Abstract
Endoplasmic reticulum aminopeptidase 1 (ERAP1) is involved in the final processing of peptide precursors to generate the N-termini of MHC class I-restricted epitopes. ERAP1 thus influences immunodominance and cytotoxic immune responses by controlling the peptide repertoire available for cell surface presentation by MHC molecules. To enable this critical role in antigen processing, ERAP1 trims peptides by a unique molecular ruler mechanism that turns on/off hydrolysis activity in a peptide-length and -sequence dependent manner. Thus unlike other aminopeptidases, ERAP1 could recognize both the N- and C-termini of peptides in order to read the substrate's length. To exemplify and validate this molecular ruler mechanism, we have carried out crystallographic studies on molecular recognition of antigenic peptide's C-terminus by ERAP1. In this report, we have determined a 2.8Å-resolution crystal structure of an intermolecular complex between the ERAP1 regulatory domain and a natural epitope's C-terminus displayed in a fusion protein. It reveals the structural details of peptide's C-termini recognition by ERAP1. ERAP1 uses specificity pockets on the regulatory domain to bind the peptide's carboxyl end and side chain of the C-terminal anchoring residue. At the same time, flexibility in length and sequence at the middle of peptides is accommodated by a kink with minimal interactions with ERAP1.
Keywords: endoplasmic reticulum aminopeptidase 1, ERAP1 regulatory domain, antigen-ERAP1 interactions, complex structure, C-terminus recognition, molecular ruler mechanism
1. Introduction
Endoplasmic reticulum aminopeptidase 1 (ERAP1) is an essential component of the immune system 1. It is involved in the final N-terminal trimming of antigenic peptides for presentation by the major histocompatibility complex class I (MHC-I) molecules in the cell-mediated immunity. ERAP1 has a high affinity for peptides with C-terminal hydrophobic residues 2. It preferentially cuts substrates of 9-16 residues in lengths using a molecular ruler mechanism that recognizes and cleaves peptide precursors of specific lengths and sequences 3. Trimming of longer precursor peptides with a variety of sequences is thought to be achieved by a concerted action of ERAP1 with another homologous enzyme named ERAP2; these two enzymes complement each other with different sequence specificities to process antigen precursors in a concerted manner 4,5. ERAP1 and ERAP2 share only 49% sequence identity but the C-terminal domain is highly conserved 6 and is unique among all aminopeptidases, suggesting a unique role of this C-terminal domain to generate appropriate size of antigens by the molecular ruler mechanism.
Even though ERAP1 is mostly retained in ER, it can also be secreted into extracellular milieu in response to inflammatory stimuli, and activates macrophages in the innate immunity 7. Further studies with murine macrophage cell line indicate that, though not taking full credits, the small peptide(s) trimmed by ERAP1 is responsible for enhanced phagocytic activity, suggesting that the enzymatic activity of secreted ERAP1 is crucial for its enhancement of phagocytosis 7. Furthermore, secretion of ERAP1 into blood circulation also plays important role in regulating blood pressure by degrading angiotensin II—a peptide that raised blood pressure 7,8.
Recent genome-wide association studies have linked mutations in ERAP1 and ERAP2 genes to various diseases, including hypertension, bacterial and viral infections and cancer. These studies reported the association of various naturally occurring single nucleotide polymorphisms (SNPs) in ERAP1 with ankylosing spondylitis (AS) and other diseases 1,9-12. Some identified SNPs, such as K528R and Q730E mutations, are situated near the substrate binding and regulatory sites and could thus affect peptide processing and trimming specificity13. Similarly studies suggested mutations of ERAP2 gene leads to an increased risk of preeclampsia with altered ERAP2 expressions 14. Inhibition of ERAP activity has also been linked to regulate innate and adaptive anti-tumor immune responses, pointing to a novel therapeutic approach against cancer 15.
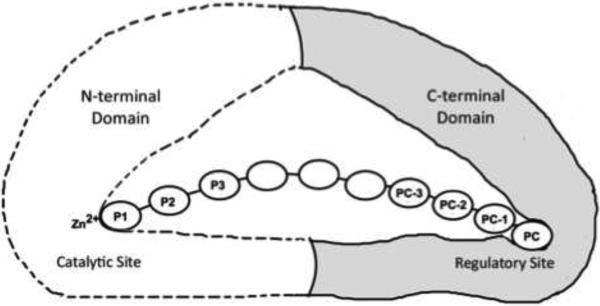
Crystal structures of apo-ERAP1 and apo-ERAP2 have provided structural basis to understand its function 16-19. Two conformations were reported for ERAP1 enzymes: an open form that allows substrate exchange, and a closed conformation with a large internal cavity. However, the actual mechanism of ERAP1 sequence and length-dependent activity is elusive since there is no structural information available for ERAP1-peptide complex. To analyze ERAP1's unique peptide recognition and substrate selection, we previously reported a 2.3Å resolution structure of the ERAP1 regulatory (ERAP1_R) domain forming a complex with a His6 peptide 20. Based on that complex structure and supporting activation assays using various His-containing peptides, we proposed a modular organization of ERAP1 for its molecular ruler mechanism 20. To function as a molecular ruler for an allosteric activation, ERAP1 contains a regulatory module that is made up of its regulatory domain unique to antigen processing enzymes ERAP1 and ERAP2. ERAP1 uses this regulatory domain to recognize and anchor the antigenic peptide's carboxyl-terminus. Since the catalytic and regulatory sites are located about 30 Å apart, a small substrate such as leucine-7-amino-4-methylcoumarin (L-AMC) or peptides shorter than 8 residues cannot concurrently reach the catalytic site and the regulatory pockets. As a result, ERAP1 stays in the lower-activity open conformation and inefficiently hydrolyses L-AMC or shorter peptides. But ERAP1 changes into the higher-activity closed conformation when a peptide longer than 9 residues can concurrently place its N-terminal and carboxyl-terminal anchors to the catalytic and regulatory sites, respectively (Figure 1). Thus the substrate-length dependent catalysis of ERAP1 is achieved by the allosteric activation of the catalytic site with a concurrent docking of the peptide's C-terminus to the ERAP1 regulatory site (Figure 1). This recognition is critical to generate a large pool of antigenic peptides with appropriate lengths, typically 8-9 residues, for MHC presentation.
