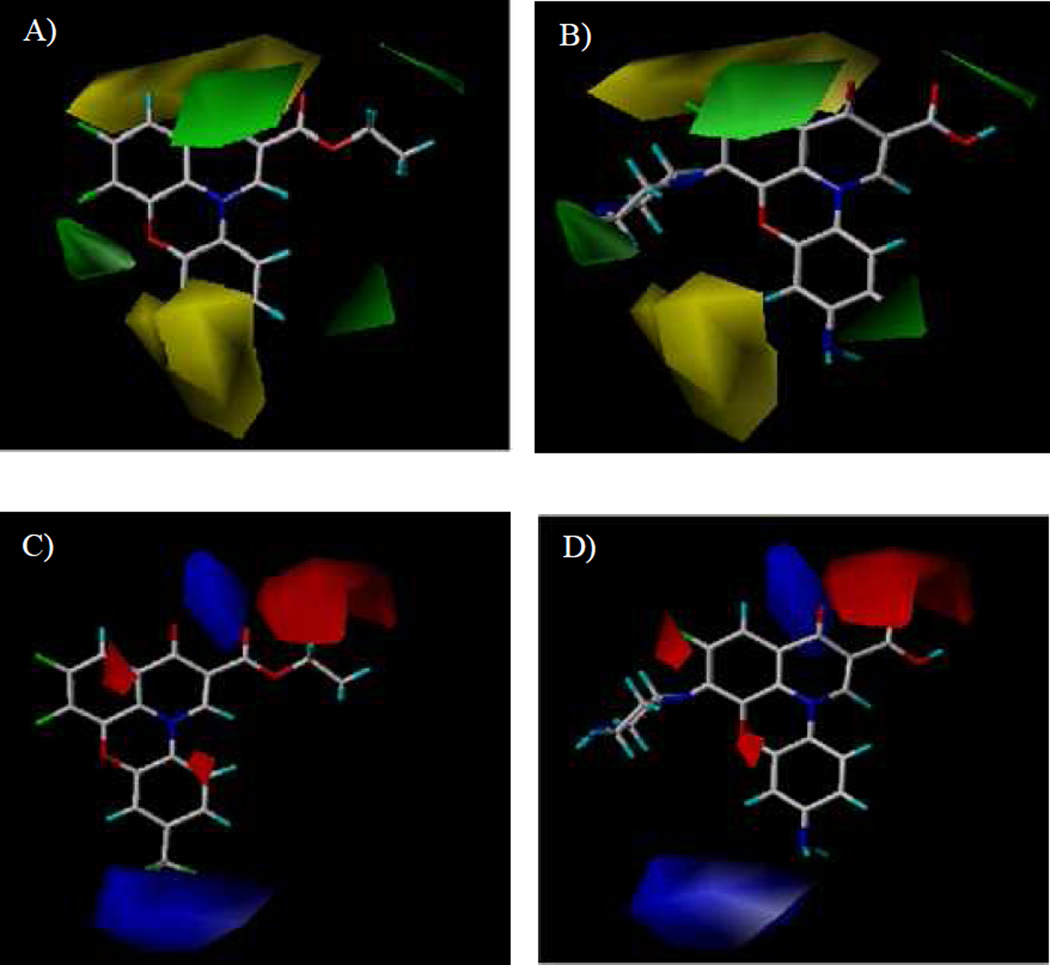
The results of the CoMFA model were analyzed and visualized using the standard deviation coefficient (StDev*Coeff) mapping option contoured by steric and electrostatic contributions. In order to probe the structure/activity correlation, the steric and electrostatic contours were mapped onto their aligned chemical structures of these fluoroquinophenoxazine molecules to identify the potential regions in which the molecules would favorably or unfavorably interact with the topoisomerase I enzyme. The representative steric and electrostatic contour maps of the most active compound 11b and the least active 6c derived from the CoMFA model are shown in Fig. 5. Briefly, the yellow areas in the steric contour maps indicate regions of steric hindrance to activity, while the green areas indicating steric contribution to potency. From the electrostatic contour maps, the regions in blue indicate positive electrostatic charge potential associated with increased activity, with the red regions show electronegative groups with increased activity.
Fig. 5.
Representative CoMFA steric and electrostatic contour maps. A). Steric contour maps with 6c; B). Steric contour maps with 11b; C). Electrostatic contour maps with 6c; and D) Electrostatic contour maps with 11b.
In Fig. 5A and Fig. 5B, one green contour was found near the piperazinyl moiety of compound 11b indicating that a moderate steric substituent would be favored at the 6 position of the quinophenoxazine scaffold. This may offer a potential explanation why the 6-substituted amino derivatives were generally more active than the 6-fluoro analogs. In addition, two yellow contours were observed near the 9 position of inactive 6c, indicating that a steric bulkiness (e.g., NO2 in 6a and 7a, CF3 in 6c, and the acetyl group in 7d) would be disfavored for activity in this area.
The CoMFA electrostatic contour maps are displayed in Fig. 5C and Fig. 5D. A large blue contour was found around the 9 position of compounds 11b and 6c, indicating that the presence of electron rich functionalities and positively charged environment at this position (e.g., NH2 vs. NO2, Ac, CN, and CF3) would be strongly favored for topoisomerase I inhibitory activity. It was also observed that a big red contour region was present around the 2 position of fluoroquinophenoxazine scaffold, suggesting that an electronegative group (e.g., COOH and COOR2) at this position may be required for activity. Finally, two red contours were found at both sides of the fused heterocyclic skeleton of 11b and 6c, suggesting that electron deficient functionalities would be favored in those regions.
3. Conclusions
On the basis of a HTS hit compound 1, a series of fluoroquinophenoxazine analogs were designed, synthesized, and evaluated as topoisomerase IA inhibitors and antibacterial agents. Among the newly produced fluoroquinophenoxazine derivatives, compounds with the 9-NH2 and/or 6-substituted amine functionalities exhibited good to excellent inhibitory activities against E. coli topoisomerase IA with IC50 values ranging from 0.24 to 3.9 µM. An illuminating SAR against E. coli topoisomerase I has also been obtained. For example, the 6-substituted amino motif significantly enhanced the topoisomerase I inhibition and the 9-NH2 functionality was the most desirable compared to some other groups evaluated. Notably, 11a bearing the 6-methylpiperazinyl and 9-amino motifs showed excellent topoisomerase I inhibition (IC50 = 0.48 µM) as well as broad spectrum antibacterial activity against all the bacteria strains tested with MICs ranging from 0.78 to 7.6 µM (SI = 3.8–37). In addition, compound 11g with the 6-bipiperidinyl lipophilic side chain represents a promising antitubercular lead with the most potent antituberculosis activity (MIC = 2.5 µM, SI = 9.8). Finally, CoMFA analysis was performed to investigate the 3D-QSAR. The constructed CoMFA model produced reasonable statistics, with q2 = 0.688 and r2 = 0.806. The predictive power of the developed model was obtained using a test set of 7 molecules, giving predictive correlation coefficient r2pred of 0.767. Collectively, this work has generated valuable SAR and critical understanding for this chemotype class of fluoroquinophenoxazine topoisomerase I inhibitors as antibacterial agents. Our developed CoMFA model can provide important structural insights toward topoisomerase I recognition and guide future structure based design and synthesis of bacterial topoisomerase I inhibitors.
