Abstract
The Hsp70 family members play an essential role in cellular protein metabolism by acting as polypeptide-binding and release factors that interact with nonnative regions of proteins at different stages of their life cycles. Hsp40 cochaperone proteins regulate complex formation between Hsp70 and client proteins. Herein, literature is reviewed that describes the mechanisms by which Hsp40 proteins interact with Hsp70 to specify its cellular functions.
INTRODUCTION
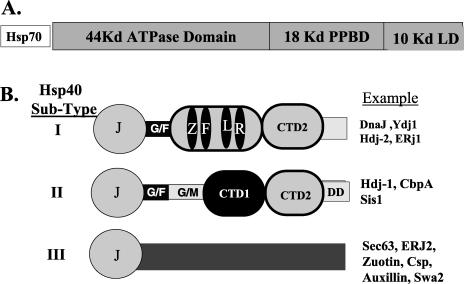
Hsp40s represent a large protein family that functions to specify the cellular action of Hsp70 chaperone proteins. The Hsp40 family is large, for example, the genomes for Saccharomyces cerevisiae and Homo sapiens encode 20 and 44 members, respectively (Cyr et al 1994; Cheetham and Caplan 1998; Venter et al 2001). Hsp40 family members have different domain structures and can be divided into 3 different subtypes (Fig 1). Type I Hsp40s are descendants of Escherichia coli DnaJ, and 12 of the 44 Human Hsp40 have a domain structure similar to that of DnaJ, the other 32 having divergent structures, with the majority populating subtype III (Venter et al 2001).
Fig 1.
Schematic diagrams of Hsp70 and members of the Hsp40 family. (A) The organization of Hsp70 into different subdomains. The 44-kDa adenosine triphosphatase (ATPase) domain represents a 44-kDa amino terminal fragment of Hsp70 that contains the ATP-binding site and retains ATPase activity. The 18-kDa polypeptide-binding domain (PPBD) represents an internal fragment of Hsp70 that functions as the PPD. The 10-kDa lid domain (LD) is a C-terminal fragment that is proposed to function as an LD that covers the PPBD and serves as a site for the binding of cochaperones. (B) Domain structure of different Hsp40 subtypes. J, J-domain; G/F, glycine-and phenylalanine-rich region; ZFLR, zinc finger–like region; G/M, glycine/methioine-rich region; CTDI, carboxyl-terminal domain I; CTDII, is carboxyl terminal domain II; DD, the dimerization domain. The examples represent Hsp40 proteins from E coli, S cerevisiae, and H sapiens
Hsp70 family members are often colocalized in the same subcellular compartment with multiple members of the Hsp40 family that have specialized individual functions. The interaction of a single Hsp70 with multiple Hsp40s generates unique Hsp70-Hsp40 pairs that facilitate specific processes at distinct locations within the cell (Caplan et al 1992a, 1992b; Ungermann et al 1994; Dey et al 1996; Liu et al 1998; Meacham et al 1999b; Gall et al 2000; Horton et al 2001). Thus, a mechanistic understanding of Hsp40 function as regulators of Hsp70 is fundamental to understanding the cell biology of molecular chaperones.
The major function of Hsp40 proteins is to regulate adenosine triphosphate (ATP)–dependent polypeptide binding by Hsp70 protein (Liberek et al 1991; Wickner et al 1991; Cyr et al 1992; Langer et al 1992; Palleros et al 1993; Szabo et al 1994) (Fig 2). Substrate release from Hsp70 is regulated by nucleotide exchange factors that are related by the E coli GrpE protein (Liberek et al 1991; Harrison et al 1997; Sondermann et al 2001). The mechanism for regulation of Hsp70 function by GrpE-like cochaperones is discussed elsewhere in previous reviews in Cell Stress and Chaperones.
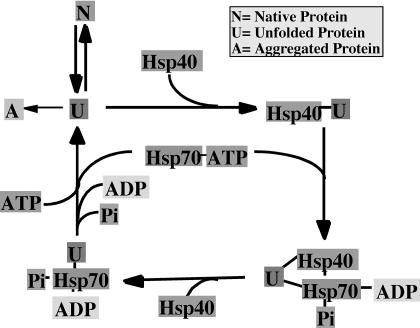
Fig 2.
