Abstract
The cochaperone GrpE functions as a nucleotide exchange factor to promote dissociation of adenosine 5′-diphosphate (ADP) from the nucleotide-binding cleft of DnaK. GrpE and the DnaJ cochaperone act in concert to control the flux of unfolded polypeptides into and out of the substrate-binding domain of DnaK by regulating the nucleotide-bound state of DnaK. DnaJ stimulates nucleotide hydrolysis, and GrpE promotes the exchange of ADP for adenosine triphosphate (ATP) and also augments peptide release from the DnaK substrate-binding domain in an ATP-independent manner. The eukaryotic cytosol does not contain GrpE per se because GrpE-like function is provided by the BAG1 protein, which acts as a nucleotide exchange factor for cytosolic Hsp70s. GrpE, which plays a prominent role in mitochondria, chloroplasts, and bacterial cytoplasms, is a fascinating molecule with an unusual quaternary structure. The long α-helices of GrpE have been hypothesized to act as a thermosensor and to be involved in the decrease in GrpE-dependent nucleotide exchange that is observed in vitro at temperatures relevant to heat shock. This review describes the molecular biology of GrpE and focuses on the structural and kinetic aspects of nucleotide exchange, peptide release, and the thermosensor hypothesis.
ANCIENT HISTORY
GrpE (for GroP-like gene E) was identified in a genetic screen for mutants that failed to propagate the bacteriophage λ in Escherichia coli (Saito and Uchida 1977). GrpE is constitutively required for E coli growth at all temperatures because it was found that grpE deletions could only survive in certain mutant dnaK backgrounds in the presence of unidentified extragenic suppressors (Ang and Georgopoulos 1989). GrpE expression is under the regulation of the σ70 and σ32 promoters, such that expression of GrpE messenger ribonucleic acid is rapidly and transiently induced in response to elevated temperatures, making it a bona fide heat shock protein (Lipinska et al 1988). The observation that adenosine triphosphate (ATP) appeared to disrupt a complex of GrpE and DnaK suggested a role for GrpE in nucleotide exchange (Zylicz et al 1987). This hypothesis was refined into a model that nucleotide exchange by GrpE promoted ATP turnover by DnaK (Liberek et al 1991). There is no evidence that GrpE possesses peptide-binding activity.
Mge1p, THE YEAST MITOCHONDRIAL GrpE
In the early 1990s, several laboratories undertook the search for cochaperones of the yeast Hsp70s. MGE1 was originally cloned based on its sequence homology to GrpE and was shown to be essential for viability in yeast (Bolliger et al 1994; Ikeda et al 1994; Laloraya et al 1994). Mge1p is localized in the mitochondria (Bolliger et al 1994; Laloraya et al 1994), but unlike GrpE, Mge1p is not heat inducible (Ikeda et al 1994). Mge1p and Ssc1p (the mitochondrial matrix Hsp70), like their counterparts GrpE and DnaK, form a complex that is dissociable by ATP (Bolliger et al 1994; Voos et al 1994), and therefore it was proposed that Mge1p acts as a nucleotide exchange factor for Ssc1p (Nakai et al 1994; Laloraya et al 1995; Westermann et al 1995). Mge1p-dependent nucleotide release from Ssc1p was demonstrated in vitro by Miao et al (1997).
Bacterial and eukaryotic GrpEs have relatively low sequence homology compared with the homology found in other chaperone families, such as the Hsp70s. For instance, there is only 34% amino acid identity between GrpE and Mge1p (Laloraya et al 1994) and 21% identity between GrpE and rat mitochondrial mt-GrpE#1 compared with 51% amino acid identity between DnaK and rat mt-Hsp70 (Naylor et al 1996). The eukaryotic homologs of GrpE are found in yeast as well as in mammalian mitochondria and chloroplasts (Naylor et al 1996; Schroda et al 2001) but have not been found in the eukaryotic endoplasmic reticulum or cytosol. However, a functional analog of GrpE, BAG1, acts as a nucleotide exchange factor in eukaryotic cytoplasm and accelerates the release of adenosine 5′-diphosphate (ADP) from cytosolic Hsp70s (see accompanying review by Höhfeld).