Figure 1. Allosteric activation of ERAP1.
ERAP1 trims peptide precursors by a molecular ruler mechanism. To form a productive complex, the peptide substrate makes a concurrent binding of its N- and C-termini to the catalytic and the regulatory sites of ERAP1, respectively. This allosteric binding changes ERAP1 into a high-activity closed conformation, and efficiently trims one residue from the peptide's N-terminus. The peptide substrate's residue positions are numbered forward from the N-terminus as P1, P2, P3, and backward from the C-terminus as peptide carboxyl-terminal PC, the penultimate PC-1, and further towards the N-terminal direction PC-2, PC-3 etc.
Due to the lack of structural information on ERAP1-substrate complexes, many questions remain to be answered. For example, how does ERAP1 recognize and bind the anchoring residues at peptides’ C-termini, and how does it accommodate peptides of different lengths? Structural studies of ERAP1 with bound peptides containing a natural carboxyl end will address these issues, and could also provide insights into the mechanism of certain mutations implicated in the pathogenesis of diseases. To address these knowledge gaps, we have carried out crystallographic analyses of interactions between the C-terminal end of a natural antigenic epitope and the ERAP1 regulatory domain. We here report a 2.8 Å resolution structure of an epitope C-terminal end bound to the ERAP1 regulatory domain. The crystallographic analyses of this complex provide a structural basis of specific recognitions of antigenic C-terminus by ERAP1. It explains how the peptide C-terminus binds specifically to the ERAP1 regulatory domain, and also provides detailed insights into the binding specificity for antigenic anchoring residues. The complex structure also reveals how ERAP1 could accommodate and process peptides with a vast variety of sequences, and to a less extent, a range of peptide lengths.
2. Materials and methods
2.1. Protein expression and purification
Baculovirus vector carrying an insert encoding for ERAP1 regulatory domain (ERAP1_R, a.a. 529-941) with IINFEKL peptide attached to the C–terminal end was constructed with a N-terminal hexa-histidine tag according to the protocols of the manufacturer (Invitrogen). The presence of ERAP1 protein and the integrity of the purified recombinant bacmid DNA were verified by PCR and sequencing. To express the protein, the bacmid DNA was transfected into Sf9 insect cells according to the manufacturer's protocols. The protein was then expressed by adding the P3 recombinant viral stock into Sf9 insect cells with an MOI of 0.8 pfu/cell, and harvested 54 hours after infection. Protein expression was confirmed by western blot using primary antibody against the hexa-histidine tag. Cell pellets were re-suspended in 50 mM NaH2PO4, pH 8.0, 300 mM NaCl and 10 mM imidazole, and lysed by freeze-thaw cycles and sonication. The supernatant was loaded onto a Ni-NTA column and washed several times with 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl, and 10–30 mM imidazole. The protein was then eluted with 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl, and 400 mM imidazole. Glycerol was added to the eluted solution to a final concentration of 16% (v/v), and then the concentrated protein was further purified through a Superdex 200 gel filtration column (Amersham Pharmacia) by AKTA purification system with a buffer containing 10 mM Tris, pH 7.5, 10 mM NaCl. A single peak for ERAP1_R domain was collected and the protein was concentrated to 5 mg/ml for crystallization.
2.2. Crystallization, data collection and structure determination
Hanging-drop vapor diffusion technique was used for initial crystallization screening at 4°C. Crystals of the intermolecular peptide-ERAP1 complex grew after initial crystallization screening using the single-chain construct. Micro-seeding method was used to improve crystal quality. The best looking crystals were formed above a well solution containing 100 mM Tris, pH 8.5 and 16% PEG8000 at 4°C in 7 days. For data collection, the crystal was cryoprotected in solution containing 100 mM Tris-HCl buffer (pH 8.0) and 30% glycerol. X-ray data were collected using the beamline X29 at National Synchrotron Light Source (NSLS) - Brookhaven. The data were processed with the Mosflm29 and the CCP4 suite 21. The structure of ERAP1_R domain in complex with IINFEKL peptide was determined by molecular replacement method using the previously published domain structure (PDB code 3RJO) 20 as the starting model, but with the C-terminal His6 peptide removed to reduce model bias. Molecular replacement was performed with Molrep and refinements were performed with the Refmac program. 5% of the total reflection data were excluded from the beginning of refinement cycles and later used to calculate the free R-factor (Rfree) for monitoring refinement progress. Rigid body and subsequent restrained refinements and model building with COOT led to the final crystallographic Rwork/Rfree of 20.7%/26.6% at 2.8 Å resolution. The X-ray data and structure refinement statistics are shown in Table 1. All the figures were drawn using PyMOL (DeLano Scientific) and labels were added using Adobe® Photoshop.
Table 1.
Crystal information, data collection and refinement statistics
| Resolution (Å)a | 53.58-2.8 (2.95-2.80) |
| Space Group | P21 |
| Cell Dimensions: | |
| a, b, c (Å) | 64.2 66.8 66.5 |
| β (°) | 110.2 |
| No. of molecules per asymmetric unit | 1 |
| I/sigma-I | 6.6 (2.3) |
| Completeness (%) | 77.0 (81.3) |
| No. of reflections | 23,937 (3,562) |
| Unique reflections | 10,113 (1,527) |
| Rsym (%)b | 11.1 (33.5) |
| Structure refinement | |
| R work c | 0.21 |
| R free d | 0.27 |
| R.m.s. deviationse | |
| Bond-lengths (Å) | 0.013 |
| Bond-angles (°) | 1.66 |
| Ramachandran plot | |
| Most favored regions (%) | 96 |
| Additional allowed regions (%) | 4 |
| Average B-factors (Å2) | |
| Main chain | 27.54 |
| Side chain | 29.21 |
| Water | 8.19 |
Numbers in parentheses refer to the outermost (highest) resolution shell.
