Abstract
R-2-hydroxyglutarate accumulates to millimolar levels in cancers with gain-of-function isocitrate dehydrogenase 1/2 mutations. These levels of R-2-hydroxyglutarate affect 2-oxoglutarate-dependent dioxygenases. Both R- and S-2-hydroxyglutarate, the other enantiomer of this metabolite, are detectible in healthy individuals, yet their physiological function remains elusive. Here we show that CD8+ T-lymphocytes accumulate 2-hydroxyglutarate in response to T-cell receptor triggering. This increases to millimolar levels in physiological oxygen conditions, via a hypoxia inducible factor 1 alpha-dependent mechanism. S-2-hydroxyglutarate predominates over R-2-hydroxyglutarate in activated T cells, and we demonstrate alterations in markers of CD8+ T-lymphocyte differentiation in response to this metabolite. Modulation of histone and DNA demethylation as well as hypoxia inducible factor 1 alpha stability mediate these effects. S-2-hydroxyglutarate treatment greatly enhances the in vivo proliferation, persistence and anti-tumour capacity of adoptively transferred CD8+ T-lymphocytes. Thus S-2-hydroxyglutarate acts as an immunometabolite that links environmental context, via a metabolic-epigenetic axis, to immune fate and function.
In response to T-cell receptor (TCR) triggering, quiescent CD8+ T-lymphocytes transition to a proliferative effector state. During this response, memory CD8+ T-lymphocytes form and can persist for the life of the organism, mounting rapid recall responses thereby providing long-term immunity. The metabolic programs of these different CD8+ T-lymphocyte states are distinct and important for function1–4. Effector CD8+ T-lymphocytes generate most ATP and biomass via glycolysis5; both naïve and memory cells rely heavily on oxidative phosphorylation 6,7. Various cytokines and transcription factors are important for the differentiation of CD8+ T-lymphocytes, and it is evident that immunological memory is influenced by epigenetic mechanisms8–12.
CD8+ T-lymphocytes traffic into severely hypoxic zones within tumours and inflammatory tissue13. The response to oxygenation mediated via the Von Hipple Lindau (VHL) and hypoxia inducible transcription factor α (HIFα) proteins, is an essential regulator of metabolism and CD8+ T-lymphocyte function14–16. Here we demonstrate that CD8+ T-lymphocytes produce 2-hydroxyglutarate (2HG) in response to TCR triggering and environmental hypoxia. Using CD8+ T-lymphocyte specific genetic deletions of VHL, HIF-1α and HIF-2α, we highlight the dependency of this metabolic feature on the HIF pathway. S-2HG constitutes the majority of the 2HG pool and we show that S-2HG alters the phenotypic and functional characteristics of CD8+ T-lymphocytes, maintaining a state of increased proliferative, survival and anti-tumour capacity.