MUTATIONAL ANALYSES OF BACTERIAL AND ORGANELLAR GrpEs
Georgopoulus et al (1994) undertook a substantial search for GrpE mutants that block phage λ growth in E coli, which led to the identification of several new mutants. These mutants blocked λ replication at nonpermissive temperatures and were temperature sensitive for E coli growth above 42°C (Georgopoulus et al 1994; Wu et al 1994). Most of the mutations were in conserved regions of GrpE although none was implicated in direct contacts to DnaK (Harrison et al 1997). The best-characterized mutation in grpE (grpE280) codes for the replacement of glycine 122 with aspartic acid within the 4-helix bundle. Mutations in λ that allow phage growth in the grpE280 mutant map to the λ P gene, which functions in localizing the E coli helicase to the λ replication complex. Classical genetic interpretation of these results was that GrpE influenced the action of the λ phage replication protein P. Published reports described the protein product of grpE280 (GrpE-G122D) as a mutation unable to support wild-type levels of λdv phage replication in vitro (Wu et al 1996), and GrpE-G122D also exhibited reduced coimmunoprecipitation efficiency with DnaK (Johnson et al 1989). However, GrpE-G122D was a capable nucleotide exchange factor in an in vitro luciferase refolding assay and also cocrystallized with the DnaK adenosine triphosphatase (ATPase) domain (Harrison et al 1997). The resulting macromolecular cocrystal structure provided clear evidence that G122D was not located at the GrpE-DnaK interface (Harrison et al 1997). Although GrpE, DnaK, and DnaJ are now better characterized functionally and structurally, the basis for some genetic and biochemical results observed for the grpE280 mutant and its gene product, GrpE-G122D, remains obscure and intriguing. Perhaps, the ability to function in a luciferase assay and interact with DnaK does not imply full activity with respect to the DnaK molecular chaperone cycle and λ phage replication.
Hydroxylamine mutagenesis of the yeast MGE1 yielded several conditional mutants (Laloraya et al 1995; Westermann et al 1995). Two of these mutants are especially interesting: the mge1–100 mutation, when mapped by sequence homology onto the crystal structure of GrpE, shows an arginine to lysine change at a position, which is important for an intramolecular contact between the β-domain and the 4-helix bundle in GrpE. This conservative substitution, which is at the position equivalent to GrpE Arg 186, no longer allows growth of yeast at 37°C (Laloraya et al 1995). Another conditional lethal mutation in mge1–3, Ala 134 to valine, was predicted by analogy of Mge1p to GrpE-DnaK to be located at the Mge1p-Ssc1p interface.
WHAT STATE OF DnaK IS THE PREFERRED SUBSTRATE FOR GrpE?
Structural studies of the bovine brain Hsc70 ATPase domain show little conformational change between ATP- and ADP-bound nucleotide states. However, solution small-angle X-ray scattering studies have shown a conformational difference between ATP- and ADP-bound states in the 60-kDa fragment of Hsc70, which encompasses the ATPase domain as well as most of the substrate-binding domain (Wilbanks et al 1995; Johnson and McKay 1999). Because release of inorganic phosphate (Pi) is relatively fast (Dekker and Pfanner 1997), it is most likely that the substrate for GrpE is DnaK bound to ADP and to polypeptide substrate, without bound inorganic phosphate. DnaJ is most likely not part of the complex, but in the absence of a complete structure of intact DnaK (or Hsp70) with DnaJ (or Hsp40), any overlap, or lack thereof, of GrpE and DnaJ interacting surfaces on DnaK cannot be determined. GrpE recognizes the ADP-bound state of DnaK, yet binds to and stabilizes an “open” conformation of the nucleotide-binding cleft of DnaK that is not compatible with high-affinity nucleotide binding (Harrison et al 1997). Surface plasmon resonance studies showed that ATP caused instantaneous dissociation of DnaK from GrpE, with a rate too rapid to be determined at 25°C (Harrison et al 1997). GrpE stimulates the dissociation of a fluorescently labeled ATP analog that is commonly used for DnaK kinetic studies, suggesting that GrpE may not discriminate against DnaK·ATP vs DnaK·ADP as a substrate (Brehmer et al 2001). Similarly, Craig and coworkers used a single-turnover assay to show that Mge1p stimulates dissociation of ATP from Ssc1p (Miao et al 1997).
