Abstract
Thymine DNA glycosylase (TDG) is a base excision repair enzyme with key functions in epige-netic regulation. Performing a critical step in a pathway for active DNA demethylation, TDG removes 5-formylcytosine and 5-carboxylcytosine, oxidized derivatives of 5-methylcytosine that are generated by TET (ten-eleven translocation) enzymes. We solved a crystal structure of TDG bound to DNA with a non-cleavable (2′-fluoroarabino) analog of 5-formyldeoxycytidine flipped into its active site, revealing how it recognizes and hydrolytically excises fC. Together with previous structural and biochemical findings, the results illustrate how TDG employs an adaptable active site to excise a broad variety of nucleobases from DNA.
Graphical abstract
Thymine DNA glycosylase (TDG) initiates base excision repair (BER) by excising chemically modified bases, specifically those arising from deamination or oxidation of 5-methylcytosine (mC).1 TDG removes T from G·T mispairs, protecting against mutations arising from deamination of mC to T.2 In a multistep pathway for active DNA demethylation, TDG excises 5-formylcytosine (fC) and 5-carboxylcytosine (caC),3-5 which are generated via oxidation of mC or 5-hydroxymethylcytosine (hmC) by TET (ten-eleven translocation) enzymes (Figure 1).4,6,7 This key epigenetic function likely explains findings that TDG is essential for embryogenesis in mice.8,9 However, our understanding of how TDG excises select forms of oxidized mC has remained limited.
Figure 1.
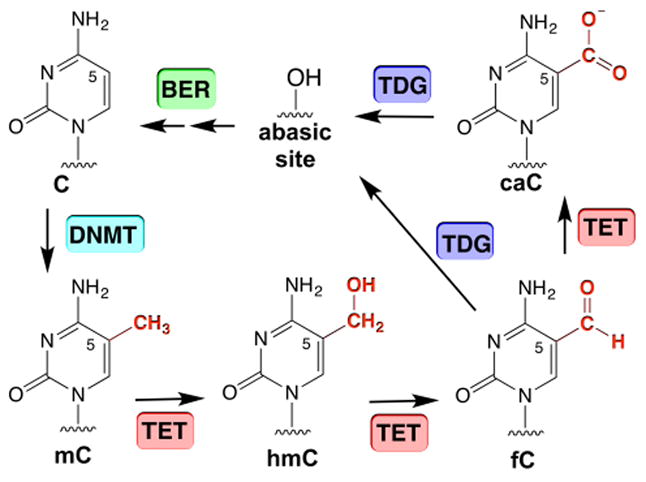
DNA cytosine methylation by DNMT and DNA demethylation via the TET-TDG-BER pathway.
To address this problem, we sought to obtain a crystal structure of TDG bound to DNA with a non-cleavable (2′-fluoroarabino) analog of 5-formyldeoxycytidine (fdC) flipped productively into its active site, giving a snapshot of the enzyme-substrate complex. Fluorination of deoxynucleotides at 2′ precludes N-glycosyl bond cleavage by DNA glycosylases, due to transition state destabilization.10-13 Indeed, 2′-F substitution precludes TDG cleavage of dT, dU, and 5-carboxyl-dC (cadC).10,14-16 These substrate analogues are synthesized in two forms, 2′-F-ribo (α) and 2′-F-arabino (β) (Chart 1), both of which halt N-glycosyl hydrolysis by TDG and other glycosylases.10,11,17 The 2′-F-β analogues are considered superior for structural studies with glycosylases, because the sugar pucker is compatible with B-form DNA and the F is less likely to disrupt binding of the nucleophile (water) because it resides on the opposite face of the sugar ring.18-21 Thus, 2′-F-β deoxynucleotides are good substrate mimics that flip into a glycosylase active site and preserve key enzyme-substrate interactions.
Chart 1.