4. Experimental
4.1. General methods for chemistry
All reagents and anhydrous solvents were purchased from Sigma-Aldrich and Fisher Scientific, and were used without further purification. All reactions were monitored either by thin-layer chromatography (TLC) or by analytical high performance liquid chromatography (HPLC) to detect the completion of reactions. TLC was performed using glass plates pre-coated with silica gel (0.25 mm, 60-Å pore size, 230–400 mesh, Sorbent Technologies, GA) impregnated with a fluorescent indicator (254 nm). TLC plates were visualized by exposure to ultraviolet light (UV). Hydrogenation reactions were performed employing domnick hunter NITROX UHP-60H hydrogen generator, USA. Microwave synthesis was performed using Biotage Initiator 8 Exp Microwave System. Compounds were purified by flash column chromatography on silica gel using a Biotage Isolera One system and a Biotage SNAP cartridge. 1H and 13C NMR spectra were obtained on a Bruker Avance DRX-400 instrument with chemical shifts (d, ppm) determined using TMS as internal standard. Coupling constants (J) are in hertz (Hz). ESI mass spectra in either positive or negative mode were provided by Varian 500-MS IT Mass Spectrometer. High-resolution mass spectra (HRMS) were obtained on an Agilent 6530 Accurate Mass Q-TOF LC/MS. The purity of compounds was determined by analytical HPLC using a Gemini, 3 µm, C18, 110 Å column (50 mm × 4.6 mm, Phenomenex) and a flow rate of 1.0 mL/min. Gradient conditions: solvent A (0.1% trifluoroacetic acid in water) and solvent B (acetonitrile): 0–2.00 min 100% A, 2.00–7.00 min 0–100% B (linear gradient), 7.00–8.00 min 100% B, UV detection at 254 and 220 nm.
4.1.1. 6-Amino-2,3-difluorophenol (2)
2,3-Difluoro-6-nitrophenol (700 mg, 4 mmol) was dissolved in ethanol (5 mL) and palladium on activated carbon (Pd/C) (84.8 mg, 20%) was added. The reaction was stirred at room temperature under H2 atmosphere (1.0 bar). After 7 h, all starting material was consumed and Pd/C was filtered through Celite. The solvent was evaporated under reduced pressure to afford 6-amino-2,3-difluorophenol (550.5 mg, 95% yield). 1H NMR (400 MHz, CDCl3): δ (ppm) 6.59 (dt, J = 9.9, 8.6 Hz, 1H), 6.47–6.31 (m, 1H), 4.27 (br s, 2H). ESI-HRMS: calc. for C6H6F2NO [M + H]+: 146.0412, found: 146.0418.
4.1.2. Diethyl 2-(((3,4-difluoro-2-hydroxyphenyl)amino)methylene)malonate (3)
6-Amino-2,3-difluorophenol (2) (550 mg, 3.8 mmol) was dissolved in ethanol (15 mL) and diethyl 2-(ethoxymethylene)malonate (819 mg, 3.8 mmol) was added. The reaction was stirred at room temperature until there was no starting material left. Ethanol was removed and the residue was purified by flash column chromatography on silica gel (EtOAc / hexane = 1 / 3 to 3 / 1) to give product 3 as a brown solid (1.04 g, 87%). 1H NMR (400 MHz, d6-DMSO): δ (ppm) 10.96 (d, J = 14.0 Hz, 1H), 8.45 (d, J = 13.9 Hz, 1H), 7.31–7.22 (m, 1H), 6.98–6.86 (m, 1H), 4.20 (q, J = 7.1 Hz, 2H), 4.12 (q, J = 7.1 Hz, 2H), 1.26 (t, J = 7.1 Hz, 3H), 1.24 (t, J = 7.1 Hz, 3H). ESI-MS: calc. for C14H14F2NO5 [M − H]−: 314.3, found: 314.3.
4.1.3. Ethyl 6,7-difluoro-8-hydroxy-4-oxo-1,4-dihydroquinoline-3-carboxylate (4)
Compound 3 (445 mg, 1.4 mmol) was added in a microwave sealed tube. Diphenyl ether (2.5 mL) was added and the tube was sealed with cap. The reaction was then set up at 250 °C in a Biotage Microwave Initiator instrument for 30 min. Hexane was added and the solid was filtered and washed with hexane. The product was then dried and obtained in 80% yield (299.4 mg), which was used for next step without further purification. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.36 (s, 1H), 7.45 (dd, J = 10.9, 7.9 Hz, 1H), 4.21 (q, J = 7.1 Hz, 2H), 1.27 (t, J = 7.1 Hz, 3H). ESI-MS: calc. for C12H8F2NO4 [M − H]−: 268.2, found: 268.2.
4.1.4. Ethyl 5,6-difluoro-9-nitro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylate (6a)
Compound 4 (269 mg, 1 mmol) and 1-chloro-2,4-dinitrobenzene (202 mg, 1 mmol) were dissolved in DMF (2 mL). NaHCO3 (252 mg, 3 mmol) was added and the reaction was stirred at 100 °C until no starting materials were detected by HPLC. The solid base was removed by filtration through Celite. The filtrate was concentrated and the residue was purified through Biotage reverse phase C18 cartridge to give 220 mg of 6a as yellow solid (yield: 57%). 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.01 (s, 1H), 8.15 (d, J = 9.6 Hz, 1H), 8.05–8.01 (m, 2H), 7.60–7.54 (m, 1H), 4.27 (q, J = 7.2 Hz, 2H), 1.32 (t, J = 7.2 Hz, 3H); 13C NMR (100 MHz, d6-DMSO): δ (ppm) 170.4, 163.7, 145.8, 142.7, 138.1, 133.2, 129.4, 124.6, 122.2, 120.8, 117.0, 113.0, 105.4, 105.2, 60.8, 14.2; ESI-MS: calc. for C18H10F2N2O6Na [M + Na]+: 411.3, found: 411.2. ESI-HRMS: calc. for C18H11F2N2O6 [M + H]+: 389.0580, found: 389.0582. HPLC purity: 100% (254 nm), tR: 6.92 min; 100% (220 nm), tR: 6.92 min. Compounds 6b–e were prepared according to the experimental procedure above for the preparation of 6a.