The VHL-HIF-1α axis regulates 2HG production
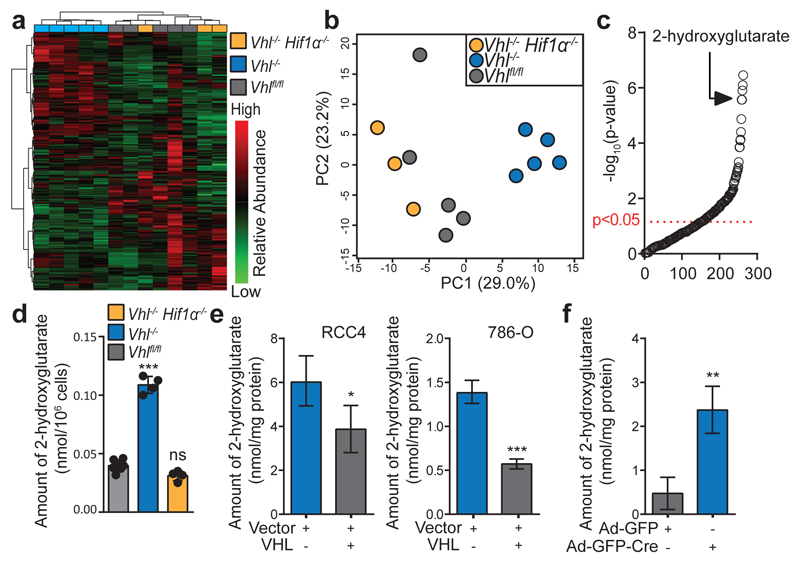
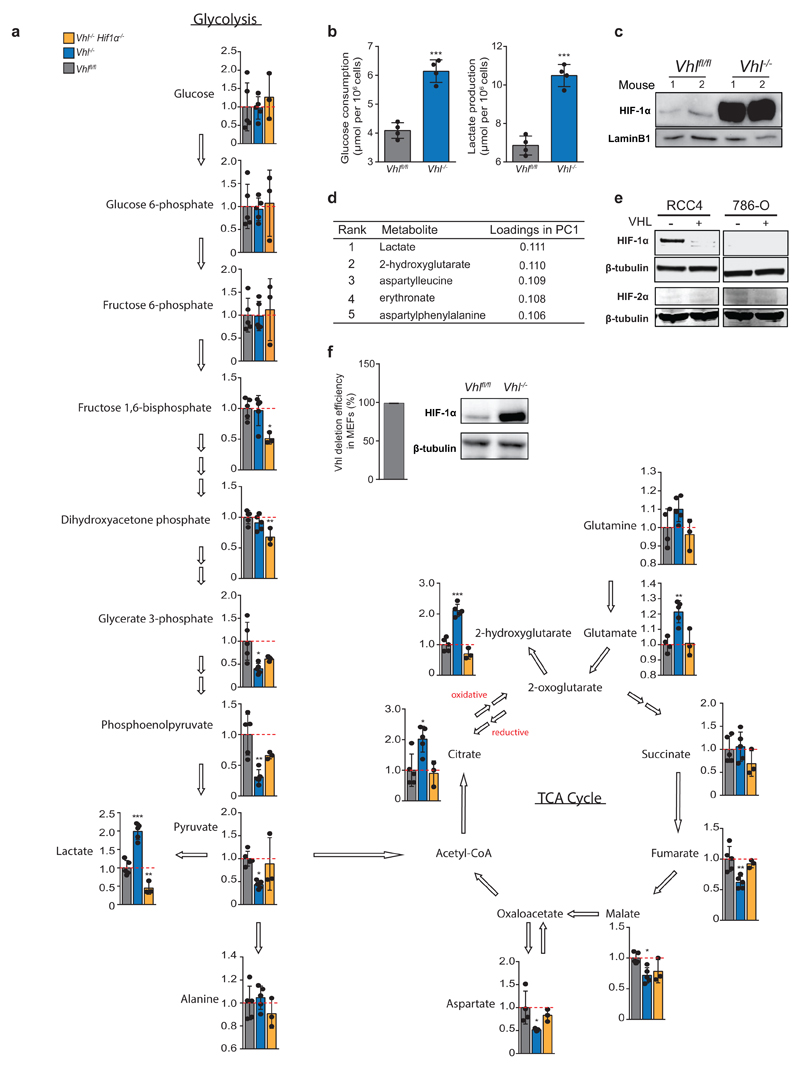
To elucidate metabolic effects of HIFα activation, we profiled the metabolome of CD8+ T-lymphocytes with low (Vhlfl/fl), or high (Vhlfl/fldlckcre referred to as Vhl-/-) HIF signalling, and HIF-1α-VHL double knockouts (Hif1αfl/flVhlfl/fldlckcre referred to as Hif1α-/-Vhl-/-) to control for a specific contribution of HIF-1α14. Unsupervised clustering and principal component analysis (Fig. 1a, b) separate Vhl-/- from Vhlfl/fl CD8+ T-lymphocytes. Vhl-/-Hif1α-/- cluster with Vhlfl/fl, indicating that HIF-1α mediates significant metabolic changes following Vhl deletion. Glycolysis is critical for sustaining effector function1–3,17 and these data indicate that VHL suppresses glycolysis via inhibition of HIF-1α, (Extended Data Fig. 1a-c).