A proposed model for Hsp40-dependent polypeptide binding and release by Hsp70. Hsp40 proteins form complexes with unfolded or nonnative proteins to prevent their aggregation. Hsp40 then delivers the unfolded protein to Hsp70. Stable Hsp70-protein complexes are then formed by a mechanism that involves Hsp40 J-domain dependent conversion of Hsp70-adenosine triphosphate (ATP) to Hsp70-adenosine diphosphate. Hsp70-protein complexes dissociate upon regeneration of Hsp70-ATP. Upon release from Hsp70, an unfolded polypeptide can fold, aggregate, or be rebound by Hsp40 and Hsp70
GENERAL MECHANISMS FOR REGULATION OF Hsp70 FUNCTION BY Hsp40
Hsp40 proteins regulate complex formation between Hsp70 and polypeptides by 3 mechanisms. First, Hsp40 proteins have evolved to contain unique classes of polypeptide-binding domains (PPDs) that bind and deliver specific clients to Hsp70 (Cyr et al 1994; Cheetham and Caplan 1998). Second, Hsp40 proteins stabilize Hsp70-polypeptide complexes by driving the conversion of Hsp70 from its ATP form to the adenosine diphosphate form (Liberek et al 1991; Cyr et al 1992; Langer et al 1992). Third, specialized members of the Hsp40 family are localized to different sites within the same cellular compartment (Brodsky and Schekman 1993; Cyr and Neupert 1996; Shen et al 2002). Interaction of Hsp70 with differentially localized Hsp40s enables different Hsp70-Hsp40 pairs to bind unique clients at these sites.
REGULATION OF Hsp70 ATPase ACTIVITY BY Hsp40
The domain in Hsp40 proteins that is responsible for regulation of Hsp70 ATPase activity is the J-domain, and it is present in all Hsp40 family members (Fig 1). The J-domain is about 75 amino acids in length and can be found at various locations within Hsp40 proteins (Caplan et al 1993). The J-domain was first identified in E coli DnaJ and contains a conserved HPD tripeptide that represents the signature motif of the Hsp40 protein family (Yochem et al 1978). The nuclear magnetic resonance (NMR) solution structure of the J-domain has been solved (Hill et al 1995; Qian et al 1996), and it is constructed from 4 α-helical regions (Fig 3A). Helix II and helix III lie in an antiparallel orientation and are separated by a solvent-exposed loop that contains the HPD motif (Fig 3A). The HPD motif plays a critical role in the regulation of Hsp70 function because mutations in it block the ability of Hsp40s to regulate Hsp70 ATPase activity (Wall et al 1994; Tsai and Douglas 1996; Kelley and Georgopoulos 1997; Mayer et al 1999).
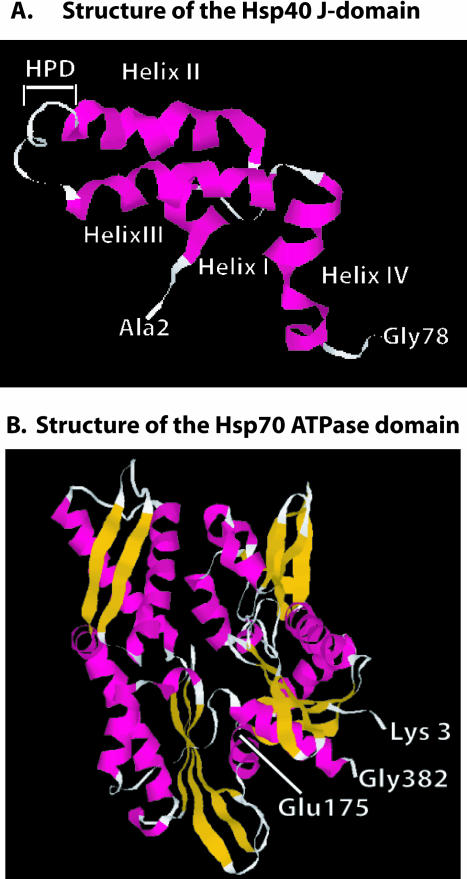
Fig 3.
Structures of the Hsp40 J-domain and the Hsp70 adenosine triphosphatase (ATPase) domain. (A) Ribbon diagram of the nuclear magnetic resonance solution structure of residues 2–77 of E coli DnaJ. HPD denotes the position of the conserved HPD motif that is found in the J-domain of all Hsp40s. D in the HPD motif corresponds to D35 in DnaJ that was mutated in experiments that identified the J-domain–binding site in DnaK (see text for details). Gly78 denotes the end of the J-domain and the beginning of the glycine- and phenylalanine-rich region. (B) Ribbon diagram of the X-ray crystal structure of the 382-residue amino-terminal fragment of Hsp70 that retains ATPase activity. Glu175 is the conserved residue in Hsp70 that is represented in E coli DnaK (Hsp70) by Glu171. This residue is denoted because it was demonstrated to function in interdomain communication between the ATPase domain and the polypeptide-binding domain of Hsp70 (see text). Glu175 is also located near R167, which is found in an acidic cleft where J-domain binding to Hsp70 is proposed to occur (see text). Gly382 denotes the terminus of ATPase domain. β-Strands are shown in gold, and α-helices are in pink. The J-domain and Hsp70 ATPase fragment structures were from PDB files 1BQZ and 1HJO, respectively, and the images shown were generated with Rasmol
The J-domain is proposed to interact with Hsp70 at an acidic groove located in the ATPase domain (Fig 3B). The putative J-domain–Hsp70 interaction site was identified by extragenic suppressor analysis of a DnaJ D35N mutant, which contains an alteration in the HPD motif that causes defective growth (Suh et al 1998). Spontaneous mutations of DnaK at R167 were observed to alleviate the growth defects caused by DnaJ D35N (Suh et al 1998). DnaK R167 maps to a solvent-exposed and highly conserved acidic groove that is located opposite to the ATP-binding cleft in DnaK (Suh et al 1998).