4.1.5. Ethyl 9-cyano-5,6-difluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylate (6b)
Yellow solid. Yield: 59%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.04 (s, 1H), 8.11 (d, J = 8.8 Hz, 1H), 7.87 (d, J = 1.6 Hz, 1H), 7.72 (dd, J = 1.6 and 8.8 Hz, 1H), 7.63–7.59 (m, 1H), 4.28 (q, J = 7.2 Hz, 2H), 1.31 (t, J = 7.2 Hz, 3H); ESI-MS: calc. for C19H10F2N2O4Na [M + Na]+: 391.3, found: 391.2. ESI-HRMS: calc. for C19H11F2N2O4 [M + H]+: 369.0681, found: 369.0684. HPLC purity: 100% (254 nm), tR: 6.73 min; 100% (220 nm), tR: 6.73 min.
4.1.6. Ethyl 5,6-difluoro-3-oxo-9-(trifluoromethyl)-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylate (6c)
Yellow solid. Yield: 10%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.05 (s, 1H), 8.12 (d, J = 8.8 Hz, 1H), 7.71 (d, J = 1.6 Hz, 1H), 7.63–7.58 (m, 2H), 4.27 (q, J = 7.2 Hz, 2H), 1.32 (t, J = 7.2 Hz, 3H); ESI-MS: calc. for C19H10F5NO4Na [M + Na]+: 434.3, found: 434.1. ESI-HRMS: calc. for C19H11F5NO4 [M + H]+: 412.0603, found: 412.0603. HPLC purity: 100% (254 nm), tR: 7.25 min; 100% (220 nm), tR: 7.25 min.
4.1.7. Ethyl 9-acetyl-5,6-difluoro-3-oxo-3H–pyrido[3,2,1-kl] phenoxazine-2-carboxylate (6d)
Dark yellow solid. Yield: 38%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.97 (s, 1H), 8.06–7.93 (m, 1H), 7.73 (d, J = 8.4 Hz, 1H), 7.69 (s, 1H), 7.55 (dd, J = 10.2, 8.0 Hz, 1H), 4.27 (q, J = 7.1 Hz, 2H), 2.58 (s, 3H), 1.33 (t, J = 7.1 Hz, 3H). ESI-MS: calc. for C20H13F2NNaO5 [M + Na]+: 408.3, found: 408.1. HPLC purity: 100% (254 nm), tR: 6.62 min; 100% (220 nm), tR: 6.62 min.
4.1.8. Ethyl 10-chloro-5,6-dif luoro-9-nitro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylate (6e)
Dark yellow solid. Yield: 40%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.13 (s, 1H), 8.48 (s, 1H), 8.12 (s, 1H), 7.69–7.60 (m, 1H), 4.30 (q, J = 7.0 Hz, 2H), 1.33 (t, J = 7.1 Hz, 3H). ESI-MS: calc. for C18H9ClF2N2NaO6 [M + Na]+: 445.0, found: 445.1. HPLC purity: 98.5% (254 nm), tR: 6.92 min; 96.9% (220 nm), tR: 6.92 min.
4.1.9. 5,6-Difluoro-9-nitro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (7a)
Compound 6a (180 mg, 0.46 mmol) was dissolved in AcOH (15 mL). Hydrochloric acid (1 N, 2 mL) was added and the reaction was stirred under reflux for 2 h. Upon completion, water was added and yellow solid was precipitated. The solid was collected by filtration and washed with water, then dried and afforded product 7a (150 mg) as yellow solid in 91% yield. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.36 (s, 1H), 8.42 (d, J = 9.2 Hz, 1H), 8.17 (d, J = 2.8 Hz, 1H), 8.09 (dd, J = 2.4 and 9.2 Hz, 1H), 7.86–7.82 (m, 1H); ESI-MS: calc. for C16H7F2N2O6 [M + H]+: 361.2, found: 361.3. ESI-HRMS: calc. for C16H7F2N2O6 [M + H]+: 361.0267, found: 361.0268. HPLC purity: 100% (254 nm), tR: 6.79 min; 100% (220 nm), tR: 6.79 min.
Compounds 7b–e were prepared according to the experimental procedure above for the preparation of 7a.
4.1.10. 9-Cyano-5,6-difluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (7b)
Yellow solid. Yield: 36%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.34 (s, 1H), 8.35 (d, J = 8.8 Hz, 1H), 7.98 (d, J = 1.6 Hz, 1H), 7.82 (d, J = 2.4 Hz, 1H), 7.78–7.75 (m, 2H); ESI-MS: calc. for C17H7F2N2O4 [M + H]+: 341.2, found: 341.1. ESI-HRMS: calc. for C17H7F2N2O4 [M + H]+: 341.0368, found: 341.0378. HPLC purity: 95.1% (254 nm), tR: 6.37 min; 97.7% (220 nm), tR: 6.37 min.
4.1.11. 5,6-Difluoro-3-oxo-9-(trifluoromethyl)-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (7c)
Yellow solid. Yield: 95%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.35 (s, 1H), 8.37 (d, J = 8.4 Hz, 1H), 7.84–7.79 (m, 2H), 7.65–7.62 (dd, J = 1.2 and 8.8 Hz, 1H); ESI-MS: calc. for C17H7F5NO4 [M + H]+: 384.2, found: 384.1. ESI-HRMS: calc. for C17H7F5NO4 [M + H]+: 384.0290, found: 384.0293. HPLC purity: 100% (254 nm), tR: 7.15 min; 100% (220 nm), tR: 7.15 min.
4.1.12. 9-Acetyl-5,6-difluoro-3-oxo-3H–pyrido[3,2,1-kl] phenoxazine-2-carboxylic acid (7d)
Yellow solid. Yield: 62%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.36 (s, 1H), 8.30 (d, J = 11.2 Hz, 1H), 7.85–7.80 (m, 3H), 2.63 (s, 3H); ESI-MS: calc. for C18H10F2NO5 [M + H]+: 358.3, found: 358.2. ESI-HRMS: calc. for C18H10F2NO5 [M + H]+: 358.0522, found: 358.0521. HPLC purity: 97.3% (254 nm), tR: 6.70 min; 97.2% (220 nm), tR: 6.70 min.
4.1.13. 10-Chloro-5,6-difluoro-9-nitro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (7e)
Brown solid. Yield: 39%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.43 (s, 1H), 8.74 (s, 1H), 8.18 (s, 1H), 7.83–7.79 (m, 1H); ESI-MS: calc. for C16H6ClF2N2O6 [M + H]+: 394.7, found: 394.9. ESI-HRMS: calc. for C16H6F2N2ClO6 [M + H]+: 394.9877, found: 394.9879. HPLC purity: 95.4% (254 nm), tR: 6.90 min; 97.0% (220 nm), tR: 6.90 min.