Rsym = Σ|I -〈I〉 /Σ(I), where I is the observed intensity and 〈I〉 is the weighted mean of the reflection intensity.
Rwork = Σ∥Fo| -Fc∥ / Σ|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
Rfree was calculated as Rwork, but with 5% of the amplitudes chosen randomly and omitted from the start of refinement.
R.m.s. deviations are deviations from ideal geometry.
Protein Data Bank accession codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with an ID code 5J5E.
3. Results and discussion
3.1. Construct design to crystallize an antigenic C-terminal end in complex with the regulatory domain of ERAP1
To study recognition of the peptide's carboxyl-terminus by the ERAP1 regulatory (ERAP1_R) domain, we took advantage of the well-studied ovalbumin-derived MHC class I-restricted epitope SIINFEKL 22. ERAP1 has been shown to efficiently trim peptide precursors containing this antigenic epitope 3,16. Co-crystallization of ERAP1 regulatory domain with antigenic peptide (SIINFEKL) has been attempted, but without success so far. Nonetheless, we have previously reported another strategy to facilitate crystallization of peptide-ERAP1_R complexes by expressing single-chain bimodular polypeptides 23. Similar to soaking protein with ligands, this approach forms a protein complex via an inter-molecular binding. At the same time, it also takes advantage of the inter-molecular interactions to facilitate crystallization. A similar approach has been applied to facilitate crystallization of DNA binding proteins by designing DNA overhangs for inter-molecular duplex hybridization 24. Furthermore, similar crystallization strategy using a single-chain construct has also been used to study protein-peptide complex in other systems 25. For this strategy of crystallization through inter-molecular complex formation, an expressing construct was generated in which the C-terminal fragment of the ovalbumin epitope, IINFEKL, is attached to the ERAP1 regulatory domain. It has been demonstrated that this C-terminal peptide fragment can interact with ERAP1 for an allosteric activation of the enzyme activity 16. Proteins of this bimodular construct have been purified to homogeneity, and diffraction quality crystals have been obtained.
Structure of the IINFEKL-ERAP1_R complex has been determined by X-ray crystallography and refined to 2.8 Å resolution, with a Rwork of 20.7% and a Rfree of 26.6%. Crystallographic data collection and refinement statistics are summarized in Table 1. The crystal has a monoclinic P21 unit cell with one molecule in the asymmetric unit. The space group and cell constants of the IINFEKL-complex crystal (P21, a = 64.2Å, b = 66.8Å, c = 66.5Å, β = 110.2°) are closely related to those of the previously reported His6-complex crystals (P21, a = 63.8Å, b = 67.3Å, c = 65.9Å, β = 110.2°) (Sui 2015). Along the screw b axis, two molecules form an inter-molecular IINFEKL-ERAP1_R complex, similar to the contacts observed previously for the His6-tagged carboxyl end 20. These intermolecular interactions are the most significant protein-protein contacts observed in these crystals. It thus appears that the ERAP1 regulatory domain has a strong binding affinity to peptide carboxyl terminus even with different sequences. This is consistent with ERAP1's critical roles of being capable of binding and processing peptide precursors with a vast variety of sequences, and to a less extent with a range of peptide lengths (see below).
3.2. Overall structure of ERAP1_R in complex with the peptide IINFEKL
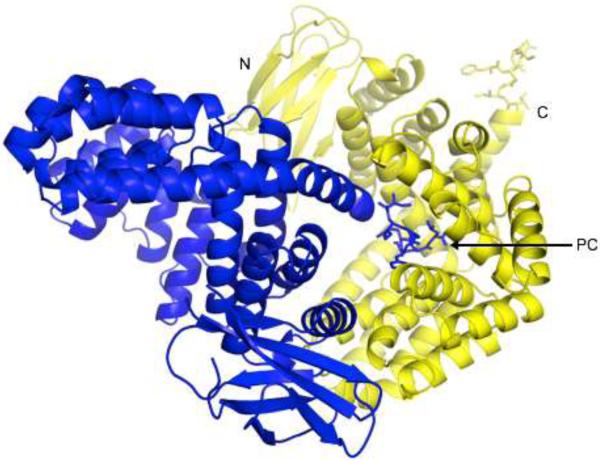
The overall structure of the ERAP1 regulatory domain in complex with the IINFEKL peptide is shown in Figure 2. The enzyme's regulatory domain is made up of two subdomains: a small beta-sandwich with two β-sheets (residues 530–611), and a larger bowl-shaped alpha-helix domain with 16 α-helices (residues 614–941) that form a concave surface (Fig. 2). This domain structure is very similar to the corresponding domain of the closed-form ERAP1 structure 16,17, with root mean square deviations (r.m.s.d.) of 1.42Å for all main-chain atoms.
Figure 2. Overall Structure of the ERAP1 regulatory domain in complex with the peptide IINFEKL.
Two ERAP1_IINFEKL molecules are shown in blue and yellow to illustrate the way they pack in the protein crystal to form the intermolecular complex. For one such complex, the IINFEKL peptide attached to the C-terminal end of the blue molecule sticks into the binding site of ERAP1 regulatory domain from the neighboring yellow molecule. For each molecule, the β-sandwich and α-helix subdomains of the ERAP1 regulatory domain are shown as a ribbon diagram, whereas the bound IINFEKL peptide is shown as a stick model. Both the N- and C-terminal ends of the yellow ERAP1 regulatory domain are labeled. Also labeled is the C-terminal end (PC) of the bound IINFEKL blue peptide.