Figure 1. VHL-HIF signalling regulates 2-hydroxyglutarate levels.
a/ Unsupervised hierarchical clustering and heat map of all detected metabolites.
b/ PCA of metabolomes. Percentage variance of each PC is in parenthesis.
c/ Metabolites ranked in order of decreasing p-value .
d/ 2HG levels in Vhlfl/fl (n=7), Vhlfl/fldlckcre (n=4) and Hif1αfl/fl Vhlfl/fldlckcre (n=4) CD8+ T-lymphocytes activated with αCD3 and αCD28 antibodies and then cultured in IL-2 for a further 5 days. Each dot represents a mouse.
e/ 2HG levels in RCC4 and 786-O cells with reconstitution of VHL; n=3.
f/ 2HG levels in MEFs with deletion of VHL; n=3.
Unpaired t-test (e-f) and one-way ANOVA (d). Error bars denote s.d.; ns = non-significant, *p<0.05, **p<0.01, ***p<0.001.
VHL loss suppresses late and increases early tricarboxylic acid (TCA) cycle intermediates (Extended Data Fig. 1a). Strikingly, 2HG is significantly enriched in Vhl-/- CD8+ T-lymphocytes (Fig. 1c and Extended Data Fig. 1d). Furthermore, increases in 2HG depend on HIF-1α when VHL is deleted (Extended Data Fig. 1a). This was validated using quantitative mass spectrometry in Vhl-/- and Vhl-/- Hif1-/- CD8+ T-lymphocytes (Fig. 1d), as well as in VHL-null cell lines, that express either HIF-1α (RCC4) or HIF-2α (786-O), reconstituted with VHL (Fig. 1e and Extended Data Fig. 1e). Deletion of Vhl in murine embryonic fibroblasts from Vhlfl/fl mice increases 2HG levels (Fig. 1f and Extended Data Fig. 1f). Hence, the VHL-HIF axis regulates 2HG levels and constitutive HIF-1α signalling underlies this effect in Vhl-null CD8+ T-lymphocytes.
HIF-1α regulates S-2HG production
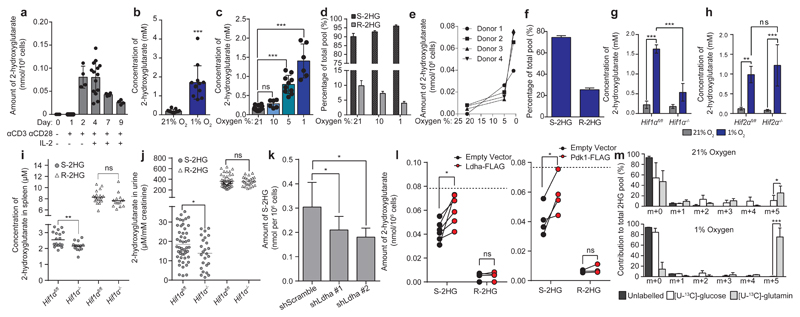
R-2HG is produced by isocitrate dehydrogenase 1/2 mutations in different cancers18–20; accumulation of S-2HG, the other enantiomer of 2HG, occurs in the context of hypoxia21,22 and mitochondrial dysfunction23,24. We thus sought to determine 2HG levels in CD8+ T-lymphocytes following activation. 2HG before activation and at sea level oxygen is undetectable, whereas levels at the same oxygenation are elevated 2-4 days after TCR stimulation (Fig. 2a).When activated CD8+ T-lymphocytes are exposed to 1% oxygen, the intracellular concentration of 2HG reaches millimolar levels (Fig. 2b and Extended Data Fig. 2a) and is proportional to the degree of oxygenation (Fig. 2c). We confirmed this using 1HNMR spectroscopy (Extended Data Fig. 2b). Given such high levels of 2HG, we sequenced25 Idh1/2, to preclude the unlikely possibility that culturing primary murine CD8+ T-lymphocytes in hypoxia gives rise to mutations known to cause R-2HG production in humans18; we found no evidence for such mutations (Extended Data Fig. 3a-e). Resolving the enantiomers of 2HG indicates that S-2HG constitutes the majority of the 2HG pool (Fig. 2d). Primary human CD8+ T-lymphocytes also accumulate S-2HG in hypoxia (Fig. 2e, f), indicating that this phenomenon is not limited to murine lymphocytes.