4.1.14. 9-Amino-5,6-difluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (1)
Compound 7a (150 mg, 0.42 mmol) was added to a mixture of AcOH / HCl (1 / 1). SnCl2 (236 mg, 1.25 mmol) was added and the reaction was stirred under reflux for 2 h. No starting material was observed in the reaction and then water was added. The large amount of solid precipitated and was collected by filtration. Product 1 was obtained in 86% yield (236 mg) as yellow solid. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.08 (s, 1H), 7.79 (d, J = 9.2 Hz, 1H), 7.76–7.73 (m, 1H), 6.48 (dd, J = 2.4 and 9.2 Hz, 1H), 6.43 (d, J = 2.4 Hz, 1H); ESI-MS: calc. for C16H9F2N2O4 [M + H]+: 331.3, found: 331.1. ESI-HRMS: calc. for C16H9F2N2O4 [M + H]+: 331.0525, found: 331.0526. HPLC purity: 100% (254 nm), tR: 6.47 min; 100% (220 nm), tR: 6.47 min.
4.1.15. Ethyl-5-fluoro-6-(4-m ethylpiperazin-1-yl)-9-nitro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylate (9)
Compound 6a (159 mg, 0.4 mmol) was dissolved in pyridine (1 mL) and 1-methylpiperazine (120 mg, 1.2 mmol) was added. The reaction was heated to 110 °C until no starting material was observed. Pyridine was removed under reduced pressure and the residue was purified by flash column chromatography on silica gel (MeOH / CH2Cl2 = 1 / 19), affording product 9 (100.1 mg) in 53% yield as orange solid. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.87 (d, J = 1.6 Hz, 1H), 8.04–7.97 (m, 2H), 7.87 (t, J = 1.2 Hz, 1H), 7.32 (dd, J = 1.2 and 8.0 Hz, 2H), 4.25 (q, J = 7.2 Hz, 2H), 3.34 (s, 4H), 3.27 (s, 4H), 2.27 (s, 3H), 1.32 (t, J = 7.2 Hz, 3H); ESI-MS: calc. for C23H22FN4O6 [M + H]+: 469.4, found: 469.3. HPLC purity: 100% (254 nm), tR: 5.48 min; 100% (220 nm), tR: 5.48 min.
4.1.16. 5-Fluoro-6-(4-methylpiperazin-1-yl)-9-nitro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (10a)
Compound 7a (108 mg, 0.3 mmol) was dissolved in pyridine (4 mL), and then the reaction was heated to 90 °C. 1-Methylpiperazine (100 µL, 0.9 mmol) was added and the reaction was stirred under nitrogen atmosphere until there was no starting material. Upon completion, pyridine was removed under reduced pressure and the residue was dissolved in ethanol (10 mL) and heated under reflux for additional 30 min. The solid was filtered and washed with water, then dried to obtain product 10a (60 mg) in 45 % yield as yellow solid. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.18 (s, 1H), 8.29 (d, J = 8.0 Hz, 1H), 8.02–7.99 (m, 2H), 7.53 (d, J = 12.6 Hz, 1H), 3.30 (br s, overlapping with H2O peak, 4H), 2.57 (br s, 4H), 2.32 (s, 3H); ESI-MS: calc. for C21H18FN4O6 [M + H]+: 441.4, found: 441.4. ESI-MS: calc. for C21H18FN4O6 [M + H]+: 441.1205, found: 441.1216. HPLC purity: 100% (254 nm), tR: 5.37 min; 100% (220 nm), tR: 5.37 min. Compounds 10b and 11a–k were prepared following the similar procedure for the preparation of 10a.
4.1.17. 5-Fluoro-9-nitro-3-oxo-6-(piperazin-1-yl)-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic a ci d (10b)
Yellow solid. Yield: 84%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.94 (s, 1H), 7.66 (d, J = 8.8 Hz, 1H), 7.45 (d, J = 12.0 Hz, 1H), 6.43–6.36 (m, 2H), 3.22 (s, 4H), 2.87 (s, 4H); ESI-MS: calc. for C20H16FN4O6 [M + H]+: 427.4, found: 427.2. ESI-HRMS: calc. for C20H16FN4O6 [M + H]+: 427.1048, found: 427.1041. HPLC purity: 100% (254 nm), tR: 5.36 min; 100% (220 nm), tR: 5.36 min.
4.1.18. 9-Amino-5-fluoro-6-(4-methylpiperazin-1-yl)-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11a)
Yellow solid. Yield: 48%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.94 (s, 1H), 7.65 (d, J = 9.2 Hz, 1H), 7.45 (d, J = 12.0 Hz, 1H), 6.43–6.36 (m, 2H), 5.77 (s, 2H), 3.31 (s, 4H), 2.57 (s, 4H), 2.32 (s, 3H); ESI-MS: calc. for C21H20FN4O4 [M + H]+: 411.4, found: 411.2. ESI-MS: calc. for C21H20FN4O4 [M + H]+: 411.1463, found: 411.1466. HPLC purity: 100% (254 nm), tR: 5.22 min; 100% (220 nm), tR: 5.22 min.
4.1.19. 9-Amino-5-fluoro-3-oxo-6-(piperazin-1-yl)-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11b)
Yellow solid. Yield: 97%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.97 (s, 1H), 7.68 (d, J = 9.2 Hz, 1H), 7.48 (d, J = 12.4 Hz, 1H), 6.44–6.38 (m, 2H), 5.77 (s, 2H), 3.22 (s, 4H), 2.86 (s, 4H); ESI-MS: calc. for C20H18FN4O4 [M + H]+: 397.4, found: 397.2. ESI-HRMS: calc. for C20H18FN4O4 [M + H]+: 397.1307, found: 397.1303. HPLC purity: 100% (254 nm), tR: 5.20 min; 100% (220 nm), tR: 5.20 min.
4.1.20. 9-Amino-5-fluoro-6-(4-methylpiperidin-1-yl)-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11c)
Yellow solid. Yield: 33%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.92 (s, 1H), 7.63 (d, J = 8.0 Hz, 1H), 7.43 (d, J = 11.6 Hz, 1H), 6.42–6.36 (m, 2H), 5.75 (s, 2H), 3.13 (t, J = 10.8 Hz, 2H), 1.71 (d, J = 11.2 Hz, 2H), 1.57 (s, 1H), 1.32–1.26 (m, 2H), 0.98 (s, 3H); 13C NMR (100 MHz, d6-DMSO): δ (ppm) 175.4, 166.4, 150.7, 144.6, 138.5, 136.0, 131.7, 125.3, 119.6, 119.5, 117.6, 112.2, 111.1, 107.0, 104.0, 101.2, 51.1, 35.1, 30.6, 22.5; ESI-MS: calc. for C22H21FN3O4 [M + H]+: 410.4, found: 410.2. ESI-HRMS: calc. for C22H21FN3O4 [M + H]+: 410.1511, found: 410.1510. HPLC purity: 100% (254 nm), tR: 6.38 min; 100% (220 nm), tR: 6.38 min.