In the crystal, the C-terminus of the IINFEKL epitope from a neighboring molecule shown in blue sticks docks into a groove on the concave surface of the alpha-helix sub-domain shown as yellow ribbon (Figure 2). Thus the crystals grew by forming a polymeric chain with a head-to-tail stacking of neighboring molecules along the 21 screw axis. This binding site is located about 29 Å away from the N-terminal catalytic zinc site in the full-length ERAP1 structures 16,17. This distance is consistent with the bidentate model of a molecular ruler that needs to bind simultaneously to both ends of peptide substrates of nine- to ten-residue in length (Figure 1). The carboxyl-end of a His6 peptide had also been shown to bind to the same site 20. This C-terminal binding site is thus likely to be the C-terminus recognition site that serves as a molecular ruler through a concurrent and allosteric binding with the N-terminal catalytic site. A 2.8 Å-resolution electron density map of the docking peptide IINFEKL is shown in Figure 3a. This inter-molecular complex structure provides us a structural foundation for analyzing specific interactions between peptide C-terminus and the ERAP1 regulatory domain.
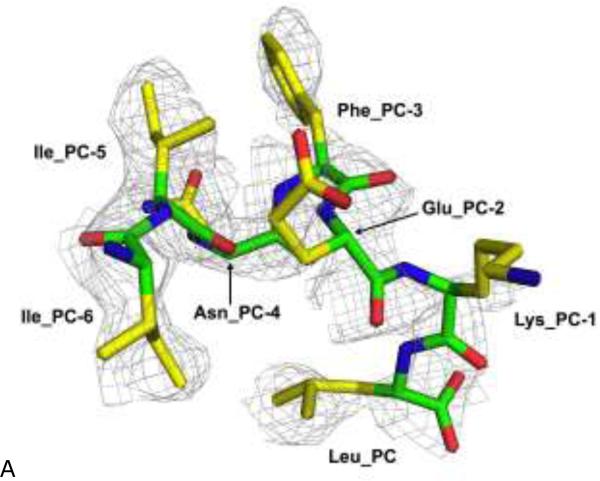
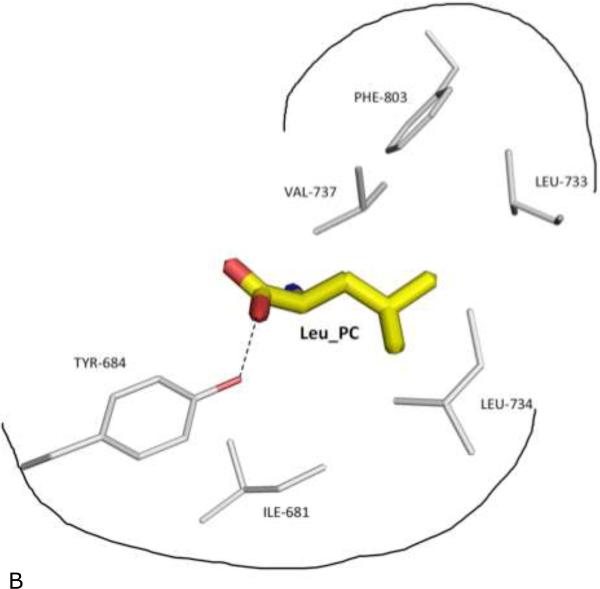
Figure 3. Recognition of peptide's C-terminus by the ERAP1 regulatory domain.
(A) The gray electron density corresponds to a refined 2Fo-Fc map at 2.8 Å resolution contoured at 1.0σ level. The bound IINFEKL peptide is shown as a thick stick model and colored by atom types: green for main-chain carbons, yellow for side-chain carbons, blue for nitrogens, and red for oxygens.
(B) Schematic outline of the specificity pocket, with surrounding side chains of ERAP1 residues shown and labeled. Also shown is Tyr-684 of ERAP1 that makes direct contact (dotted hydrogen bond) with the PC carboxylate end.
3.3. Binding site of ERAP1_R for peptide's carboxyl-end
The binding site for the carboxyl-end of IINFEKL is located between the conserved residues Tyr684 and Arg841 of the ERAP1 regulatory domain (Figure 3b). This binding cleft carries a positive surface charge, surrounded by additional basic residues Arg807 and Lys685 (Figures 3b and 4a). Using the same notation as in Figure 1, the IINFEKL peptide residue positions are denoted backward from the C-terminus as Peptide Carboxyl-terminal PC position (Leu), penultimate PC-1 position (Lys), and further towards N-terminal direction PC-2 position (Glu), PC-3 position (Phe) etc. In the IINFEKL-ERAP1_R complex, the peptide's C-terminus makes various hydrogen-bonded and hydrophobic interactions with the carboxyl-end recognition site on the ERAP1 regulatory domain (Figure 3b). In addition, the peptide utilizes its PC α-carboxylate group to form a salt bridge with the Arg807 (Figure 3b). Meanwhile, the PC carboxylate group also interacts with the conserved Tyr684 via a direct hydrogen bond. In addition, the conserved Arg841 makes a direct hydrogen-bond contact with peptide's PC-3 main chain carbonyl group. Similarly, Lys685 makes a direct hydrogen-bonding with peptide's PC-1 main chain carbonyl (Figure 3b). Interestingly, in the recently reported ERAP2-analog complex structure, the conserved residue equivalent to ERAP1's Arg841 (Arg864 in ERAP2) also makes a hydrogen bond contact with the carboxyl end of the DG025 peptide analog 26.