TERTIARY AND QUATERNARY STRUCTURE OF GrpE AND GrpE-DnaKATPase
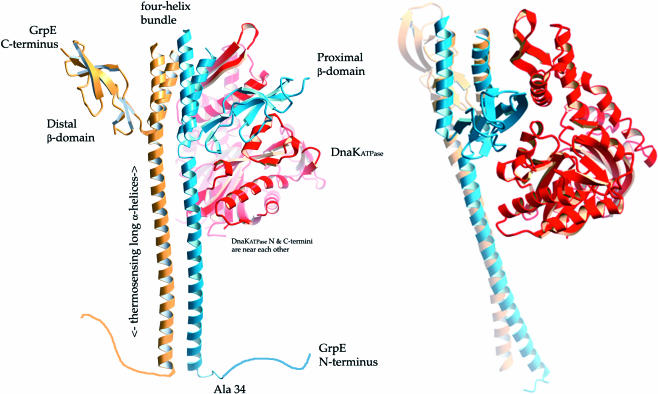
Before the solution of the cocrystal structure of N-terminally truncated GrpE and the ATPase domain of DnaK in 1997, there were no high-resolution structural data on GrpE. Hydrodynamic experiments revealed that GrpE and DnaK formed a complex with a 2:1 stoichiometry (Schonfeld et al 1995), and it had been predicted that GrpE possessed a coiled-coil domain (Wu et al 1994). Dimeric GrpE has a striking and asymmetric appearance (see Fig 1). A monomer of GrpE consists of an unstructured region not present in the crystal structure (Mehl et al 2001; Gelinas et al 2002), a long α-helix followed by a short α-helix, and then a compact β-sheet domain at the C terminus. In the dimer, the 2 long α-helices appear to be almost directly parallel and do not bury side chains in the same fashion as coiled-coils (neither “knobs into holes” nor “ridges into grooves”). The packing of the side chains is slightly staggered as a result of 2 direct repeats of a heptad-hendecad (7-11-7-11) spacing of buried hydrophobic residues, which practically abolishes super-helical twist (Lupas 1996; Offer et al 2002). The long and short α-helices form a 4-helix bundle from which the compact β-sheet domains protrude like the arms of a cactus. In the cocrystal, GrpE is an asymmetric dimer that is bent toward the DnaK ATPase domain as a result of the break in the long α-helix in the GrpE monomer proximal to DnaK. The nucleotide-binding cleft of the DnaK ATPase domain is in an open conformation because binding of GrpE positions a β-sheet domain at the mouth of the nucleotide-binding cleft. This change from the closed, nucleotide-bound conformation occurs by a simple rigid body motion of a single domain of DnaK (domain IIB), which moves outward by 14°. Domain IIB contributes 3 side chains to binding of the adenine and ribose rings of the nucleotide, and these contacts are disrupted by the GrpE-induced conformational change, resulting in nucleotide dissociation. The magnitude of the GrpE-induced acceleration of nucleotide release is similar to that of other nucleotide exchange factors, but there is 1 clear mechanistic difference. Classical small guanosine triphosphatase (GTPase)–associated guanine nucleotide exchange factors (GEFs) enhance nucleotide dissociation by directly disrupting the ionic interactions used by the GTPase to stabilize the phosphates of the nucleotide (Renault et al 2001). GrpE, however, does not directly contact the nucleotide-binding pocket of DnaK; the closest approach that any GrpE side chain makes to the nucleotide-binding pocket of DnaK is about 15Å.
Fig 1.
Ribbon drawings of dimeric GrpE bound to the adenosine triphosphatase (ATPase) domain of DnaK (PDB: 1DKG). GrpE monomers are colored blue and gold, and the ATPase domain is colored red. In the cocrystal structure, the connecting loops at the top of the 4-helix bundle were too disordered to be modeled. The unstructured N-terminal 33 amino acids of GrpE are drawn as a random coil. These residues have been implicated in interactions with the DnaK substrate-binding domain and were removed by proteolysis before crystallization. GrpE amino acids 34–39 were either unstructured (blue monomer) or too disordered to be modeled (gold monomer) in the cocrystal structure. The bottom of the long α-helices corresponds to residue 40 of GrpE. It can be seen that the compact β-sheet domain on the GrpE monomer proximal to DnaK sits at the top of the DnaK nucleotide-binding cleft, wedging it open. The figure on right is rotated by ∼70° about the vertical axis with respect to the figure on the left. These figures are rendered with “fog” to provide depth cueing. This figure was made with Molscript (Kraulis et al 1991), converted to a POVRAY-style format (D. Jeruzlami, unpublished software), and rendered with POVRAY (www.povray.org).