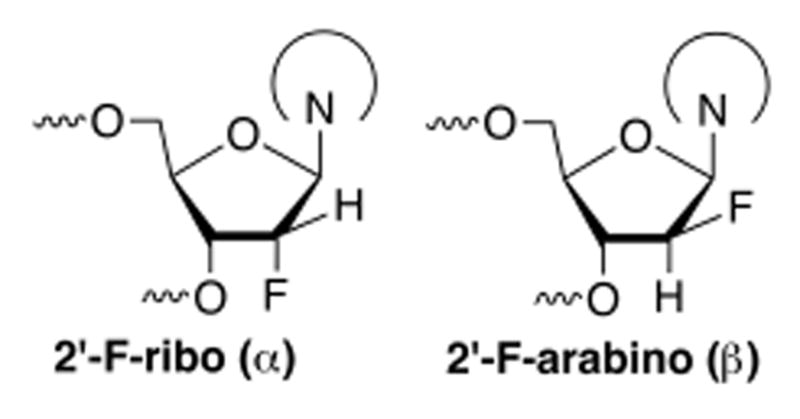
2′-F substitutions in 2′-deoxynucleotides
We prepared oligodeoxynucleotides (ODNs) containing 2′-F-β-fdC (or fdCF) using phosphoramidite 4, which was synthesized in a manner similar to that previously reported (Scheme 1).22-24 The 2′-fluoronucleoside core was prepared using a strategy described by Damha and more recently by He.22,23 The protecting group scheme developed by Carell was employed to introduce 5-formylcytosine.24 Using ODNs containing fdCF, we prepared duplex DNA containing a G·fC base pair for crystallization with TDG. As expected, we find no detectable TDG hydrolysis of fdCF, even after 48 h (Figure S1).
Scheme 1. Synthesis of the phosphoramidite for 2′-fluoroarabino-5-formyldeoxycytidinea.
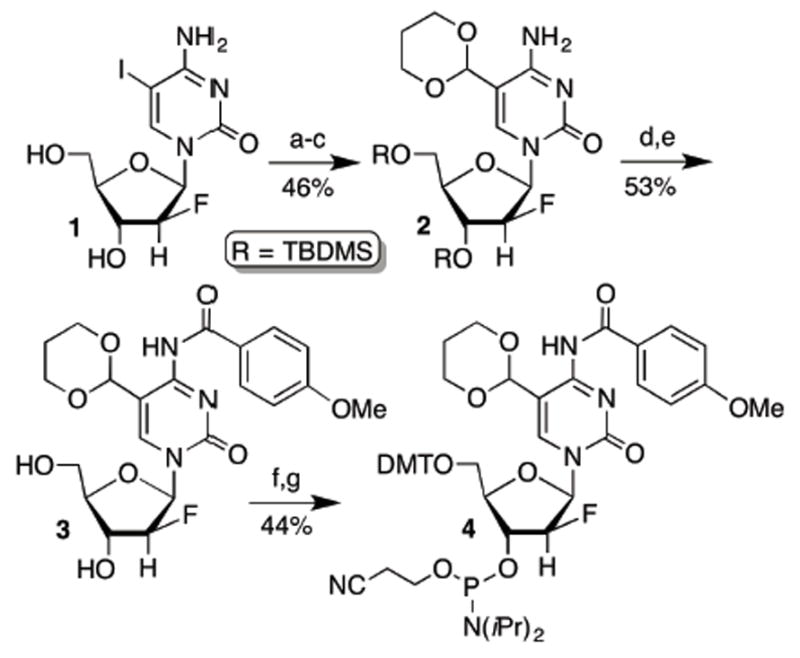
aKey: a) TBSCl b) Pd2(dba)3, PPh3, Bu3SnH, CO c) 1,3-Propanediol, triethyl orthoformate, TiCl4 d) p-Methoxybenzoyl chloride e) HF•pyridine f) DMTCl g) Phosphitylation.
Following our improved approach,21,25 we crystallized TDG82-308 bound to G·fC DNA and solved a structure at 2.20 Å resolution (Supplementary Table S1). TDG82-308 contains residues 82-308 of human TDG (410 total residues), including the catalytic domain and an N-terminal regulatory region that mediates interactions with other proteins, is subject to post-translational modifications, and is disordered.21 We recently reported a structure (1.54 Å) of TDG82-308 bound to a G·U mispair (2′-F-β-dU analog),21 revealing several key interactions that were not observed for TDG111-308 (or TDGcat), the construct that had been used for all previous DNA-bound structures of TDG. Importantly, the glycosylase activity of TDG82-308 is equivalent to that of full-length TDG, for G·fC pairs (Figure S2) and for G·T mispairs.21
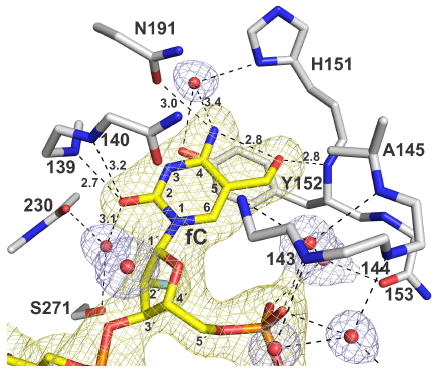
The structure here reveals key enzyme-substrate (E ·S) interactions that enable TDG to recognize and excise fC from DNA (Figure 2). Remarkably, TDG provides a relatively short (2.8 Å) hydrogen bond from the Tyr152 backbone N-H to the fC formyl oxygen, a contact that is equivalent in length to the intramolecular hydrogen bond involving the formyl oxygen and exocyclic amine of fC. This is the only polar contact provided to the fC formyl oxygen, and it is likely important for substrate binding and catalysis. Indeed, the fC-Tyr152 contact likely accounts for the ability of TDG to bind tightly to DNA containing fC but not mC or hmC.16 The methyl of Ala145 forms a nonpolar contact with the fC formyl carbon, which might help position fC to interact with the Tyr152 backbone; the A145G mutation causes a threefold loss in the maximal rate of fC excision.26
Figure 2.