Figure 2. Hypoxic induction of 2-hydroxyglutarate depends on HIF-1α in CD8+ T-lymphocytes.
a/ 2HG in naïve and activated cells, n≥4 mice per time point.
b/ 2HG in cells cultured at 21% or 1% oxygen 48 h; n=12 mice.
c/ 2HG in cells cultured at 21% (n=15), 10% (n=6), 5% (n=10) or 1% (n=6) oxygen for 48 h.
d/ Enantioselective MS for S- and R-2HG.
e/ 2HG in human cells( n=4 healthy donors).
f/ Enantioselective MS for S- and R-2HG from cells in e.
g-h/ 2HG in Hif1αfl/fl and Hif1αfl/fl dlckcre (g) or Hif2αfl/fl and Hif2αfl/fl dlckcre (h) cells, cultured as in b; n=4 mice per genotype.
i/ 2HG in splenic extracts from Hif1αfl/fl (n=20) and Hif1αfl/fl dlckcre (n=15) mice.
j/ 2HG in urine from Hif1αfl/fl (n=49) and Hif1αfl/fl dlckcre (n=25) mice.
k/. S-2HG in cells with shLdha. n=4 individual transductions.
l/ Ldha-FLAG (n=6) or Pdk1-FLAG (n=4) over-expression in Hif1αfl/fl dlckcre cells. The dotted line represents S-2HG levels in Hif1αfl/fl cells n=4/6 individual tranductions.
m/ 13C-isotopologue profile of 2HG in cells cultured as in b ; n=7 mice per condition.
Unpaired t-test ( i, j). Paired t-test (b, l, m). One-way ANOVA (c, k) and two-way ANOVA (g, h). Error bars denote s.d.; each dot in a, b, c, i and j represents a. ns = non-significant, *p<0.05, **p<0.01, ***p<0.001.
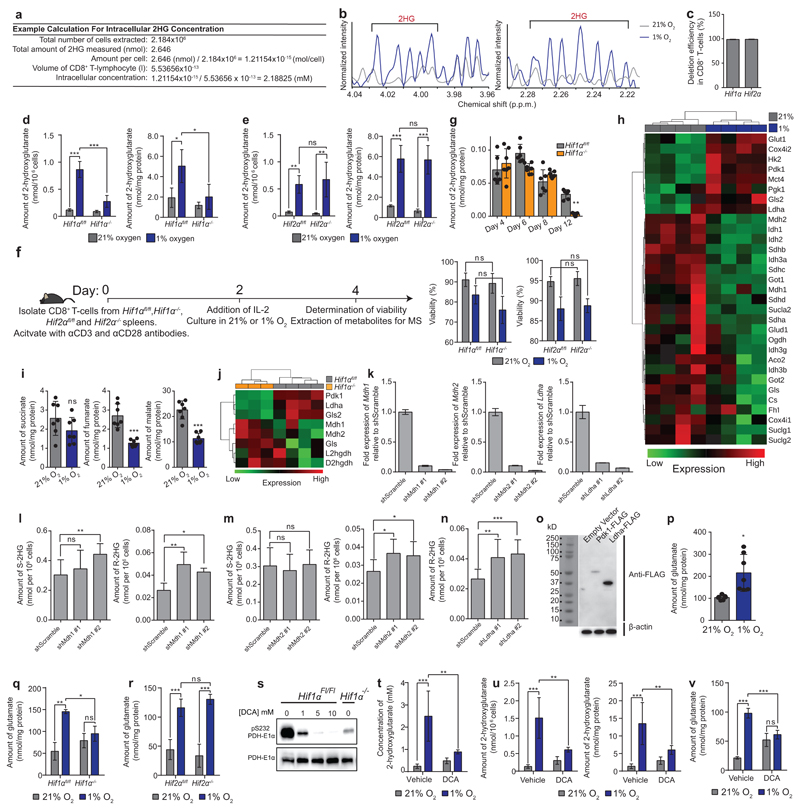
We carried out deletion of loxP-flanked Hif1α or Epas1 (referred to as Hif2α) genes in CD8+ T-lymphocytes, with cre recombinase expressed under the distal Lck promoter (dlckcre)26 (Extended Data Fig. 2c). 2HG accumulation is abolished in Hif1αfl/fl dlckcre (referred to as Hif1α-/-), but not Hif2αfl/fl dlckcre (referred to as Hif2α-/-) cells under hypoxia (Fig. 2g, h and Extended Data Fig. 2d, e), with no difference in viability (Extended Data Fig. 2f). We next examined 2HG levels in vivo, in the murine spleen. There is more R-2HG than S-2HG (Fig. 2i); furthermore, the levels of S-2HG are significantly decreased in spleens of Hif1αfl/fl dlckcre mice (Fig. 2i), There is also a slight decrease in the levels of R-2HG (Fig. 2i). 