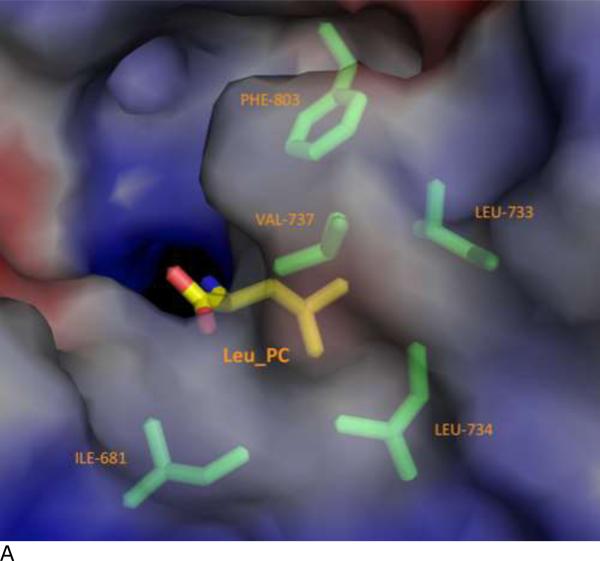
Figure 4. Specificity pocket on ERAP1 regulatory domain for binding peptide's PC anchor.
(A) The C terminus binding cleft of ERAP1 is shown as solvent accessible surfaces colored by electrostatic potentials: from red to blue for negatively charged to positively charged areas. The peptide's Leu anchor at PC position (LEU_PC) is shown as a thick stick model colored by atom types: yellow for carbons, blue for nitrogen, and red for oxygens. ERAP1 residues surrounding the LEU_PC side chain are shown as thin stick models colored by atom types: green for carbons, blue for nitrogens, and red for oxygens.
(B) Schematic outline of the specificity pocket, with surrounding side chains of ERAP1 residues shown and labeled. Also shown is Tyr-684 of ERAP1 that makes direct contact (dotted hydrogen bond) with the PC carboxylate end.
3.4. Additional interactions between the peptide's side chains and the ERAP1_R
At the PC position of IINFEKL, the leucine side chain tucks into a hydrophobic pocket surrounded by Ile681, Leu733, Leu734, Val737, and Phe803 (Fig. 3b and Figure 4). In contrast, side chains at other positions point into the bulk solvent inside the large internal binding cavity, with minimal contact with ERAP1 (Figure 3a). This would explain how ERAP1 could accommodate a vast array of peptide sequences for processing. One close contact from peptide other than the PC anchor is the side chain of the PC-1 lysine that projects into the bulk solvent and forms a hydrogen-bond with the side chain of Gln881 of ERAP1. However, the closest contacts with the middle of the peptide come between the peptide PC-4 Asn and Gln730 of ERAP1: Gln730 of ERAP1 makes hydrogen-bond contacts with the side chain and main chain atoms of PC-4 Asn. Interestingly, a mutation at Gln730 has been reported to be associated with disease 27. It appears that the disease-related mutation Q730E would interfere with the natural hydrogen-bond interaction and thus affects processing of antigens by ERAP1.
3.5. Specificity pocket of ERAP1_R for peptide's C-terminal (PC) anchoring side chain
It has long been recognized that MHC-I restricted antigens has canonical sequence motifs with dominant residues at certain positions 28. Apparently selective bindings of peptides by MHC molecules and antigen processing enzymes contribute to the sequence bias of presented antigens. These so-called anchor residues have been shown to lock into complementary specificity pockets located inside the binding cleft of MHC-I molecules 29. Similarly specificity pockets are expected in the ERAP1 binding cleft to interact with peptides’ anchoring site chains. Indeed, ERAP1 prefers a hydrophobic anchoring residue at the PC end 2,3, and leucine is one of the dominant hydrophobic residues at the peptide's carboxyl-end (PC position). In the complex structure with IINFEKL peptide, the side chain of the PC Leu fits in a hydrophobic pocket at conserved residues Leu734 and Phe803, surrounded by additional non-conserved hydrophobic residues Ile681, Leu733, and Val737 (Fig. 3b and Figure 4). Thus this PC specificity pocket in ERAP1 plays critical role to accommodate the substrate C-terminal anchoring residue for the molecular ruler mechanism. The hydrophobic pocket of ERAP1 regulatory domain also provides some insights into the fact that secreted ERAP1 could trim angiotensin II peptide (DRVYIHPF-OH) that contains a hydrophobic carboxyl residue. It is worth noting that this PC anchoring pocket of ERAP1 is comprised of polymorphic residues Ile681, Leu 733, and Val737, for which the corresponding residues in ERAP2 are Glu704, Lys756, and Asp760, respectively. It is thus conceivable that ERAP2 would complement ERAP1 by processing antigen precursors with a hydrophilic anchoring residue at the PC position. Indeed, peptide with a Lys anchor at the PC position has been shown to be a good substrate for ERAP2 26.
The IINFEKL complex structure provides the first structural details of peptide's C-terminus recognition by ERAP1. In a recent report of complex structures of the related ERAP2 enzyme, only peptide's N-terminus was found to bind to the catalytic site of the aminopeptidase. No electron density was observed for the C-terminal end of peptide analogs in those ERAP2 complexes 26. This could reflect mechanistic differences between ERAP1 and ERAP2. However, it is also possible that the lack of C-terminus binding in the ERAP2 complexes is due to the unnatural peptide analogs used. One peptide analog in those structures has an unnatural C-terminal end: an amide group rather than a carboxylate end. Thus that ERAP2-analog structures may not reflect the binding mode of a natural substrate peptide which allows allosteric bindings to both the N-catalytic and C-regulatory sites for the substrate catalysis (Figure 1). The other analog used in the same study is a 9-residue peptide which has been shown to be a trimming product rather than a cleavable substrate of ERAP1 4. As Mpakali et al. pointed out, the peptide's N-terminal end of the product peptide was positioned at the catalytic site in a canonical orientation as would be expected for catalysis, yet a cleavage did not occur. This indicates that the ERAP2-product complex was trapped at a nonproductive conformation. One plausible interpretation is that the length of that product peptide is too short for a concurrent binding of the peptide's N- and C-terminal ends to the catalytic and a regulatory site of ERAP2, respectively. This would explain why no electron density for peptide's C-terminus was observed in the ERAP2-product complex 26.