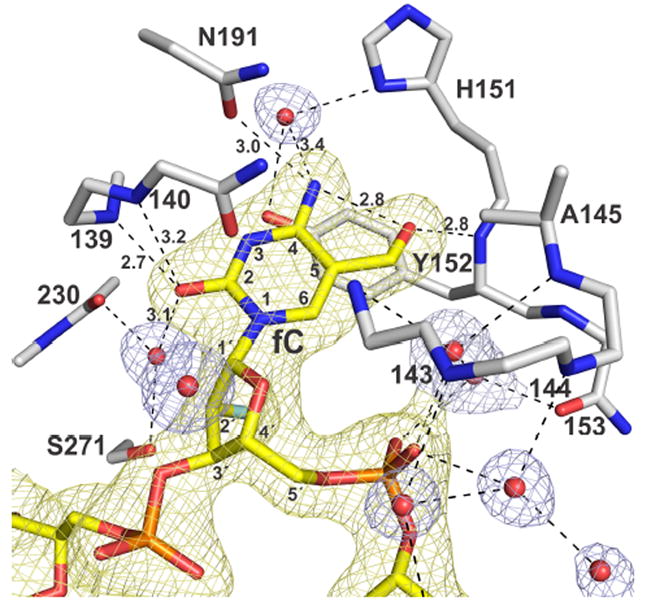
Structure of TDG82-308 bound to G·fC DNA (PDB ID: 5T2W). TDG residues are white (N blue, O red), the dfCF-containing DNA is yellow (2′-F is cyan), and water molecules are red spheres. A 2Fo-Fc electron density map, contoured at 1.0 σ, is shown for DNA (yellow) and water molecules (blue) but not the enzyme (for clarity). Dashed lines are hydrogen bonds, with interatomic distances (Å) shown for contacts to fC (all others are ≤3.5 Å).
TDG contacts other regions of fC that might not confer specificity but could mediate fC binding and/or excision. The N4H2 of fC is contacted by Asn191 and a water molecule. While Asn191 may contribute to binding fC in the E ·S complex, it is dispensable for the chemical step, as the N191A mutation does not decrease the rate of fC excision.26 No contacts are provided to N3 of fC, consistent with findings that fC excision is not acid-catalyzed.26 TDG provides two backbone N-H contacts to fC-O2, one fairly short (2.7 Å). fC-O2 is also contacted by a water molecule, bound by a backbone N-H and Ser271. Thus, the important fC contacts involve backbone groups and water molecules, rather than side chains. Notably, MUG, the bacterial homolog of TDG, also removes fC and caC,27 but most active site residues are not conserved. Our results suggest that MUG employs similar interactions (backbone, water) to excise fC.
The new structure also reveals how TDG binds the putative nucleophilic water molecule in the G·fC E ·S complex (Figure 3). Notably, the mechanism is the same as that seen for a TDG-G·U E ·S complex.15,21 The nucleophile is bound by Asn140 and the backbone oxygen of Thr197. Supporting an essential catalytic role for Asn140,14,28 the N140A mutation causes a 16,000-fold decrease in fC glycosylase activity (Figure S3), consistent with the large effect of this mutation on TDG activity for other substrates (G·U, G·FU, G·BrU).14 The Asn140 side chain is positioned by Thr197 and a backbone oxygen. The structure reveals a distance of 4.0 Å between the nucleophile and the nascent electrophile, C1′ of fdCF. The proximity and relative position of the nucleophile and electrophile for TDG (G·fC, G·U) are nearly identical to that observed in an E ·S complex for the related enzyme, UNG (in a 1.8 Å resolution structure).29
Figure 3.