In complementary studies on Hsp40 and Hsp70 interactions, chemical shift mapping was used to demonstrate that amino acid residues located in helix II and the HPD motif of the J-domain form an interface with the Hsp70 ATPase domain (Greene et al 1998). In addition, a synthetic peptide that contains the HPD motif and helix II is sufficient to inhibit functional interactions between Hsp40 and Hsp70 (Tsai and Douglas 1996). Thus, helix II and the HPD motif of the J-domain appear to bind an acidic cleft in the Hsp70 ATPase domain and thereby stimulate Hsp70 ATPase activity.
Studies on the molecular dynamics of J-domain–Hsp70 interactions demonstrate that the J-domain exists in a dynamic ensemble of conformations that is constrained upon binding to Hsp70 (Landry 2003). Dynamic flexibility was observed in several regions of the J-domain, and helix II was observed to bend upon interacting with Hsp70 (Landry 2003). The influence that J-domain binding has on the conformation of the Hsp70 ATPase domain has not been investigated. Nonetheless, the dynamic nature of the J-domain structure and its constraint upon interaction with Hsp70 suggests that the details of the J-domain–Hsp70 complex formation play a critical role in regulation of Hsp70 ATPase activity (Landry 2003).
The ability of the J-domain to stimulate Hsp70 ATPase activity is enhanced by the presence of peptides bound in the polypeptide-binding site of Hsp70 (Bukau and Horwich 1998). Because ATP hydrolysis leads to a conformational change in Hsp70 that stabilizes Hsp70-polypeptide complexes, a mechanism for interdomain communication between the ATPase and PPD of Hsp70 appears to exist (Buchberger et al 1994; Montgomery et al 1999). The Bukau group has identified E171 of the DnaK ATPase domain as a residue that is dispensable for ATP hydrolysis but is required for DnaK to refold model substrates (Buchberger et al 1994). DnaK E171 is conserved as E175 in mammalian forms of Hsp70 (Fig 3B) and is part of the ATP-binding pocket that contacts bound Mg2+-ATP (Flaherty et al 1990). DnaK E171 is proposed to form part of a hinge that facilitates movements in the ATP-binding pocket that drive conformational changes in the PPD that regulate substrate binding and release (Buchberger et al 1994). Interestingly, amino acid E171 is located near the acidic groove that contains R167 in the J-domain–binding site in Hsp70. Thus, J-domain binding to Hsp70 has the potential to influence the conformation of the hinge that controls communication between the Hsp70 ATPase and PPDs.
Hsp40-DEPENDENT LOADING OF Hsp70 WITH NONNATIVE PROTEIN
Results from biochemical studies carried out with Hsp40 fragments demonstrate that the J-domain alone is sufficient to stimulate Hsp70 ATPase activity (Wall et al 1994). However, the J-domain needs to be attached to a functional PPD to promote complex formation between Hsp70 and nonnative proteins (Ungewickell et al 1995; Minami et al 1996; Hartl and Hayer-Hartl 2002). Presently, the mechanism by which different Hsp40s function to bind and deliver nonnative proteins to Hsp70 is unclear. Hsp40s can bind substrates independent of Hsp70 and enhance the ability of Hsp70s to bind nonnative proteins (Langer et al 1992). Thus, Hsp40s are proposed to bind nonnative proteins before Hsp70 (Fig 2), but whether this is always the case is not clear. Because Hsp70 and Hsp40s are capable of simultaneously binding different regions on nonnative proteins, the formation of an Hsp40-polypeptide-Hsp70 ternary complex is proposed to represent an important intermediate in the Hsp70 polypeptide-binding and release cycle (Han and Christen 2003). Hsp40-polypeptide-Hsp70 ternary complexes have been isolated (Hartl and Hayer-Hartl 2002), and the formation of such complexes appears to facilitate substrate transfer from Hsp40 to Hsp70 (Han and Christen 2003).
POLYPEPTIDE BINDING BY Hsp40S
Type I and type II Hsp40s function as ATP-independent chaperones that bind nonnative polypeptides and protect cells from stress by preventing protein aggregation (Cheetham and Caplan 1998). Type I and type II Hsp40s can form complexes with newly synthesized proteins and, therefore, are proposed to assist Hsp70 in cotranslational protein folding (Frydman et al 1994; Hartl and Hayer-Hartl 2002). Purified type I Hsp40s can function independent of Hsp70 to directly bind nonnative proteins and suppress protein aggregation (Langer et al 1992; Cyr 1995; Meacham et al 1999b). Type II Hsp40s can directly interact with nonnative proteins to maintain them in a folding-competent conformation (Freeman and Morimoto 1996; Lee et al 2002). However, type II Hsp40s are not equivalent to type I Hsp40s as chaperones because they must function with Hsp70 to suppress the aggregation of model proteins (Minami et al 1996; Lu and Cyr 1998b; Muchowski et al 2000).