KINETICS OF NUCLEOTIDE EXCHANGE
GrpE function is analogous to that of other well-known nucleotide exchange factors, such as the elongation factor EFTs and RCC1, which are GEFs for the small GTPases EFTu and Ran, respectively, with many similarities with respect to their kinetics. For EFTs and RCC1, as well as for GrpE, transient ternary complexes of exchange factor-nucleotide hydrolase-nucleotide diphosphate can be detected (Klebe et al 1995; Neuhofen et al 1996; Packschies et al 1997; Gromadski et al 2002). Thermodynamic balance requires that the nucleotide exchange factors and the nucleotide mutually reduce affinity for the nucleotide hydrolase, and this has been experimentally confirmed: GrpE reduces the affinity of DnaK for ADP by 200-fold, and ADP reduces the affinity of GrpE for DnaK by 200-fold (Packschies et al 1997). One hallmark of the transient kinetic studies of GrpE-DnaK-ADP associations is the hyperbolic appearance of the plot of the observed dissociation rate constant of a fluorescently labeled ADP analog vs concentration of GrpE. This implies an associative displacement mechanism in which the initial collision complex between GrpE and DnaK·ADP subsequently undergoes an isomerization that results in nucleotide release, accelerating the rate of ADP release from DnaK 5000-fold (Packschies et al 1997). An associative displacement mechanism has also been seen in the actomyosin system (Bagshaw et al 1974) and in the Rho-GDS system (Hutchinson and Eccleston 2000). In a quirk of biology, actin (a structural homolog of the DnaK ATPase domain) itself acts as a nucleotide exchange factor for myosin.
GrpE-DnaK INTERACTIONS
GrpE and DnaK form a high-affinity complex that is resistant to high salt but readily disrupted by ATP (Zylicz et al 1987). Two dissociation constants that are in reasonably good agreement have been reported for the GrpE-DnaK complex: a Kd of 30 nM determined by surface plasmon resonance (Harrison et al 1997) and a Kd of 1 nM determined by transient kinetics (Packschies et al 1997). The crystal structure of GrpE-DnaK revealed an extended protein-protein interface that buries over 2800 Å2 of surface area (Harrison et al 1997), a substantial interaction interface that is consistent with the observed high-affinity interactions of GrpE and DnaK (Janin 1995; Lo Conte et al 1999). The bulk of the GrpE-DnaK contacts is made by the proximal GrpE β-domain, which is wedged between both sides of the nucleotide-binding cleft. Additional areas of GrpE-DnaK contact are from the proximal half of the 4-helix bundle of GrpE to domain IIB of DnaK (the so-called GrpE signature motif area of DnaK; see below) and from the top half of the proximal long α-helix to several exposed loops of DnaK. The latter region includes the DnaK Glu 28 loop, which has been shown to be necessary for GrpE binding (Buchberger et al 1994). As stated earlier, none of the known temperature-sensitive mutations of GrpE is implicated directly in GrpE-DnaK binding at the intermolecular interface observed in the cocrystal, and only 1 of the temperature-sensitive mutations of the yeast mitochondrial Mge1p (mge1–3: Ala 134 to valine) is appropriately positioned to make functionally significant contacts to Ssc1p, based on sequence homology.
A GrpE signature motif of DnaK has been identified by structural comparison of 12 Hsp70 homologs (using both existing crystal structures of DnaK and Hsc70 ATPase domains and homology models). The extended loop of DnaK at the top of domain IIB appears to be a hallmark of DnaK-like molecules that are capable of binding GrpE (Brehmer et al 2001). This extended loop of DnaK immediately follows several amino acids in a β-strand that make potentially significant intermolecular contacts (Asn 282, Pro 284, and Tyr 285 of DnaK) to side chains of GrpE located on the proximal half of the 4-helix bundle (Arg 104, Glu 107, and Val 108). GrpE will neither bind to nor stimulate nucleotide dissociation from the cytosolic Hsp70s, nor from HscA, the bacterial Hsp70-like molecule found in E coli, which lacks this loop (Brehmer et al 2001).