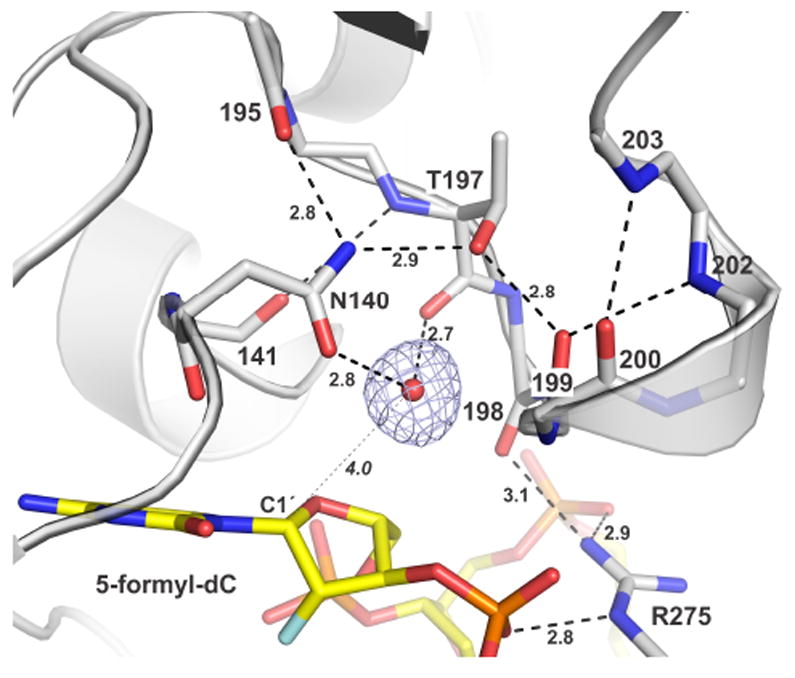
Binding of the nucleophilic water molecule. TDG is in cartoon and stick format (white), the fdC nucleotide is yellow, and the nucleophile (water) is a red sphere (other waters omitted for clarity). A 2Fo-Fc map, contoured at 1.0 σ, is shown for the nucleophile. Dashed lines are H bonds, some with interatomic distances (Å). The distance from the nucleophile to C1′ of fdCF (4.0 Å) is indicated.
We next consider implications for the mechanism of fC excision (Figure 4). All studies to date of deoxynucleotide N-glycosyl hydrolysis, using transition-state analysis, indicate a stepwise mechanism whereby N-glycosyl bond cleavage yields a short-lived intermediate, followed by nucleophile addition.30-32 These studies included UNG, which catalyzes departure of a U anion.30,33 This precedent, together with previous structural and biochemical studies of TDG, suggests that TDG could also employ a stepwise mechanism, featuring expulsion of an anionic leaving group. The evidence includes findings that (i) TDG activity (log kmax) depends on N1 acidity (pKa) of the leaving group base,34 (ii) TDG excision of fC, U, and T does not involve acid catalysis (to activate the base for departure),26,35 and (iii), TDG binds but does not activate the nucleophile (Figure 3),15,21,26,35 consistent with nucleophile addition following N-glycosyl bond cleavage. The calculated N1 acidities of U and fC are nearly identical,26 and TDG activity is similar (twofold higher) for U relative to fC (37 °C).3,35 Previous structures indicate TDG could stabilize a departing U anion via hydrogen bonds to O2 and O4.15,21 The structure here indicates a departing fC anion could be stabilized by hydrogen bonds to O2 and the formyl oxygen (Figures 2, 4).
Figure 4.
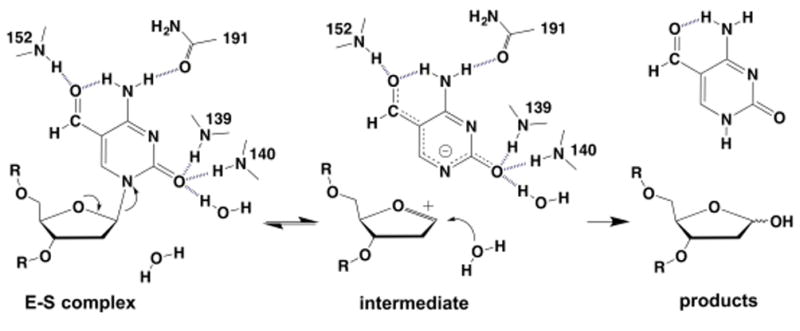
Potential stepwise mechanism for TDG excision of fC from DNA. Interactions in the E ·S complex are those observed in the structure reported here. Rupture of the N-glycosyl bond yields a putative short-lived intermediate, with an fC anion and oxacarbenium ion; addition of the nucleophile (water) gives products (fC and an abasic site).