Type III Hsp40s do not appear to be general chaperones and have evolved to contain PPDs that recognize specific substrates (Cheetham and Caplan 1998). For example, the yeast Hsp40 Swa2 and mammalian axullin contain a J-domain and a clathrin-binding domain that cooperate to facilitate Hsp70-dependent uncoating of clathrin-coated vesicles (Ungewickell et al 1995; Gall et al 2000). Most of the mechanistic studies on Hsp40 chaperone function have been carried out with type I and type II Hsp40s. Thus, the remainder of this section of the article will review data that concern the mechanism by which type I and type II Hsp40 function as polypeptide-binding proteins.
Because Hsp40s and Hsp70s can bind different regions of the same protein, they are likely to exhibit differences in substrate specificity or binding affinity. Indeed, DnaJ and Dnak have been observed to activate the bacteriophage P1 RepA protein in E coli by binding it at distinct sites (Kim et al 2002). DnaJ recognizes a 21-residue stretch in RepA, SKLWELFQLDYRVLLQHHALR, that is located between amino acids 180 and 200 of this 286–amino acid protein (Kim et al 2002). This DnaJ-binding motif is enriched with a mixture of hydrophobic and charged residues, with the longest stretch of hydrophobic amino acids being 3 residues long. The DnaK-binding site on P1 was located between residues 36 and 49, RLGVFVPKPSKSKG, and is thus distinct from the DnaJ-binding site (Kim et al 2002). These data are consistent with the model put forth by Christen where Hsp70 and Hsp40 can bind protein targets at different sites simultaneously (Han and Christen 2003).
In a study designed to define motifs in nonnative proteins that are recognized by type I Hsp40s, the Bukau group used purified DnaJ to screen cellulose-bound peptide arrays that displayed 1633 different peptides derived from 14 different protein sequences (Rudiger et al 2001). This analysis identified a motif recognized by DnaJ that had a core of 8 residues and was enriched in aromatic and large hydrophobic amino acids and arginine (Rudiger et al 2001). Residues 1910–2000 in the P1 RepA DnaJ-binding motif, RVLLQHHALR, closely resemble those predicted to represent the peptide-binding motif of DnaJ (Rudiger et al 2001).
The substrate specificity of yeast type I and type II Hsp40s was recently compared side by side in studies that used purified Ydj1 and Sis1 to biopan a 7-mer phage peptide display library (Fan et al 2004). Ydj1 and Sis1 were both found to select sets of peptides that were enriched in aromatic and bulky hydrophobic amino acids. However, the groups of peptides that were selected by Ydj1 and Sis1 exhibited differences in the enrichment of specific amino acids. Ydj1 preferred peptides that had a hydrophobic stretch of 3–4 residues, but peptides selected by Sis1 did not contain a patch of hydrophobic residues (Fan et al 2004). Thus, the PPDs of Ydj1 and Sis1 can bind hydrophobic regions exposed by nonnative proteins and exhibit some overlapping substrate specificity, but they are selective (Fan et al 2004).
Because Ydj1 and DnaJ are both type I Hsp40s, they would be expected to exhibit similar substrate selectivity. Indeed, DnaJ and Ydj1 both bind peptides enriched in the aromatic amino acids F, W, and Y; the large hydrophobics I and L; and the polar residue H. In addition, Ydj1 and DnaJ appear to exclude the amino acids P and K from the peptides they select (Rudiger et al 2001; Fan et al 2004). Thus, type I Hsp40s DnaJ and Ydj1 share a conserved domain structure and exhibit similar substrate specificity. Investigators have also examined whether peptide recognition by Hsp40s relies on side-chain interactions or involves backbone recognition (Rudiger et al 2001; Bischofberger et al 2003). To address this question, the ability of DnaJ to interact with peptides consisting of l- and d-amino acids was examined. DnaJ was found to bind both d- and l-peptides (Rudiger et al 2001; Bischofberger et al 2003). Thus, substrate recognition by type I Hsp40s is proposed to rely exclusively on side-chain recognition (Rudiger et al 2001). Substrate binding by Hsp70 involves both side-chain and backbone contacts (Zhu et al 1996). Therefore, there is a clear distinction in the mechanism for substrate recognition by Hsp70 and Hsp40. The ability of Hsp40 to recognize amino acid side chains appears to enable it to scan substrates for hydrophobic surfaces and make the initial contacts with proteins that are subsequently targeted to Hsp70 (Rudiger et al 2001).
PPDS OF Hsp40S
The mechanism by which Hsp40s function to bind nonnative polypeptides is not clear. However, there is evidence that suggests that structurally distinct domains located within the central regions of type I and type II Hsp40s function in polypeptide binding (Fig 1). In type I Hsp40 proteins, a protein module that lies between the J-domain and carboxyl-terminal domain II (CTDII), which contains a conserved zinc fingerlike region (ZFLR), has been implicated as a component of the polypeptide-binding site (Banecki et al 1996; Szabo et al 1996; Lu and Cyr 1998a). In type II Hsp40s the ZFLR protein module has been replaced by a glycine- and methionine-rich region (G/M) and CTDI (Fig 1). Studies on the yeast Hsp40 Sis1 suggest that CTDI functions as a component of the type II Hsp40 PPD (Lee et al 2002).