4.1.21. 9-Amino-5-fluoro-6-morpholino-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11d)
Yellow solid. Yield: 71%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.93 (s, 1H), 7.64 (d, J = 8.8 Hz, 1H), 7.45 (d, J = 12.0 Hz, 1H), 6.43–6.37 (m, 2H), 5.75 (s, 2H), 3.69 (s, 4H), 3.28 (s, 4H); ESI-MS: calc. for C20H17FN3O5 [M + H]+: 398.4, found: 398.2. ESI-HRMS: calc. for C20H17FN3O5 [M + H]+: 398.1147, found: 398.1146. HPLC purity: 100% (254 nm), tR: 6.38 min; 100% (220 nm), tR: 6.38 min.
4.1.22. 9-Amino-5-fluoro-3-oxo-6-(phenethylamino)-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11e)
Yellow solid. Yield: 55%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.90 (s, 1H), 7.61 (d, J = 9.2 Hz, 1H), 7.45 (d, J = 13.2 Hz, 1H), 7.29–7.18 (m, 5H), 6.43–6.37 (m, 2H), 6.15 (br s, 1H), 5.76 (br s, 2H), 3.68–3.61 (m, 2H), 2.89–2.85 (m, 2H); ESI-MS: calc. for C24H18FN3O4 [M + H]+: 432.4, found: 432.1. ESI-HRMS: calc. for C24H19FN3O4 [M + H]+: 432.1354, found: 432.1362. HPLC purity: 100% (254 nm), tR: 7.07 min; 100% (220 nm), tR: 7.07 min.
4.1.23. 6-(((3s,5s,7s)-Adamantan-1-yl)amino)-9-amino-5-fluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11f)
Brown solid. Yield: 29%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.00 (s, 1H), 7.71 (d, J = 9.1 Hz, 1H), 7.54 (d, J = 10.7 Hz, 1H), 6.50–6.38 (m, 2H), 5.81 (s, 2H), 4.34 (s, 1H), 2.09–2.04 (m, 3H), 1.85 (s, 6H), 1.65–1.55 (m, 6H). ESI-MS: calc. for C26H23FN3O4 [M − H]+: 460.2, found: 460.0. HPLC purity: 85.2% (254 nm), tR: 7.41 min; 91.1% (220 nm), tR: 7.41 min.
4.1.24. 6-([1,4’-Bipiperidin]-1’-yl)-9-amino-5-fluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11g)
Yellow solid. Yield: 64%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.98 (s, 1H), 7.68 (d, J = 4.0 Hz, 1H), 7.49 (d, J = 9.6 Hz, 1H), 6.45–6.41 (m, 2H), 5.77 (br s, 2H), 3.20–3.15 (m, 1H), 1.84–1.82 (m, 3H), 1.64–1.41 (m, 14H), 1.06–1.04 (m, 1H); ESI-MS: calc. for C26H28FN4O4 [M + H]+: 479.5, found: 479.3. ESI-HRMS: calc. for C26H28FN4O4 [M + H]+: 479.2089, found: 479.2093. HPLC purity: 100% (254 nm), tR: 5.53 min; 100% (220 nm), tR: 5.53 min.
4.1.25. 9-Amino-5-fluoro-6-((2-morpholinoethyl)amino)-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11h)
Yellow solid. Yield: 74%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.90 (s, 1H), 8.55 (s, 1H), 7.46 (d, J = 6.0 Hz, 1H), 7.39–7.34 (m, 2H), 6.42–6.37 (m, 2H), 5.95 (s, 1H), 5.78 (br s, 2H), 2.57 (s, 2H), 2.44 (br s, 6H); ESI-MS: calc. for C22H22FN4O5 [M + H]+: 441.4, found: 441.3. ESI-MS: calc. for C22H22FN4O5 [M + H]+: 441.1569, found: 441.1579. HPLC purity: 97.2% (254 nm), tR: 5.22 min; 99.3% (220 nm), tR: 5.22 min.
4.1.26. 9-Amino-6-(cyclohexylamino)-5-fluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11i)
Brown solid. Yield: 74%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.86 (s, 1H), 7.58–7.56 (m, 1H), 7.43–7.40 (m, 1H), 6.40 (s, 1H), 5.72 (s, 2H), 5.40–5.38 (m, 1H), 1.92 (s, 3H), 1.73–1.67 (m, 3H), 1.31–1.22 (m, 6H); ESI-MS: calc. for C22H21FN3O4 [M + H]+: 410.4, found: 410.3. ESI-HRMS: calc. for C22H21FN3O4 [M + H]+: 410.1511, found: 410.1494. HPLC purity: 100% (254 nm), tR: 7.27 min; 100% (220 nm), tR: 7.27 min.
4.1.27. 9-Amino-6-(cyclopentylamino)-5-fluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11j)
Black solid. Yield: 63%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.91 (s, 1H), 7.62 (d, J = 8.8 Hz, 1H), 7.47 (d, J = 12.4 Hz, 1H), 6.46–6.41 (m, 2H), 5.73 (s, 2H), 5.53 (d, J = 2.4 Hz, 1H), 4.30 (s, 1H), 1.93 (br s, 4H), 1.73 (br s, 4H); ESI-MS: calc. for C21H19FN3O4 [M + H]+: 396.4, found: 396.1. ESI-HRMS: calc. for C21H19FN3O4 [M + H]+: 396.1354, found: 396.1358. HPLC purity: 96.7% (254 nm), tR: 7.07 min; 96.0% (220 nm), tR: 7.06 min.
4.1.28. 9-Amino-5-fluoro-6-(hexylamino)-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (11k)
Yellow solid. Yield: 49%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.88 (s, 1H), 7.64 (s, 1H), 7.44 (s, 1H), 6.56–6.36 (m, 2H), 5.88–5.50 (m, 2H), 1.57 (s, 2H), 1.27 (s, 7H), 0.84 (s, 4H); ESI-MS: calc. for C22H23FN3O4 [M + H]+: 412.4, found: 412.2. ESI-HRMS: calc. for C22H23FN3O4 [M + H]+: 412.1667, found: 412.1672. HPLC purity: 100% (254 nm), tR: 7.47 min; 100% (220 nm), tR: 7.47 min.