As discussed above, to form a productive complex in the ERAP1 or ERAP2 molecular ruler, the peptide substrates need to have an appropriate length for concurrent bindings to both N-catalytic and C-regulatory sites (Figure 1). Natural antigenic precursors also need to have regular amino- and carboxyl-ends. Thus to study a productive complex structure of ERAP1 or ERAP2, the challenge in the field is to prepare a complex with substrate peptides containing natural N- and C-termini, with appropriate anchor residues at both ends, and with more than 8-9 residues in length. A recent study described a slower editing activity of MHC-I bound peptides by ERAP1-ERAP2 heterodimers 30. Nonetheless, that MHC-I assisted activity is different from the molecular ruler mechanism studied in this report since it has been well demonstrated that purified ERAP1 and/or ERAP2 enzyme is able to efficiently process antigenic precursors in the absence of MHC molecules 2,3,16,20.
3.6. Comparing the IINFEKL epitope binding to the His6-peptide binding
Comparing to the IINFEKL complex with previously reported His6 complex, the ERAP1_R domain in both complexes has an essentially identical structure. The rmsd between these two domain structures is 0.39Å for all main-chain atoms of 406 residues. Binding mode of IINFEKL is also similar to that observed for the His6 peptide 20. Both peptides utilize their α-carboxylate group to form salt bridges with the Arg807 (Figure 3b). In addition, the conserved Arg-841 also makes a direct hydrogen-bond contact with IINFEKL peptide's PC-3 main-chain carbonyl group (Figure 3b). In the His6 complex, Arg-841 also makes a similar hydrogen-bond contact with peptide PC-2 main-chain carbonyl (Figure 5). At the PC position of IINFEKL, leucine side chain tucks into the specificity pocket surrounded by Ile681, Leu733, Leu734, Val737, and Phe803 (Fig. 3b and Figure 4). This binding pocket has also been shown to be able to accommodate the PC histidine in the His6 complex 20.
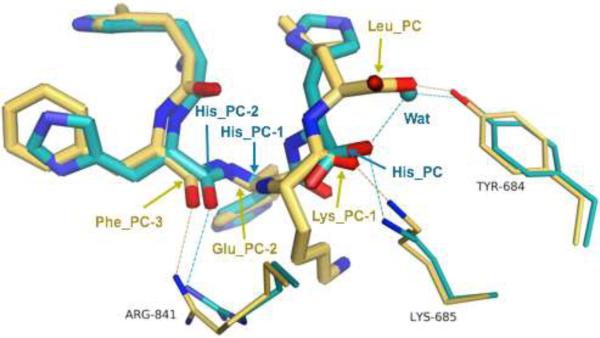
Figure 5. Superimposition of IINFEKL and His6-peptide structures bound by the ERAP1 regulatory domain.
The two complex structures with IINFEKL peptide and His6-peptide are superimposed by all the main-chain atoms of the ERAP1 regulatory domains (408 residues). Bound peptides are shown as thick stick models, while side chains of selected ERAP1 residues are shown as skinny stick models. Atoms are colored by atom types: gold for carbons of IINFEKL complex, cyan for carbons of His6 complex, blue for nitrogens, red for oxygens, and cyan sphere for a bound water molecule in the His6 complex. From left to right, the last four residues of the IINFEKL peptide are labeled in gold (PC-3 to PC), whereas to fill the same space in the His6 complex only three residues of the peptide are labeled in cyan (PC-2 to PC) plus a water molecule (Wat). Note the different locations and orientations of the carboxyl ends (PC) for these two peptides. Side chains of ERAP1 are labeled in black. Dotted lines represent hydrogen-bond interactions.
However, when backbone of the bound IINFEKL peptide is compared to that of the His6 peptide 20, substantial conformational differences are found at the peptide's last few residues: especially from the PC-2 to the carboxyl-terminal PC positions. At the very end (PC), the carboxylate group has shifted by more than 3 Å between the two bound peptides (Figure 5). The main-chain traces at the Cα of the PC-1 residues also differ by about 3.7Å. Part of this shift is to accommodate the length-difference: IINFEKL peptide is one-residue longer than the His6-peptide. As a result, the carboxylate end of IINFEKL forms a direct hydrogen-bond contact with the conserved residue Tyr684, in contrast to a water-mediated interaction in the His6 complex 20. Furthermore Lys685 makes a direct hydrogen-bonding with the PC-1 main-chain carbonyl of IINFEKL, whereas it makes a direct hydrogen-bond with the PC carboxyl group of the His6-peptide (Figure 5). Nonetheless, for both peptides, side chain of the peptide's PC anchoring residue tucks into the same specific pocket discussed above to accommodate the PC anchoring side chain of antigenic substrates.