The ZFLR of type I Hsp40s is a centrally located and cysteine-rich protein module that consists of 4 repeated Cys-X-X-Cys-X-Gly-X-Gly motifs that function in pairs, and each pair binds a single molecule of zinc. The NMR structure of a 79-residue ZFLR fragment of E coli DnaJ (Martinez-Yamout et al 2000) depicts this domain to have a novel fold with an overall V-shaped, extended β-hairpin topology (Fig 4A). The conformation of the cysteine residues coordinated to zinc ion resembles that of a rubredoxin knuckle, but there are differences in the hydrogen-bonding patterns of these 2 different metal-binding motifs (Martinez-Yamout et al 2000).
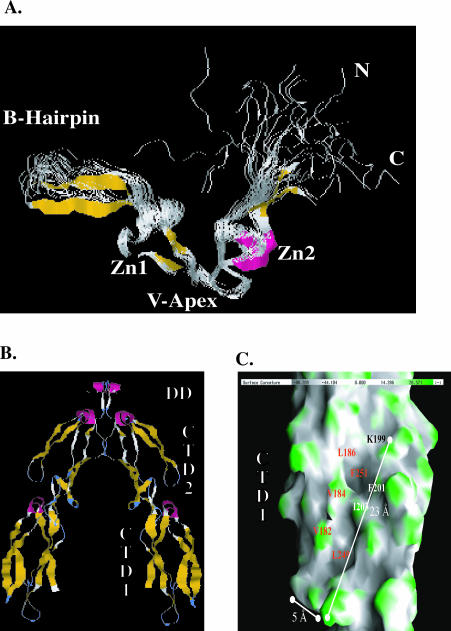
Fig 4.
Structure of regions in type I and type II Hsp40s that are involved in chaperone function. (A) The nuclear magnetic resonance solution structure of the zinc finger–like region (ZFLR) from E coli DnaJ. A ribbon diagram depicting the nuclear magnetic resonance solution structure of a protein fragment that corresponds to Gly131 to Ser209 of E coli DnaJ. This image was rendered from PDB file 1EXK. β-Strands are shown in gold, and α-helices are in pink. Zn1 and Zn2 denote the 2 regions in the ZFLR where zinc is bound. N and C denote the position of resides 8 and 79 of DnaJ 131–209. This diagram represents 20 structures. (B) Ribbon diagram of the Sis1 (171–352) dimer. Sis1 (171–352) is truncated at the end of the glycine- and methionine-rich region (G/M) region, and this model shows residues 180–352. A and B represent the monomers that form the Sis1 (171–352) dimer, which has a 2-fold axis. Subdomains present in the Sis1 (171–352) monomer are labeled. Carboxyl-terminal domain I (CTDI) corresponds to residues 180–254. CTDII corresponds to residues 255–341. The Sis1 dimerization domain (DD) lies between residues 341 and 352. β-Strands are shown in gold, and α-helices are in pink. (C) An enlarged view of the surface of CTDI as depicted in monomer B of (B). The surface shown depicts contours on CTDI with concave areas in gray and convex areas in green. A surface hydrophobic groove that contains 2 shallow depressions is visible. Solvent-exposed residues that line the depression are denoted. F251 and L249 form the base of the individual depressions. Panels in (B) and (C) were rendered from PDB file 1C3G with Rasmol and GRASP software packages, respectively
The V-shaped groove in the DnaJ ZFLR has the potential to be involved in protein-protein interactions (Martinez-Yamout et al 2000), and fragments of E coli DnaJ that contain the ZFLR are capable of directly interacting with at least some nonnative proteins (Szabo et al 1996). However, whether the Hsp40 ZFLR plays a direct or indirect role in substrate binding is not clear. This is the case because Hsp40 ZFLR mutants that exhibit defects in protein-folding activity do not exhibit defects in polypeptide binding (Lu and Cyr 1998a). In addition, deletion of the ZFLR from DnaJ does not abolish substrate binding (Banecki et al 1996). A proteolytic fragment of Ydj1, Ydj1 (179–384), which lacks the J-domain and the first zinc-binding module of the ZFLR is capable of suppressing protein aggregation and therefore retains the chaperone function of Ydj1 (Lu and Cyr 1998a). On the basis of these data, it was suggested that the ZFLR and adjacent C-terminal regions were components of the type I Hsp40 PPD (Lu and Cyr 1998a).