THE BACTERIAL MOLECULAR THERMOSENSOR
Up to a point, the rates of reactions generally increase with increasing temperature. Reactions for which the rate increases more slowly than expected or even decreases as the temperature increases when plotted as the log of the observed rate constant vs the inverse of temperature are said to be “non-Arrhenius.” Such observations imply that 1 of the components in the reaction is undergoing either a gradual or a sharp temperature-dependent conformational change, which compromises its activity (Gutfreund 1998). Christen and coworkers (2001) discovered that E coli GrpE exhibited non-Arrhenius behavior in that it was less active as a nucleotide exchange factor at higher temperatures than would be predicted from an Arrhenius plot. It is important to note that this study assayed DnaK activity, with GrpE added as a cofactor, rather than directly assaying GrpE-DnaK binding. The assays used (1) intrinsic fluorescence of DnaK Trp 102, which is sensitive to nucleotide state but not to GrpE binding, (2) release of a fluorescently labeled nucleotide analog, and (3) release of a fluorescently labeled peptide from the DnaK substrate-binding domain. Decreases in the rates of these 3 assays, which would reflect a decrease in the rate of ATP hydrolysis or ADP dissociation (or both), were observed only in the presence of GrpE, and not in the presence of DnaJ, and were fully reversible as the temperature was decreased (Grimshaw et al 2001).
Thermodynamic analyses of E coli GrpE temperature-induced protein unfolding revealed that GrpE has 2 thermal transitions. One of these transitions initiates unfolding around 35°C and has a midpoint (Tm) of ∼50°C, a temperature range that is relevant to both heat shock and the observed non-Arrhenius behavior of GrpE (Grimshaw et al 2001). Subsequently, that thermal transition was assigned to the long α-helices of GrpE. These α-helices undergo a helix-to-coil transition and do not exhibit any concentration dependence to the midpoint of the unfolding transition, which indicates that dimeric GrpE is not dissociating into unfolded monomers during this helix-to-coil transition. The temperature-induced unfolding of the 4-helix bundle was assigned to the second thermal transition observed in the DSC scan, which has a midpoint of ∼75°C (Gelinas et al 2002). The melting of the β-domains has also been assigned to this transition (Gelinas et al, personal communication).
These results raise 3 interesting questions. Why is the rate of GrpE catalysis of nucleotide exchange downregulated by temperature? What keeps the unfolded long α-helices of GrpE from becoming a substrate for DnaK at elevated temperatures? Why is GrpE activity apparently controlled by unfolding and inactivation at high temperatures, rather than by transcriptional repression? Although there are no immediate answers to the first 2 questions, the answer to the last question may be that heat shock–induced transcriptional overexpression of DnaK at higher temperatures is deleterious in the absence of compensating amounts of GrpE.
THERMOSENSORS
Reinstein and coworkers (2001) reported that Thermus thermophilus GrpE also exhibited non-Arrhenius behavior at temperatures that are relevant to the upper temperature range of T thermophilus growth. Their thermodynamic studies of the temperature-induced protein unfolding of T thermophilus GrpE and various constructs thereof led them to conclude that the β-domains of T thermophilus GrpE were unfolding at the relevant temperatures (Tm ∼90°C) (Groemping and Reinstein 2001; Groemping et al 2001), an observation that is distinct from the results seen with E coli GrpE (Gelinas et al 2002). One possible explanation for the difference between the thermodynamic properties of E coli GrpE and T thermophilus GrpE stems from the different thermodynamic pressures exerted on thermophiles compared with mesophiles. In addition, the different oligomeric organization of the T thermophilus DnaK molecular chaperone cycle may require T thermophilus GrpE to have modified functionalities (Schlee and Reinstein 2002).
The yeast mitochondrial GrpE, Mge1p, is required for growth in S cerevisiae. Mge1p is not a heat shock protein even though its cognate binding partner, Ssc1p (the mitochondrial Hsp70 mtHsp70), is upregulated by heat shock. An immediate, but as of yet unanswered, question is whether mitochondrial and chloroplastic GrpEs also possess a putative “thermosensing” activity that influences the cognate Hsp70s.