3.7. Flexibility (promiscuity) to accommodate peptides with various lengths and sequences
To process a large pool of antigen precursors, ERAP1 would need to bind common features of all substrates with various sequences. This has been proposed to be achieved by binding peptides’ N- and C- termini into two discrete sites: the N-terminal catalytic sites and the C-terminal recognition sites, respectively (Figure 1) 20. Meanwhile, to accommodate a range of peptide lengths of about 9 to 16 residues with various sequences, flexibility could be accomplished by kinking or bulging extra residues at the middle of peptides in the large internal cavity of the closed ERAP1 conformation 16,17. In support of this binding mechanism as a molecular ruler, the C-termini of IINFEKL and His6-tag dock into the same surface cleft at the ERAP1 regulatory domain. Both peptides utilize the same specificity pocket to bind their PC anchoring side chains (Figure 5). This is particularly significant since IINFEKL is one residue longer than His6-peptide, indicating that anchoring of the PC side chains to the Leu734/Phe803 specificity pocket is independent of peptide lengths. Instead it is likely to be the result of a simultaneous recognition by ERAP1 of peptides’ carboxylate group and the PC anchoring side chains. In fact, packing of PC side-chains into the specificity pocket appears to be the main determinant of C-terminus binding, with some wiggling room for the carboxylate-group through direct contacts or water mediated interactions (Figure 5). Flexibility to accommodate various peptide lengths is further achieved by taking a kink and/or turn at the middle of peptide. The main-chain trace of IINFEKL takes a bit bulge at the PC-1 and PC-2 positions when compared the conformation of the His6-peptide (Figure 5). In addition, the amino group of the PC-2 Glu makes a hydrogen bond contact with the carbonyl group of the PC-5 Ile to form a 310 turn in the IINFEKL structure. On the other hand, there are minimal interactions between ERAP1 and the middle part of peptides. Such promiscuity is necessary for ERAP1 to be able to process a wide variety of peptides for generating a wide spectrum of epitopes that are different not only in lengths but also in sequences.
4. Conclusions
The IINFEKL-ERAP1 complex structure reported here allows the first crystallographic analyses of specific ERAP1 recognition of an antigenic peptide's C-terminus. To act as the gatekeeper with a molecular ruler for generating peptides with an appropriate length for MHC presentation, ERAP1 is equipped with three major capacities: 1) a catalytic site to bind and trim peptide from its N-terminus, 2) a recognition site to bind peptide's C-terminus for a peptide length-dependent allosteric activation of substrate N-terminal trimming, 3) a large internal substrate cavity to accommodate and process a large pool of peptide precursors with various lengths and sequences. This peptide-ERAP1 complex structure allows a direct visualization of antigenic peptide's C-terminus binding to the ERAP1 regulatory domain.
To accommodate a variety of peptide sequence, ERAP1 uses critical residues, mainly conserved residues, for recognizing common features of peptides. For examples, residues Tyr684 and Arg841 interact with carboxylate end and main chain oxygen atoms of both the IINFEKL and His6 peptides. Interestingly these two residues are conserved in ERAP1 and ERPA2, and could thus serve as constant contact sites for antigenic precursors. In addition, Arg807 makes hydrogen-bond contacts with the carboxylate group. Also as shown in Figure 5, Lys685 makes additional hydrogen-bond contacts with peptide main chain, either with the PC carboxylate of the IINFEKL peptide, or with the PC-1 carbonyl of the His6 peptide. These interactions with constant features of peptides apparently allows ERAP1 to process antigen precursors in a pretty much sequence-independent manner, except the anchor side chains at the N- and C-termini (P1 and PC positions in Figure 1). Additional flexibility and promiscuity to accommodate antigenic precursors of various lengths and sequences comes from the bulky internal substrate cavity of ERAP1 that has essentially no direct contacts with the middle part of the peptide substrates.
Highlights.
Structure of ERAP1 regulatory domain in complex with antigenic peptide is reported.
The structure reveals binding details of ERAP1 with antigenic peptide's C-terminus.
The structure reveals the role of ERAP1 in binding peptide with various lengths.
Acknowledgments
We thank Dr. Howard Robinson for assistance on data collection, Suchita Pande for helpful discussions. This work was supported by Grants AI068831 and AI078134 from the NIH.
Abbreviations
- ER
endoplasmic reticulum
- ERAP1
endoplasmic reticulum aminopeptidase 1
- ERAP1_R
ERAP1 regulatory domain
- MHC-I
major histocompatibility complex class I
- TAP
transporter associated with antigen processing
- rmsd
root mean square deviation/displacement
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
L.S. completed the structure determination and analyses.
A.G. collected the x-ray data and carried out the initial crystallographic study.
All contributed to the writing of the manuscript.
References
- 1.Fruci D, Romania P, D'Alicandro V, Locatelli F. Endoplasmic reticulum aminopeptidase 1 function and its pathogenic role in regulating innate and adaptive immunity in cancer and major histocompatibility complex class I- associated autoimmune diseases. Tissue antigens. 2014;84(2):177–186. doi: 10.1111/tan.12410. [DOI] [PubMed] [Google Scholar]
- 2.Evnouchidou I, Momburg F, Papakyriakou A, Chroni A, Leondiadis L, Chang SC, Stratikos E. The internal sequence of the peptide-substrate determines its N-terminus trimming by ERAP1. PLoS One. 2008;3(11):e3658. doi: 10.1371/journal.pone.0003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(47):17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, van Endert PM. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nature immunology. 2005;6(7):689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 5.Lorente E, Barriga A, Johnstone C, Mir C, Jiménez M, López D. Concerted in vitro trimming of viral HLA-B27-restricted ligands by human ERAP1 and ERAP2 aminopeptidases. PloS one. 2013;8(11):e79596. doi: 10.1371/journal.pone.0079596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanioka T, Hattori A, Masuda S, Nomura Y, Nakayama H, Mizutani S, Tsujimoto M. Human Leukocyte-derived Arginine Aminopeptidase The Third member of the oxytocinase subfamily of aminopeptidases. Journal of Biological Chemistry. 2003;278(34):32275–32283. doi: 10.1074/jbc.M305076200. [DOI] [PubMed] [Google Scholar]