Study of the yeast Hsp40 Sis1 has localized the polypeptide-binding site of type II Hsp40s to a C-terminal fragment that contains residues 171–352 of this 352–amino acid protein (Lu and Cyr 1998b). The X-ray crystal structure of Sis1 171–352 was solved (Sha and Cyr 1999; Sha et al 2000), and it depicts a homodimer that has a cystallographic 2-fold axis (Fig 4B). Sis1 171–352 monomers are elongated and constructed from 2 barrellike domains that have similar folds and a mostly β-structure. Sis1 dimerizes through a short C-terminal α-helical domain. The Sis1 dimer has a wishbone shape, and there is a cleft that separates the arms of the 2 elongated monomers. CTDI on each monomer contains a shallow depression that is lined by highly conserved, solvent-exposed hydrophobic residues (Fig 4C). Sis1 is purified from yeast as a dimer (Luke et al 1991), and monomeric forms of Sis1, which lack its dimerization domain, can stimulate the ATPase activity of Hsp70 but are unable to cooperate with Hsp70 to refold denatured luciferease (Sha et al 2000). Monomeric Sis1 is also defective in maintaining denatured luciferease in a folding-competent conformation (Sha et al 2000). Mutational analysis of the residues that line the hydrophobic depression in Sis1 has identified K199, F201, and F251 as amino acids that are essential for cell viability and are required for Sis1 to bind model substrates and cooperate with Hsp70 to refold model substrates (Lee et al 2002). A hydrophobic depression similar to the one in CTDI is also found in CTDII, but it is occupied by intramolecular protein-protein interactions and, therefore, may not be available for substrate binding (Sha et al 2000). These collective data suggest that type II Hsp40s function as divalent chaperones that use a solvent-exposed hydrophobic patch located on CTDI to bind nonnative proteins.
MECHANISMS FOR SPECIFICATION OF Hsp70 FUNCTION BY TYPE I AND TYPE II Hsp40S
As stated above, a single Hsp70 protein can interact with multiple members of the Hsp40 family to form unique Hsp70-Hsp40 couples that have unique functions. In the case of type III Hsp40s, specification of Hsp70 function occurs through the binding of specific clients via their unique PPDs (Cyr et al 1994; Cheetham and Caplan 1998). However, because type I and type II Hsp40s appear to have overlapping substrate specificity, how they specify Hsp70 is not clear.
To investigate this question, investigators have used yeast as a model system and examined the functional relationships between Ydj1 and Sis1 that occur with the cytosolic Hsp70 Ssa1–4 and Hsp70 Ssb1–2 proteins. Biochemical studies suggest that Ydj1 and Sis1 interact with Hsp70 Ssa proteins but not with members of the Hsp70 Ssb protein family (Cyr et al 1992, 1994; Cyr and Douglas 1994; Cyr 1995). Genetic studies indicate that Ydj1 and Sis1 have specific functional properties that enable them to direct Hsp70 Ssa proteins to facilitate different cellular processes. For example, the overexpression of Sis1 can complement the slow-growth phenotype of ydj1Δ strains, but Ydj1 cannot complement the lethal phenotype of sis1Δ strains (Caplan and Douglas 1991; Luke et al 1991). In addition, the cellular functions of Ydj1 and Sis1 are different. Ydj1 and its human homolog Hdj2 function on the cytoplasmic face of the endoplasmic reticulum to promote membrane protein folding and protect cells from stress (Caplan et al 1992b; Meacham et al 1999b). Ydj1 is known to be required for proper folding of the insulinase-like protease Axl1 (Meacham et al 1999a) and regulation of cyclin 3 phophporylation and ubiquitination (Yaglom et al 1996). In addition, Ydj1 is more efficient than Sis1 in maintaining hormone receptors in ligand-binding competent conformations (Fliss et al 1999). In contrast, Sis1 is found in association with translating ribosomes and is required to facilitate the assembly of translation initiation complexes (Zhong and Arndt 1993; Horton et al 2001). Sis1, but not Ydj1, is required for the maintenance of the prion [RNQ+] (Sondheimer et al 2001; Lopez et al 2003).
Examination of the domain structures of Ydj1 and Sis1 reveals 2 structural differences. First, the glycine-and phenylalanine-rich region (G/F) of Ydj1 and Sis1 are different, with that of Sis1 containing a 10-residue-long insert (Lopez et al 2003). Second, as mentioned above, the protein modules located in the middle of Ydj1 and Sis1 are different (Fig 1). Thus, it is plausible that either the G/F domain or the chaperone modules of Ydj1 and Sis1 serve to specify their in vivo functions.
In tests of the latter hypothesis, chimeric forms of Ydj1 and Sis1 were constructed in which the chaperone modules were swapped to form YSY and SYS (Fan et al 2004). Purified SYS and YSY were found to exhibit protein-folding activity and substrate specificity that mimicked that of Ydj1 and Sis1, respectively (Fan et al 2004). In in vivo studies YSY exhibited a gain of function and, unlike Ydj1, could complement the lethal phenotype of sis1Δ and promote the propagation of the yeast prion [RNQ1+]. SYS exhibited a loss of function and was unable to maintain [RNQ1+]. These in vitro and in vivo data suggest that the chaperone modules of Ydj1 and Sis1 are exchangeable and that they help specify Hsp70s cellular functions (Fan et al 2004).