AUGMENTATION OF PEPTIDE RELEASE BY GrpE
Biochemical evidence hinted at a larger role for GrpE as a cochaperone that promotes substrate release from the DnaK substrate-binding domain (Langer et al 1992; Szabo et al 1994; Jordan and McMacken 1995). A GrpE fragment (34–197) lacking the proteolytically susceptible N terminus was found to be active in a luciferase refolding assay but did not promote the release of a tightly bound substrate (reduced carboxymethylated α-lactalbumin) from DnaK in an ATP-dependent manner. This suggests that GrpE plays a role in the release of tightly bound substrates, but that this activity is dispensable for some substrates that might be more weakly bound (Harrison et al 1997). Mally and Witt (2001) reported that GrpE accelerates the release of a fluorescently labeled peptide from the DnaK substrate-binding domain in an ATP-dependent manner. For the 2 peptides used in the study, the researchers concluded that GrpE contributed more to the rates of peptide release from the DnaK substrate-binding domain than did the addition of ATP to the reactions and noted that GrpE did not promote substrate release from the isolated DnaK substrate-binding domain. It was also observed that GrpE 89–197, a fragment that lacks the unstructured N terminus and the long α-helices, could promote nucleotide dissociation but not substrate dissociation (Mehl et al 2001).
CONCLUSIONS
GrpE-DnaK interactions provide an unusual and fascinating example of biological regulation. GrpE is a structurally unique nucleotide exchange factor that is functionally distinct from other known GEFs because it has a secondary function of augmenting peptide release from the DnaK substrate-binding domain. Interestingly, the activity of GrpE is hypothesized to be thermally regulated during heat shock. Future experiments are required to characterize the relationship between the temperature-dependent structural changes within GrpE and its temperature-dependent regulation of DnaK. A true understanding of this behavior will also require a high-resolution structure of DnaK interacting with the N terminus of GrpE.
Acknowledgments
The author is indebted to Susan Lyman, Amy Gelinas, Tyler Cutforth, and Nick Rhind for critical reading of the manuscript and gratefully acknowledges David Jeruzalmi for his unpublished figure-making software patch.
REFERENCES
- Ang D, Georgopoulos C. The heat-shock-regulated grpE gene of Escherichia coli is required for bacterial growth at all temperatures, but is dispensable in certain mutant backgrounds. J Bacteriol. 1989;171:2748–2755. doi: 10.1128/jb.171.5.2748-2755.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw CR, Eccleston JF, Eckstein F, Goody RS, Gutfreund H, Trentham DR. Magnesium ion-dependent adenosine-triphosphatase of myosin—2-step processes of adenosine-triphosphate association and adenosine-diphosphate dissociation. Biochem J. 1974;141:351–364. doi: 10.1042/bj1410351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolliger L, Deloche O, and Glick BS. et al. 1994 A mitochondrial homolog of bacterial GrpE interacts with mitochondrial Hsp70 and is essential for viability. EMBO J. 13:1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehmer D, Rudiger S, and Gassler CS. et al. 2001 Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol. 8:427–432. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Schroder H, Buttner M, Valencia A, Bukau B. A conserved loop in the ATPase domain of the DnaK chaperone is essential for stable binding of GrpE. Nat Struct Biol. 1994;1:95–101. doi: 10.1038/nsb0294-95. [DOI] [PubMed] [Google Scholar]
- Dekker PJT, Pfanner N. Role of mitochondrial GrpE and phosphate in the ATPase cycle of matrix Hsp70. J Mol Biol. 1997;270:321–327. doi: 10.1006/jmbi.1997.1131. [DOI] [PubMed] [Google Scholar]
- Gelinas AD, Langsetmo K, Toth J, Bethoney KA, Stafford WF, Harrison CJ. A structure-based interpretation of E. coli GrpE thermodynamic properties. J Mol Biol. 2002;323:131–142. doi: 10.1016/s0022-2836(02)00915-4. [DOI] [PubMed] [Google Scholar]