It is of interest to compare the interactions that TDG forms with fC to those for caC and U (Figure 5). In terms of overall protein fold, the structure of TDG82-308-G·fC is closer to a 1.54 Å structure of TDG82-308-G·U (percentile based spread or p.b.s.36 of 0.20 Å, backbone atoms), than it is to a lower resolution (3.0 Å) structure of TDG111-308-G·caC (p.b.s. of 0.53 Å).16 This is due likely to differences in TDG construct, crystallization conditions, and structural resolution for the G·fC versus G·caC structures. Nevertheless, TDG interactions with fC are far more similar to those observed for caC than for U. Indeed, nearly all contacts observed in a TDG structure with caC are seen here for fC (Figure 5A), although water molecules are not observed in the caC structure. An exception is the putative contact (2.6 Å) from Asn191-Oδ1 to caC-N3,16 which might facilitate acid-catalyzed caC excision.26 This contact is not seen for fC, which is excised without need for acid-catalysis.26 Notably, the N191A mutant retains full glycosylase activity for fC excision but lacks detectable activity for caC excision.26 The structure here reveals water-mediated contacts to fC, and some of these are likely relevant to caC recognition. Water molecules might also form carboxyl-specific contacts with caC, but testing this idea must await an improved structure for caC-bound TDG.
Figure 5.
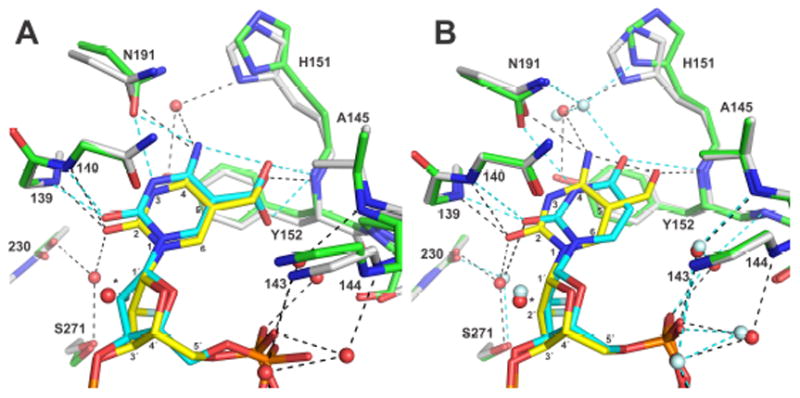
Superposition of structures to compare TDG interactions with (A) fC versus caC and (B) fC versus U (all are 2′-F-β analogs). (A) For the TDG82-308-G·fC structure, DNA is yellow, protein is white, water molecules are red spheres, and hydrogen bonds are black dashes (distances ≤3.5 Å). For TDG111-308-G·caC (PDB ID: 3UOB), DNA is cyan, protein is green, and hydrogen bonds are cyan dashes (no waters are observed). (B) For TDG82-308-G·fC the coloring is the same as in panel A. For TDG82-308-G·U (PDB ID: 5HF7), DNA is cyan, protein is green, water molecules are light cyan, and hydrogen bonds are cyan. In both panels, the putative nucleophilic water molecule(s) is marked (*).
In contrast with caC, the active-site position and contacts for U differ from that of fC (Figure 5B). While the Tyr152 backbone N-H contacts the formyl oxygen of fC, it contacts O4 of U. Although Asn191 is well aligned in the two structures, its side chain oxygen contacts the exocyclic N4 of fC compared to N3 of U. In addition, the position of the water molecule that contacts fC-N4 differs from that which contacts U-O4. On the other hand, TDG forms similar contacts to the O2 position of both fC and U. Together with previous studies, the structure here reveals how TDG employs an adaptable active site to excise a variety of modified bases, including deaminated and oxidized derivatives of mC.
Acknowledgments
Funding Sources
Supported by NIH grants GM-072711 (A.C.D.), and GM-063028 and GM-054996 (M.M.G).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Supplementary Figures, Supplementary Table S1, and experimental methods
Accession Code
A structure is in the Protein Data Bank (PDB ID: 5T2W)
The authors declare no competing financial interest.