4.1.29. 9-acetamido-5,6-difluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (12)
To the solution of 1 (33 mg, 0.1 mmol) and pyridine (1 mL), acetic anhydride (12.2 mg, 0.12 mmol) was added. The reaction was stirred at 80 °C for 3 h, then at 100 °C until no starting material was detected by HPLC. The solid was filtered and dried to give 34.1 mg of 12 in 92% yield. Yellow solid. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 10.31 (s, 1H), 9.20 (s, 1H), 8.05 (d, J = 9.2 Hz, 1H), 7.78 (t, J = 8.0 Hz, 1H), 7.66 (s, 1H), 7.29 (d, J = 7.6 Hz, 1H), 2.06 (s, 3H); ESI-MS: calc. for C18H11F2N2O5 [M + H]+: 373.3, found: 373.2. ESI-MS: calc. for C18H11F2N2O5 [M + H]+: 373.0631, found: 373.0638. HPLC purity: 98.6% (254 nm), tR: 6.42 min; 99.1% (220 nm), tR: 6.42 min. Compounds 14 and 15a–b were prepared at 70 °C following the similar procedure for the preparation of 10a
4.1.30. (S)-9-amino-6-(3-((tert-butoxycarbonyl)amino)pyrrolidin-1-yl)-5-fluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (15a)
Yellow solid. Yield: 86%. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.77 (s, 1H), 7.49 (d, J = 9.6 Hz, 1H), 7.31 (d, J = 14.0 Hz, 1H), 7.17 (d, J = 5.6 Hz, 1H), 6.38 (dd, J = 2.4 and 9.2 Hz, 2H), 6.29 (d, J = 2.0 Hz, 1H), 5.67 (s, 2H), 3.82–3.80 (m, 5H), 2.05–2.04 (m, 1H), 1.83 (br s, 1H), 1.38 (s, 9H); ESI-MS: calc. for C25H26FN4O6 [M + H]+: 497.5, found: 497.3. ESI-HRMS: calc. for C25H26FN4O6 [M + H]+: 497.1831, found: 497.1831. HPLC purity: 100% (254 nm), tR: 6.94 min; 100% (220 nm), tR: 6.94 min.
4.1.31. (S)-6-(3-aminopyrroli din-1-yl)-5-fluoro-9-nitro-3-oxo-3H–pyrido[3,2,1-kl] phenoxazine-2-carboxyl ic acid (16)
Compound 16 was prepared following the same procedure for the synthesis of 7. Brown solid. Yield: 32% (two steps). 1H NMR (400 MHz, d6-DMSO): δ (ppm) 9.10 (s, 1H), 8.41 (s, 2H), 8.25 (d, J = 9.3 Hz, 1H), 8.09 (s, 1H), 8.01 (d, J = 7.2 Hz, 1H), 7.48 (d, J = 13.6 Hz, 1H), 4.07 – 3.74 (m, 5H), 2.28 (s, 1H), 2.08 (s, 1H). ESI-MS: calc. for C20H16FN4O6 [M + H]+: 427.4, found: 427.1. HPLC purity: 100% (254 nm), tR: 5.45 min; 100% (220 nm), tR: 5.46 min.
4.1.32. (S)-9-amino-6-(3-aminopyrrolidin-1-yl)-5-fluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (17a)
Compound 15a (85 mg, 0.17 mmol) was dissolved in 10 mL of hydrochloric acid (1 N). The reaction was stirred under reflux for 2.5 h. The solvent was removed and ethanol (10 mL) was added. The mixture was heated under reflux for 30 min. The solid was collected by filtration and dried to yield 52 mg of 17a in 78% yield. Yellow solid. 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.94 (s, 1H), 8.24 (s, 3H), 7.65 (d, J = 9.2 Hz, 1H), 7.50 (d, J = 13.6 Hz, 1H), 6.46–6.40 (m, 2H), 3.99–3.85 (m, 5H), 2.34–2.26 (m, 1H), 2.09–1.98 (m, 1H); ESI-MS: calc. for C20H18FN4O4 [M + H]+: 397.4, found: 397.1. ESI-MS: calc. for C20H18FN4O4 [M + H]+: 397.1307, found: 397.1298. HPLC purity: 100% (254 nm), tR: 5.20 min; 100% (220 nm), tR: 5.20 min.
4.1.33. (R)-9-amino-6-(3-aminopyrrolidin-1-yl)-5-fluoro-3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acid (17b)
Compound 17b was prepared using the same procedure for the synthesis of 17a. Brown solid. Yield: 48% (two steps). 1H NMR (400 MHz, d6-DMSO): δ (ppm) 8.88 (s, 1H), 8.31 (s, 3H), 7.59 (d, J = 9.2 Hz, 1H), 7.43 (d, J = 13.2 Hz, 1H), 6.42 (d, J = 9.2 Hz, 1H), 6.38 (s, 1H), 3.98–3.88 (m, 2H), 3.73–3.68 (m, 3H), 2.30–2.25 (m, 1H), 2.06–2.03 (m, 1H); ESI-MS: calc. for C20H18FN4O4 [M + H]+: 397.4, found: 397.2. ESI-HRMS: calc. for C20H18FN4O4 [M + H]+: 397.1307, found: 397.1302. HPLC purity: 100% (254 nm), tR: 5.24 min; 100% (220 nm), tR: 5.24 min.
4.2. Target-based biochemical assays
Topoisomerase enzymes
Recombinant E. coli topoisomerase I and gyrase expressed in E. coli were purified as described previously [24, 25]. Human topoisomerase I and topoisomerase IIα were purchased from TopoGen (Buena vista, CO, USA).
4.2.1. E. coli topoisomerase I relaxation activity inhibition assay
The relaxation activity of E. coli topoisomerase I was assayed in a buffer containing 10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 0.1 mg/mL gelatin, and 0.5 mM MgCl2. Half microliter from the appropriate stock solutions of compounds dissolved in the solvent (DMSO) or the solvent alone (control) was mixed with 9.5 µL of the reaction buffer containing 10 ng of enzyme before the addition of 10 µL of reaction buffer containing 200 ng of supercoiled pBAD/Thio plasmid DNA purified by cesium chloride gradient as substrate. Following incubation at 37 °C for 30 min, the reactions were terminated by the addition of 4 µL of a stop buffer (50% glycerol, 50 mM EDTA, and 0.5% (v/v) bromophenol blue), and analyzed by agarose gel electrophoresis. The gels were stained in ethidium bromide and photographed under UV light.