- Georgopoulus C, Liberek K, Zylicz M, and Ang D 1994 Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response. In: The Biology of Heat Shock Proteins and Molecular Chaperones, ed Morimoto R, Tissieres A, Georgopolous C. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 209–249. [Google Scholar]
- Grimshaw JP, Jelesarov I, Schonfeld HJ, Christen P. Reversible thermal transition in GrpE, the nucleotide exchange factor of the DnaK heat-shock system. J Biol Chem. 2001;276:6098–6104. doi: 10.1074/jbc.M009290200. [DOI] [PubMed] [Google Scholar]
- Groemping Y, Klostermeier D, Herrmann C, Veit T, Seidel R, Reinstein J. Regulation of ATPase and chaperone cycle of DnaK from Thermus thermophilus by the nucleotide exchange factor GrpE. J Mol Biol. 2001;305:1173–1183. doi: 10.1006/jmbi.2000.4373. [DOI] [PubMed] [Google Scholar]
- Groemping Y, Reinstein J. Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J Mol Biol. 2001;314:167–178. doi: 10.1006/jmbi.2001.5116. [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Wieden HJ, Rodnina MV. Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry. 2002;41:162–169. doi: 10.1021/bi015712w. [DOI] [PubMed] [Google Scholar]
- Gutfreund H 1998 Kinetics for the Life Sciences. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- Hutchinson JP, Eccleston JF. Mechanism of nucleotide release from rho by the gdp dissociation stimulator protein. Biochemistry. 2000;39:11348–11359. doi: 10.1021/bi0007573. [DOI] [PubMed] [Google Scholar]
- Ikeda E, Yoshida S, Mitsuzawa H, Uno I, Tohe A. YGE1 is a yeast homolog of Escherichia coli GrpE and is required for maintenance of mitochondrial functions. FEBS Lett. 1994;339:265–268. doi: 10.1016/0014-5793(94)80428-1. [DOI] [PubMed] [Google Scholar]
- Janin J. Principles of protein-protein recognition from structure to thermodynamics. Biochimie. 1995;77:497–505. doi: 10.1016/0300-9084(96)88166-1. [DOI] [PubMed] [Google Scholar]
- Johnson C, Chandrasekhar GN, Georgopoulos C. Escherichia coli DnaK and GrpE heat shock proteins interact both in vivo and in vitro. J Bacteriol. 1989;171:1590–1596. doi: 10.1128/jb.171.3.1590-1596.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ER, McKay DB. Mapping the role of active site residues for transducing an atp-induced conformational change in the bovine 70-kda heat shock cognate protein. Biochemistry. 1999;38:10823–10830. doi: 10.1021/bi990816g. [DOI] [PubMed] [Google Scholar]
- Jordan R, McMacken R. Modulation of the ATPase activity of the molecular chaperone DnaK by peptides and the DnaJ and GrpE heat shock proteins. J Biol Chem. 1995;270:4563–4569. doi: 10.1074/jbc.270.9.4563. [DOI] [PubMed] [Google Scholar]
- Klebe C, Prinz H, Wittinghofer A, Goody RS. The kinetic mechanism of Ran-nucleotide exchange catalyzed by RCC1. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- Kraulis P. Molscript: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Laloraya S, Dekker PJT, Voos W, Craig EA, Pfanner N. Mitochondrial GrpE moldulates the function of the matrix Hsp70 in translocation and maturation of preproteins. Mol Cell Biol. 1995;15:7098–7105. doi: 10.1128/mcb.15.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S, Gambill BD, Craig EA. A role for a eukaryotic GrpE-related protein, Mge1p, in protein translocation. Proc Natl Acad Sci U S A. 1994;91:6481–6485. doi: 10.1073/pnas.91.14.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska B, King J, Ang D, Georgopolous C. Sequence analysis and transcriptional regulation of the Escherichia coli grpE gene, encoding a heat shock protein. Nucleic Acids Res. 1988;16:7545–7562. doi: 10.1093/nar/16.15.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:177–198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Lupas A. Coiled-coils: new structures and functions. Trends Biochem Sci. 1996;21:379–382. [PubMed] [Google Scholar]
- Mally A, Witt SN. GrpE accelerates peptide binding and release from the high affinity state of DnaK. Nat Struct Biol. 2001;8:254–257. doi: 10.1038/85002. [DOI] [PubMed] [Google Scholar]