To determine whether the G/F regions of type I and type II Hsp40s help specify Hsp70 functions, the Craig group has carried out a number of complementation studies with Hsp40 fragments (Yan and Craig 1999). In these studies, which were conducted with a sis1Δ strain, the G/F region of Sis1, but not that of Ydj1, was shown to be important for suppression of the inviability caused by the loss of Sis1 function (Yan and Craig 1999). In addition, Sis1Δ G/F was demonstrated to be defective at [RNQ1+] maintenance (Sondheimer et al 2001). The G/F region of Sis1 contains a 10-residue insert GHAFSNEDAF that corresponds to amino acids 102–112 that are not present in the G/F region of Ydj1 (Lopez et al 2003). When the G/M region is deleted from the Sis1 chaperone module (see Fig 1), residues N108I and D110G in the G/F region become important for Sis1 in vivo function (Lopez et al 2003). Thus, the Sis1 G/F rich region is clearly important for its in vivo function, and it plays an important role in modulating the conformation of at least some substrates.
How the G/F domain functions to help specify Hsp40 action is not established, and a direct interaction between the G/F domain and a substrate protein has not been demonstrated. However, the G/F region is enriched in hydrophobic residues and lies adjacent to regions in Hsp40s that are involved in substrate binding. Thus, it is possible that the G/F region operates as a component of the Hsp40 PPD. On the other hand, the G/F region may have evolved to mediate interactions between Hsp40 and Hsp70 that are important for the conformational maturation of different substrate proteins. Nonetheless, it is clear that the G/F domain and chaperone modules of type I and type II Hsp40 both act to specify Hsp70 cellular functions.
Acknowledgments
Work in D.M.C.'s laboratory was supported by the National Institutes of Health.
REFERENCES
- Banecki B, Liberek K, Wall D, Wawrzynow A, Georgopoulos C, Bertoli E, Tanfani F, Zylicz M. Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J Biol Chem. 1996;271:14840–14848. doi: 10.1074/jbc.271.25.14840. [DOI] [PubMed] [Google Scholar]
- Bischofberger P, Han W, Feifel B, Schonfeld HJ, Christen P. d-Peptides as inhibitors of the DnaK/DnaJ/GrpE chaperone system. J Biol Chem. 2003;278:19044–19047. doi: 10.1074/jbc.M300922200. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Valencia A, McMacken R, Sander C, Bukau B. The chaperone function of DnaK requires the coupling of ATPase activity with substrate binding through residue E171. EMBO J. 1994;13:1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992a;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. Eukaryotic homologues of Escherichia coli dnaJ: a diverse protein family that functions with hsp70 stress proteins. Mol Biol Cell. 1993;4:555–563. doi: 10.1091/mbc.4.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ, Douglas MG. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ, Tsai J, Casey PJ, Douglas MG. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J Biol Chem. 1992b;267:18890–18895. [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr DM. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Douglas MG. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- Cyr DM, Langer T, Douglas MG. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- Cyr DM, Neupert W. Roles for hsp70 in protein translocation across membranes of organelles. Exs. 1996;77:25–40. doi: 10.1007/978-3-0348-9088-5_3. [DOI] [PubMed] [Google Scholar]
- Dey B, Caplan AJ, Boschelli F. The Ydj1 molecular chaperone facilitates formation of active p60v-src in yeast. Mol Biol Cell. 1996;7:91–100. doi: 10.1091/mbc.7.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CY, Lee S, and Cyr DM 2004 Type I and type II Hsp40s contain exchangeable chaperone modules that specify Hsp70 function. Mol Biol Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Fliss AE, Rao J, Melville MW, Cheetham ME, Caplan AJ. Domain requirements of DnaJ-like (Hsp40) molecular chaperones in the activation of a steroid hormone receptor. J Biol Chem. 1999;274:34045–34052. doi: 10.1074/jbc.274.48.34045. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Morimoto RI. The human cytosolic molecular chaperones hsp90 hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Gall WE, Higginbotham MA, Chen C, Ingram MF, Cyr DM, Graham TR. The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr Biol. 2000;10:1349–1358. doi: 10.1016/s0960-9822(00)00771-5. [DOI] [PubMed] [Google Scholar]
- Greene MK, Maskos K, Landry SJ. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci U S A. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Christen P. Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J Biol Chem. 2003;278:19038–19043. doi: 10.1074/jbc.M300756200. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hill RB, Flanagan JM, Prestegard JH. 1H and 15N magnetic resonance assignments secondary structure and tertiary fold of Escherichia coli DnaJ(1–78) Biochemistry. 1995;34:5587–5596. doi: 10.1021/bi00016a033. [DOI] [PubMed] [Google Scholar]
- Horton LE, James P, Craig EA, Hensold JO. The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J Biol Chem. 2001;276:14426–14433. doi: 10.1074/jbc.M100266200. [DOI] [PubMed] [Google Scholar]