4.2.2. DNA gyrase supercoiling inhibition assay
DNA gyrase supercoiling assays were carried out by mixing the compounds and the enzyme in a similar manner as above (EcTopI relaxation inhibition assay) but in a gyrase assay buffer (35 mM Tris-HCl, 24 mM KCl, 4 mM MgCl2, 2 mM DTT, 1.75 mM ATP, 5 mM spermidine, 0.1 mg/mL BSA, 6.5 % glycerol at pH 7.5), followed by the addition of 300 ng of relaxed covalently closed circular DNA (New England Biolabs, Ipswich, MA, USA) to a final reaction volume of 20 µL. The samples were incubated at 37 °C for 30 minutes before being terminated by the addition of a buffer containing 5% SDS, 0.25% bromophenol blue, and 25% glycerol. The reactions were then analyzed by agarose gel electrophoresis.
4.2.3. Human topoisomerase I relaxation inhibition assay
Human topoisomerase I relaxation assays were carried out with 0.5 U of enzyme in reaction buffer supplied by the manufacturer. The enzyme was mixed with the indicated concentration of compound dissolved before 200 ng of supercoiled pBAD/Thio plasmid DNA was added in the same buffer, for a final volume of 20 µL. Following incubation at 37 °C for 30 minutes, the reactions were terminated with a buffer containing 5% SDS, 0.25% bromophenol blue, and 25% glycerol, and analyzed by agarose gel electrophoresis.
4.2.4. Human topoisomerase IIα decatenation inhibition assay
Human Topoisomerase IIα assays were carried out by adding the compounds to 185 ng of kinetoplast DNA (kDNA, from TopoGen) in the buffer supplied by the manufacturer before the addition of 2 U of the enzyme. The samples were incubated for 15 minutes at 37 °C before the addition of 4 µL of a stop buffer containing 5% sarkosyl, 0.25% bromophenol blue, and 25% glycerol. The reactions were then analyzed by electrophoresis in 1% agarose gels containing 0.5 µg/mL ethidium bromide before being photographed under UV light.
4.3. Cell-based assays
The minimum inhibitory concentrations (MIC) of the compounds were determined against E. coli and B. subtilis in cation-adjusted Mueller-Hinton Broth according to standard microdilution protocol [26].
MICs of compounds against M. tuberculosis were determined by a modified microplate Alamar blue assay (MABA) [27]. Vero cell cytotoxicity assay was performed as previously described [27].
4.4. CoMFA modeling
4.4.1. Dataset
All the synthesized and tested fluoroquinophenoxazine derivatives were used for CoMFA study. The topoisomerase I inhibitory activity (IC50, µM) from biochemical enzyme assay was converted to pIC50 values for correlation purpose (pIC50 = – logIC50). The total compound set is divided into two subsets: a training set of 21 compounds for generating 3D-QSAR models and a test set of 7 compounds for validating the quality of the model (Table 2). The compound selections of training and test sets were done manually so that compounds ranging from weak, moderate, to strong topoisomerase I inhibitory activities were present in both sets and were in approximately equal proportions.
4.4.2. Conformational model analysis and molecular alignment
In the 3D-QSAR studies, alignment rule and biological conformation selection are two important factors to construct reliable models. For both training and test set molecules, conformational models representing their available conformational space were calculated. All the molecules were subjected to produce a maximum of 255 conformations within 20 kcal/mol in energy from global minimum. Due to the relatively rigid structural feature of these molecules, the core structure of quinophenoxazine was used for the alignment.
4.4.3. CoMFA model generation
CoMFA was performed using the QSAR module of SYBYL-X [28]. The steric and electrostatic field energies were calculated using the Lennard-Jones and the Coulomb potentials, respectively, with a 1/r distance-dependent dielectric constant in all intersections of a regularly spaced (0.2 nm) grid. The electrostatic fields were computed using Gasteiger-Huckel charge calculation methods. A sp3 hybridized carbon atom with a radius of 1.53 Å and a charge of +1.0 was used as a probe to calculate the steric and electrostatic energies between the probe and the molecules using the Tripos force field. The standard parameters implemented in SYBYL-X were used. The truncation for both steric and electrostatic energies was set to 30 kcal/mol.
4.4.4. Partial least square (PLS) analysis
PLS methodology [29] was used for 3D-QSAR analysis. The cross-validation analysis [30, 31] was performed using the leave one out (LOO) methods in which one compound is removed from the dataset and its activity is then predicted using the model derived from the rest of the dataset. The cross validated r2 that resulted in the optimum number of components and the lowest standard error of prediction were considered for further analysis. To speed up the analysis and reduce noise, a minimum filter value of 2.00 kcal/mol was used. A final analysis was performed to calculate conventional r2 using the optimum number of components obtained from the cross-validation analysis.
Supplementary Material
Highlights.
Fluoroquinophenoxazines were synthesized as bacterial topoisomerase IA inhibitors.
Some derivatives showed excellent inhibitory activity against topoisomerase IA.
CoMFA analysis was performed to investigate the 3D-QSAR of this chemical series.
The constructed CoMFA model produced reasonable and good statistics.
Acknowledgments
This project was supported in part by the National Institutes of Health grants (P20GM103466 and R15AI092315 to DS, R01AI069313 to YT). MICs against M. tuberculosis and Vero cell cytotoxicity were provided by the service available at the Institute for Tuberculosis Research at University of Illinois at Chicago. NSC648059 was provided by the NCI Developmental Therapeutics Program.