- Mehl AF, Heskett LD, Neal KM. A GrpE mutant containing the Nh(2)-terminal “tail” region is able to displace bound polypeptide substrate from DnaK. Biochem Biophys Res Commun. 2001;282:562–569. doi: 10.1006/bbrc.2001.4567. [DOI] [PubMed] [Google Scholar]
- Miao BJ, Davis JE, Craig EA. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J Mol Biol. 1997;265:541–552. doi: 10.1006/jmbi.1996.0762. [DOI] [PubMed] [Google Scholar]
- Nakai M, Kato Y, Ikeda E, Tohe A, Endo T. YGE1p, a eukaryotic GrpE homolog, is localized in the mitochondrial matrix and interacts with mitochondrial Hsp70. Biochem Biophys Res Commun. 1994;200:435–442. doi: 10.1006/bbrc.1994.1468. [DOI] [PubMed] [Google Scholar]
- Naylor DJ, Hoogenraad NJ, Hoj PB. Isolation and characterisation of a cdna encoding rat mitochondrial GrpE, a stress-inducible nucleotide-exchange factor of ubiquitous appearance in mammalian organs. FEBS Lett. 1996;396:181–188. doi: 10.1016/0014-5793(96)01100-3. [DOI] [PubMed] [Google Scholar]
- Neuhofen S, Theyssen H, Reinstein J, Trommer WE, Vogel PD. Nucleotide binding to the heat-shock protein DnaK as studied by ESR spectroscopy. Eur J Biochem. 1996;240:78–82. doi: 10.1111/j.1432-1033.1996.0078h.x. [DOI] [PubMed] [Google Scholar]
- Offer G, Hicks MR, Woolfson DN. Generalized Crick equations for modeling noncanonical coiled coils. J Struct Biol. 2002;137:41–53. doi: 10.1006/jsbi.2002.4448. [DOI] [PubMed] [Google Scholar]
- Packschies L, Theyssen H, Buchberger A, Bukau B, Goody RS, Reinstein J. GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism. Biochemistry. 1997;36:3417–3422. doi: 10.1021/bi962835l. [DOI] [PubMed] [Google Scholar]
- Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- Saito H, Uchida H. Initiation of DNA-replication of bacteriophage-lambda in Escherichia coli-K12. J Mol Biol. 1977;113:1–25. doi: 10.1016/0022-2836(77)90038-9. [DOI] [PubMed] [Google Scholar]
- Schlee S, Reinstein J. The DnaK/ClpB chaperone system from Thermus thermophilus. Cell Mol Life Sci. 2002;59:1598–1606. doi: 10.1007/PL00012486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonfeld HJ, Schmidt D, Shroder H, Bukau B. The DnaK chaperone system of Escherichia coli: quaternary structures and interactions of the DnaK and GrpE components. J Biol Chem. 1995;270:2183–2189. doi: 10.1074/jbc.270.5.2183. [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Whitelegge JP, Beck CF, Wollman FA. The chloroplastic GrpE homolog of chlamydomonas: two isoforms generated by differential splicing. Plant Cell. 2001;13:2823–2839. doi: 10.1105/tpc.010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Langer T, Shroder H, Flanagan J, Bukau B, Hartl FU. The atp hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system-DnaK, DnaJ, and GrpE. Proc Natl Acad Sci U S A. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Gambill BD, Laloraya S, Ang D, Craig EA, Pfanner N. Mitochondrial GrpE is present in a complex with Hsp70 and preproteins in transit across membranes. Mol Cell Biol. 1994;14:6627–6634. doi: 10.1128/mcb.14.10.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B, Prip-Buus C, Neupert W, Schwarz E. The role of the GrpE homologue, Mge1p, in mediating protein import and protein folding in mitochondria. EMBO J. 1995;14:3452–3460. doi: 10.1002/j.1460-2075.1995.tb07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbanks SM, Chen L, Tsuruta H, Hodgson KO, McKay DB. Solution small-angle x-ray scattering study of the molecular chaperone Hsc70 and its subfragments. Biochemistry. 1995;34:12095–12106. doi: 10.1021/bi00038a002. [DOI] [PubMed] [Google Scholar]
- Wu B, Ang D, Snavely M, Georgopoulos C. Isolation and characterization of point mutations in the Escherichia coli GrpE heat shock gene. J Bacteriol. 1994;176:6965–6973. doi: 10.1128/jb.176.22.6965-6973.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Wawrzynov A, Zylicz M, Georgopoulos C. Structure-function analysis of the Escherichia coli GrpE heat shock protein. EMBO J. 1996;15:4806–4816. [PMC free article] [PubMed] [Google Scholar]
- Zylicz M, Ang D, Georgopolous C. The grpE protein of Escherichia coli. J Biol Chem. 1987;262:17437–17442. [PubMed] [Google Scholar]