References
- 1.Bellacosa A, Drohat AC. DNA Repair (Amst) 2015;32:33–42. doi: 10.1016/j.dnarep.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neddermann P, Jiricny J. J Biol Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 3.Maiti A, Drohat AC. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drohat AC, Coey CT. Chem Rev. 2016;116:12711–12729. doi: 10.1021/acs.chemrev.6b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. Angew Chem Int Ed Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 8.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, Macdougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A, Schar P. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 10.Scharer OD, Kawate T, Gallinari P, Jiricny J, Verdine GL. Proc Natl Acad Sci U S A. 1997;94:4878–4883. doi: 10.1073/pnas.94.10.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stivers JT, Pankiewicz KW, Watanabe KA. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 12.Barrett TE, Scharer OD, Savva R, Brown T, Jiricny J, Verdine GL, Pearl LH. EMBO J. 1999;18:6599–6609. doi: 10.1093/emboj/18.23.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chepanoske CL, Porello SL, Fujiwara T, Sugiyama H, David SS. Nucleic Acids Res. 1999;27:3197–3204. doi: 10.1093/nar/27.15.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiti A, Morgan MT, Drohat AC. J Biol Chem. 2009;284:36680–36688. doi: 10.1074/jbc.M109.062356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiti A, Noon MS, Mackerell AD, Jr, Pozharski E, Drohat AC. Proc Natl Acad Sci U S A. 2012;109:8091–8096. doi: 10.1073/pnas.1201010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Nat Chem Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroder AS, Kotljarova O, Parsa E, Iwan K, Raddaoui N, Carell T. Org Lett. 2016;18:4368–4371. doi: 10.1021/acs.orglett.6b02110. [DOI] [PubMed] [Google Scholar]
- 18.Berger I, Tereshko V, Ikeda H, Marquez V, Egli M. Nucl Acids Res. 1998;26:2473–2480. doi: 10.1093/nar/26.10.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Verdine GL. Proc Natl Acad Sci U S A. 2009;106:18497–18502. doi: 10.1073/pnas.0902908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S, Bowman BR, Ueno Y, Wang S, Verdine GL. J Am Chem Soc. 2008;130:11570–11571. doi: 10.1021/ja8025328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coey CT, Malik SS, Pidugu LS, Varney KM, Pozharski E, Drohat AC. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilds CJ, Damha MJ. Nucl Acids Res. 2000;28:3625–3635. doi: 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai Q, Lu X, Zhang L, He C. Tetrahedron. 2012;68:5145–5151. doi: 10.1016/j.tet.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroder AS, Steinbacher J, Steigenberger B, Gnerlich FA, Schiesser S, Pfaffeneder T, Carell T. Angew Chem Int Ed Engl. 2014;53:315–318. doi: 10.1002/anie.201308469. [DOI] [PubMed] [Google Scholar]
- 25.Malik SS, Coey CT, Varney KM, Pozharski E, Drohat AC. Nucleic Acids Res. 2015;43:9541–9552. doi: 10.1093/nar/gkv890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maiti A, Michelson AZ, Armwood CJ, Lee JK, Drohat AC. J Am Chem Soc. 2013;135:15813–15822. doi: 10.1021/ja406444x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morera S, Grin I, Vigouroux A, Couve S, Henriot V, Saparbaev M, Ishchenko AA. Nucleic Acids Res. 2012;40:9917–9926. doi: 10.1093/nar/gks714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardeland U, Bentele M, Jiricny J, Schar P. J Biol Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 29.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Proc Natl Acad Sci USA. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werner RM, Stivers JT. Biochemistry. 2000;39:14054–14064. doi: 10.1021/bi0018178. [DOI] [PubMed] [Google Scholar]
- 31.McCann JAB, Berti PJ. J Am Chem Soc. 2008;130:5789–5797. doi: 10.1021/ja711363s. [DOI] [PubMed] [Google Scholar]
- 32.Drohat AC, Maiti A. Org Biomol Chem. 2014;12:8367–8378. doi: 10.1039/c4ob01063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drohat AC, Stivers JT. J Am Chem Soc. 2000;122:1840–1841. [Google Scholar]
- 34.Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. J Am Chem Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maiti A, Drohat AC. DNA Repair. 2011;10:545–553. doi: 10.1016/j.dnarep.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pozharski E. Acta Crystallogr D Biol Crystallogr. 2010;66:970–978. doi: 10.1107/S0907444910027927. [DOI] [PubMed] [Google Scholar]