Abbreviations
- SAR
structure-activity relationship
- HTS
high-throughput screening
- QSAR
quantitative structure-activity relationship
- CoMFA
comparative molecular field analysis
- MIC
minimum inhibitory concentration
- SI
selectivity index
- 3D-QSAR
three dimensional QSAR
- PA
predicted activity
- TLC
thin-layer chromatography
- HPLC
high performance liquid chromatography
- MABA
microplate Alamar blue assay
- PLS
partial least square
- LOO
leave one out
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
Appendix A: Supplementary data
Supplementary data related to this article can be found at
References
- 1.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olivier S, Qasim AK, Kurt WK, Yves P. Apoptosis induced by topoisomerase inhibitors. Curr. Med Chem. Anticancer Agents. 2003;3:271–290. doi: 10.2174/1568011033482378. [DOI] [PubMed] [Google Scholar]
- 4.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pommier Y. Drugging topoisomerases: Lessons and challenges. ACS Chem. Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tse-Dinh Y-C. Targeting bacterial topoisomerase I to meet the challenge of finding new antibiotics. Future Med. Chem. 2015;7:459–471. doi: 10.4155/fmc.14.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tse-Dinh Y-C. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucl. Acids Res. 2009;37:731–737. doi: 10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin H, Annamalai T, Bansod P, Tse-Dinh Y-C, Sun D. Synthesis and antibacterial evaluation of anziaic acid and its analogues as topoisomerase I inhibitors. MedChemComm. 2013;4:1613–1618. doi: 10.1039/C3MD00238A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng L, Maddox MM, Alam MZ, Tsutsumi LS, Narula G, Bruhn DF, Wu X, Sandhaus S, Lee RB, Simmons CJ, Tse-Dinh Y-C, Hurdle JG, Lee RE, Sun D. Synthesis, structure-activity relationship studies, and antibacterial evaluation of 4-chromanones and chalcones, as well as olympicin A and derivatives. J. Med. Chem. 2014;57:8398–8420. doi: 10.1021/jm500853v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karl D, Muhammad M. Fluoroquinolones: Action and resistance. Curr. Top. Med. Chem. 2003;3:249–282. doi: 10.2174/1568026033452537. [DOI] [PubMed] [Google Scholar]
- 11.Rádl S, Zikán V. Synthesis and antimicrobial activity of some 3-oxo-3H–pyrido[3,2,1-kl]phenoxazine-2-carboxylic acids. Collect. Czech. Chem. Commun. 1989;54:506–515. [Google Scholar]
- 12.Chu DTW, Maleczka RE. Synthesis of 4-oxo-4H–quino[2,3,4-i,j][1,4]-benoxazine-5-carboxylic acid derivatives. J. Heterocyclic Chem. 1987;24:453–456. [Google Scholar]
- 13.Kang D-H, Kim J-S, Jung M-J, Lee E-S, Jahng Y, Kwon Y, Na Y. New insight for fluoroquinophenoxazine derivatives as possibly new potent topoisomerase I inhibitor. Bioorg. Med. Chem. Lett. 2008;18:1520–1524. doi: 10.1016/j.bmcl.2007.12.053. [DOI] [PubMed] [Google Scholar]
- 14.Permana PA, Snapka RM, Shen LL, Chu DTW, Clement JJ, Plattner JJ. Quinobenoxazines: A class of novel antitumor quinolones and potent mammalian DNA topoisomerase II catalytic inhibitors. Biochemistry. 1994;33:11333–11339. doi: 10.1021/bi00203a031. [DOI] [PubMed] [Google Scholar]
- 15.Fan J-Y, Sun D, Yu H, Kerwin SM, Hurley LH. Self-assembly of a quinobenzoxazine-Mg2+ complex on DNA: A new paradigm for the structure of a drug-DNA complex and implications for the structure of the quinolone bacterial gyrase-DNA complex. J. Med. Chem. 1995;38:408–424. doi: 10.1021/jm00003a003. [DOI] [PubMed] [Google Scholar]
- 16.Duan W, Rangan A, Vankayalapati H, Kim M-Y, Zeng Q, Sun D, Han H, Fedoroff OY, Nishioka D, Rha SY, Izbicka E, Von Hoff DD, Hurley LH. Design and synthesis of fluoroquinophenoxazines that interact with human telomeric G-quadruplexes and their biological effects. Mol. Cancer Ther. 2001;1:103–120. [PubMed] [Google Scholar]
- 17.Smith CJ, Ali A, Chen L, Hammond ML, Anderson MS, Chen Y, Eveland SS, Guo Q, Hyland SA, Milot DP, Sparrow CP, Wright SD, Sinclair PJ. 2-Arylbenzoxazoles as CETP inhibitors: Substitution of the benzoxazole moiety. Bioorg. Med. Chem. Lett. 2010;20:346–349. doi: 10.1016/j.bmcl.2009.10.099. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Q, Kwok Y, Kerwin SM, Mangold G, Hurley LH. Design of new topoisomerase II inhibitors based upon a quinobenzoxazine self-assembly model. J. Med. Chem. 1998;41:4273–4278. doi: 10.1021/jm980265c. [DOI] [PubMed] [Google Scholar]
- 19.Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli . Mol. Microbiol. 2002;45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 21.Ravishankar S, Ambady A, Awasthy D, Mudugal NV, Menasinakai S, Jatheendranath S, Guptha S, Sharma S, Balakrishnan G, Nandishaiah R, Ramachandran V, Eyermann CJ, Reck F, Rudrapatna S, Sambandamurthy VK, Sharma UK. Genetic and chemical validation identifies Mycobacterium tuberculosis topoisomerase I as an attractive anti-tubercular target. Tuberculosis. 2015;95:589–598. doi: 10.1016/j.tube.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed W, Menon S, Godbole AA, Karthik PVDNB, Nagaraja V. Conditional silencing of topoisomerase I gene of Mycobacterium tuberculosis validates its essentiality for cell survival. FEMS Microbiol. Lett. 2014;353:116–123. doi: 10.1111/1574-6968.12412. [DOI] [PubMed] [Google Scholar]
- 23.Cramer RD, Patterson DE, Bunce JD. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988;110:5959–5967. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- 24.Narula G, Annamalai T, Aedo S, Cheng B, Sorokin E, Wong A, Tse-Dinh Y-C. The strictly conserved Arg-321 residue in the active site of Escherichia coli topoisomerase I plays a critical role in DNA rejoining. J. Biol. Chem. 2011;286:18673–18680. doi: 10.1074/jbc.M111.229450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallett P, Grimshaw AJ, Wigley DB, Maxwell A. Cloning of the DNA gyrase genes under tac promoter control: overproduction of the gyrase A and B proteins. Gene. 1990;93:139–142. doi: 10.1016/0378-1119(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 26.Andrews JM. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 27.Falzari K, Zhu Z, Pan D, Liu H, Hongmanee P, Franzblau SG. In vitro and in vivo activities of macrolide derivatives against Mycobacterium tuberculosis . Antimicrob. Agents Chemother. 2005;49:1447–1454. doi: 10.1128/AAC.49.4.1447-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.SYBYL-X/QSAR. Molecular Modelling Software. St. Louis, Missouri, 63144, USA: Tripos International, 1699 South Hanley Rd; [Google Scholar]
- 29.Masand VH, Mahajan DT, Alafeefy AM, Bukhari SNA, Elsayed NN. Optimization of antiproliferative activity of substituted phenyl 4-(2-oxoimidazolidin-1-yl) benzenesulfonates: QSAR and CoMFA analyses. Eur. J. Pharm. Sci. 2015;77:230–237. doi: 10.1016/j.ejps.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Podlogar BL, Poda GI, Demeter DA, Zhang SP, Carson JR, Neilson LA, Reitz AB, Ferguson DM. Synthesis and evaluation of 4-(N,N-diarylamino)piperidines with high selectivity to the delta-opioid receptor: a combined 3D-QSAR and ligand docking study. Drug Des. Discov. 2000;17:34–50. [PubMed] [Google Scholar]
- 31.Ståhle L, Wold S. Partial least squares analysis with cross-validation for the two-class problem: A Monte Carlo study. J. Chemom. 1987;1:185–196. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.