- 7.Goto Y, Ogawa K, Hattori A, Tsujimoto M. Secretion of endoplasmic reticulum aminopeptidase 1 is involved in the activation of macrophages induced by lipopolysaccharide and interferon-γ. Journal of Biological Chemistry. 2011;286(24):21906–21914. doi: 10.1074/jbc.M111.239111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto N, Nakayama J, Yamakawa-Kobayashi K, Hamaguchi H, Miyazaki R, Arinami T. Identification of 33 polymorphisms in the adipocyte-derived leucine aminopeptidase (ALAP) gene and possible association with hypertension. Human mutation. 2002;19(3):251–257. doi: 10.1002/humu.10047. [DOI] [PubMed] [Google Scholar]
- 9.Alvarez-Navarro C, de Castro JAL. ERAP1 structure, function and pathogenetic role in ankylosing spondylitis and other MHC-associated diseases. Molecular immunology. 2014;57(1):12–21. doi: 10.1016/j.molimm.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Cortes A, Pulit SL, Leo PJ, Pointon JJ, Robinson PC, Weisman MH, Chiocchia G. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nature communications. 2015;6 doi: 10.1038/ncomms8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenna TJ, Robinson PC, Haroon N. Endoplasmic reticulum aminopeptidases in the pathogenesis of ankylosing spondylitis. Rheumatology. 2015;54(9):1549–1556. doi: 10.1093/rheumatology/kev218. [DOI] [PubMed] [Google Scholar]
- 12.International Genetics of Ankylosing Spondylitis Consortium Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nature genetics. 2013;45(7):730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamogiannos A, Koumantou D, Papakyriakou A, Stratikos E. Effects of polymorphic variation on the mechanism of Endoplasmic Reticulum Aminopeptidase 1. Molecular immunology. 2015;67(2):426–435. doi: 10.1016/j.molimm.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Hill LD, Hilliard DD, York TP, Srinivas S, Kusanovic JP, Gomez R, Strauss JF. Fetal ERAP2 variation is associated with preeclampsia in African Americans in a case-control study. BMC medical genetics. 2011;12(1):1. doi: 10.1186/1471-2350-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fruci D, Locatelli F, Cifaldi L. ERAAP modulation: A possible novel strategy for cancer immunotherapy?. Oncoimmunology. 2012;1(1):81–82. doi: 10.4161/onci.1.1.17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen TT, Chang SC, Evnouchidou I, York IA, Zikos C, Rock KL, Stern LJ. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nature structural & molecular biology. 2011;18(5):604–613. doi: 10.1038/nsmb.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kochan G, Krojer T, Harvey D, Fischer R, Chen L, Vollmar M, Wordsworth P. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proceedings of the National Academy of Sciences. 2011;108(19):7745–7750. doi: 10.1073/pnas.1101262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birtley JR, Saridakis E, Stratikos E, Mavridis IM. The crystal structure of human endoplasmic reticulum aminopeptidase 2 reveals the atomic basis for distinct roles in antigen processing. Biochemistry. 2011;51(1):286–295. doi: 10.1021/bi201230p. [DOI] [PubMed] [Google Scholar]
- 19.Stratikos E, Stern LJ. Antigenic peptide trimming by ER aminopeptidases—Insights from structural studies. Molecular immunology. 2013;55(3):212–219. doi: 10.1016/j.molimm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandhi A, Lakshminarasimhan D, Sun Y, Guo HC. Structural insights into the molecular ruler mechanism of the endoplasmic reticulum aminopeptidase ERAP1. Scientific reports. 2011;1:186. doi: 10.1038/srep00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collaborative CP. The CCP4 suite: programs for protein crystallography. Acta crystallographica. Section D, Biological crystallography. 1994;50(Pt 5):760. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Rötzschke O, Falk K, Stevanovic S, Jung G, Walden P, Rammensee HG. Exact prediction of a natural T cell epitope. European journal of immunology. 1991;21(11):2891–2894. doi: 10.1002/eji.1830211136. [DOI] [PubMed] [Google Scholar]
- 23.Sui L, Gandhi A, Guo HC. Single-Chain Expression and Crystallization of an Antigenic C-Terminus in Complex with the Regulatory Domain of ER Aminopeptidase 1. Crystal Structure Theory and Applications. 2015;4(04):47. [Google Scholar]
- 24.Xu QS, Kucera RB, Roberts RJ, Guo HC. An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site. Structure. 2004;12(9):1741–1747. doi: 10.1016/j.str.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Karthikeyan S, Leung T, Ladias JAA. Structural Basis of the Na+/H+ Exchanger Regulatory Factor PDZ1 Interaction with the Carboxyl-terminal Region of the Cystic Fibrosis Transmembrane Conductance Regulator. Journal of Biological Chemistry. 2001;276(23):19683–19686. doi: 10.1074/jbc.C100154200. [DOI] [PubMed] [Google Scholar]
- 26.Mpakali A, Giastas P, Mathioudakis N, Mavridis IM, Saridakis E, Stratikos E. Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (ER) Aminopeptidase 2. Journal of Biological Chemistry. 2015;290(43):26021–26032. doi: 10.1074/jbc.M115.685909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey D, Pointon JJ, Evans DM, Karaderi T, Farrar C, Appleton LH, Wordsworth BP. Investigating the genetic association between ERAP1 and ankylosing spondylitis. Human molecular genetics. 2009;18(21):4204–4212. doi: 10.1093/hmg/ddp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rammensee HG, Bachmann J, Emmerich NPN, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3-4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 29.Guo HC, Madden DR, Silver ML, Jardetzky TS, Gorga JC, Strominger JL, Wiley DC. Comparison of the P2 specificity pocket in three human histocompatibility antigens: HLA-A* 6801, HLA-A* 0201, and HLA-B* 2705. Proceedings of the National Academy of Sciences. 1993;90(17):8053–8057. doi: 10.1073/pnas.90.17.8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Li L, Weimershaus M, Evnouchidou I, van Endert P, Bouvier M. ERAP1-ERAP2 dimers trim MHC I-bound precursor peptides; implications for understanding peptide editing. Sci. Rep. 2016;6:28902. doi: 10.1038/srep28902. [DOI] [PMC free article] [PubMed] [Google Scholar]