- Kelley WL, Georgopoulos C. The T/t common exon of simian virus 40 JC and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc Natl Acad Sci U S A. 1997;94:3679–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Sharma S, Hoskins JR, Wickner S. Interaction of the DnaK and DnaJ chaperone system with a native substrate P1 RepA. J Biol Chem. 2002;277:44778–44783. doi: 10.1074/jbc.M206176200. [DOI] [PubMed] [Google Scholar]
- Landry SJ. Structure and energetics of an allele-specific genetic interaction between dnaJ and dnaK: correlation of nuclear magnetic resonance chemical shift perturbations in the J-domain of Hsp40/DnaJ with binding affinity for the ATPase domain of Hsp70/DnaK. Biochemistry. 2003;42:4926–4936. doi: 10.1021/bi027070y. [DOI] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Lee S, Fan CY, Younger JM, Ren H, Cyr DM. Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. J Biol Chem. 2002;277:21675–21682. doi: 10.1074/jbc.M111075200. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS, Kuo SR, Makhov AM, Cyr DM, Griffith JD, Broker TR, Chow LT. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J Biol Chem. 1998;273:30704–30712. doi: 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ(+)] Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998a;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998b;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- Luke MM, Sutton A, Arndt KT. Characterization of SIS1 a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Yamout M, Legge GB, Zhang O, Wright PE, Dyson HJ. Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ. J Mol Biol. 2000;300:805–818. doi: 10.1006/jmbi.2000.3923. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Laufen T, Paal K, McCarty JS, Bukau B. Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J Mol Biol. 1999;289:1131–1144. doi: 10.1006/jmbi.1999.2844. [DOI] [PubMed] [Google Scholar]
- Meacham GC, Browne BL, Zhang W, Kellermayer R, Bedwell DM, Cyr DM. Mutations in the yeast Hsp40 chaperone protein Ydj1 cause defects in Axl1 biogenesis and pro-a-factor processing. J Biol Chem. 1999a;274:34396–34402. doi: 10.1074/jbc.274.48.34396. [DOI] [PubMed] [Google Scholar]
- Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999b;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y, Hohfeld J, Ohtsuka K, Hartl FU. Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog Hsp40. J Biol Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- Montgomery DL, Morimoto RI, Gierasch LM. Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone DnaK which alter substrate affinity or interdomain coupling. J Mol Biol. 1999;286:915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci U S A. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleros DR, Reid KL, Shi L, Welch WJ, Fink AL. ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- Qian YQ, Patel D, Hartl FU, McColl DJ. Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J Mol Biol. 1996;260:224–235. doi: 10.1006/jmbi.1996.0394. [DOI] [PubMed] [Google Scholar]
- Rudiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha B, Cyr D. Purification crystallization and preliminary X-ray crystallographic studies of S. cerevisiae Hsp40 Sis1. Acta Crystallogr D Biol Crystallogr. 1999;55:1234–1236. doi: 10.1107/s090744499900476x. [DOI] [PubMed] [Google Scholar]
- Sha B, Lee S, Cyr DM. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Struct Fold Des. 2000;8:799–807. doi: 10.1016/s0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- Shen Y, Meunier L, Hendershot LM. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J Biol Chem. 2002;277:15947–15956. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. Interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. Proc Natl Acad Sci U S A. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK DnaJ and GrpE. Proc Natl Acad Sci U S A. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Douglas MG. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- Ungermann C, Neupert W, Cyr DM. The role of Hsp70 in conferring unidirectionality on protein translocation into mitochondria. Science. 1994;266:1250–1253. doi: 10.1126/science.7973708. [DOI] [PubMed] [Google Scholar]
- Ungewickell E, Ungewickell H, and Holstein SE. et al. 1995 Role of auxilin in uncoating clathrin-coated vesicles. Nature. 378:632–635. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, and Myers EW. et al. 2001 The sequence of the human genome. Science. 291:1304–1351. [DOI] [PubMed] [Google Scholar]
- Wall D, Zylicz M, Georgopoulos C. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- Wickner S, Hoskins J, McKenney K. Monomerization of RepA dimers by heat shock proteins activates binding to DNA replication origin. Proc Natl Acad Sci U S A. 1991;88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaglom JA, Goldberg AL, Finley D, Sherman MY. The molecular chaperone Ydj1 is required for the p34CDC28-dependent phosphorylation of the cyclin Cln3 that signals its degradation. Mol Cell Biol. 1996;16:3679–3684. doi: 10.1128/mcb.16.7.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Craig EA. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Uchida H, Sunshine M, Saito H, Georgopoulos CP, Feiss M. Genetic analysis of two genes dnaJ and dnaK necessary for Escherichia coli and bacteriophage lambda DNA replication. Mol Gen Genet. 1978;164:9–14. doi: 10.1007/BF00267593. [DOI] [PubMed] [Google Scholar]
- Zhong T, Arndt KT. The yeast SIS1 protein a DnaJ homolog is required for the initiation of translation. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]