2HG is present in urine of healthy individuals and is elevated in patients with 2HG acidurias27. Hif1αfl/fl dlckcre mice have lower levels of S-2HG in urine (Fig. 2j) indicating that HIF-1α in the T-lymphocyte (CD4+ and CD8+) compartment makes a contribution to S-2HG production in vivo. Activated Hif1α-/- CD8+ T-lymphocytes in 21% oxygen have lower 2HG at extended time points (Extended Data Fig. 2g), indicating a contribution of HIF-1α in non-hypoxic conditions also.
We next sought to determine the metabolic route by which HIF-1α promotes S-2HG production in CD8+ T-lymphocytes. Transcriptionally, hypoxic CD8+ T-lymphocytes show induction of glycolysis and suppression of the TCA cycle (Extended Data Fig. 2h). Moreover, TCA cycle intermediates are decreased (Extended Data Fig. 2i). Recent reports implicate lactate and malate dehydrogenases (Ldha and Mdh1/2), as enzymatic sources of S-2HG in hypoxia21,22. In Hif1α-/- CD8+ T-lymphocytes, the hypoxic expression of these enzymes suggests that Mdh1 and Mdh2 are unlikely to mediate the hypoxia-induced accumulation of S-2HG (Extended Data Fig. 2j). Confirming this, knockdown of Mdh1 or Mdh2 does not decrease S-2HG in hypoxia (Extended Data Fig. 2k-m); knockdown of Mdh1 leads to marginal increases in S-2HG (Extended Data Fig. 2l). Knockdown of Mdh1 or Mdh2 increases R-2HG levels (Extended Data Fig. 2l, m). Knockdown of Ldha (Extended Data Fig. 2k) decreases S-2HG (Fig. 2k), whilst also increasing R-2HG levels in hypoxic CD8+ T-lymphocytes (Extended Data Fig. 2n).Over-expression of Ldha in hypoxic Hif1α-/- CD8+ T-lymphocytes (Extended Data Fig. 2o) rescues S-2HG production (Fig. 2l). Consistent with this, Ldha expression in hypoxic CD8+ T-lymphocytes is HIF-1α-dependent (Extended Data Fig. 2j).
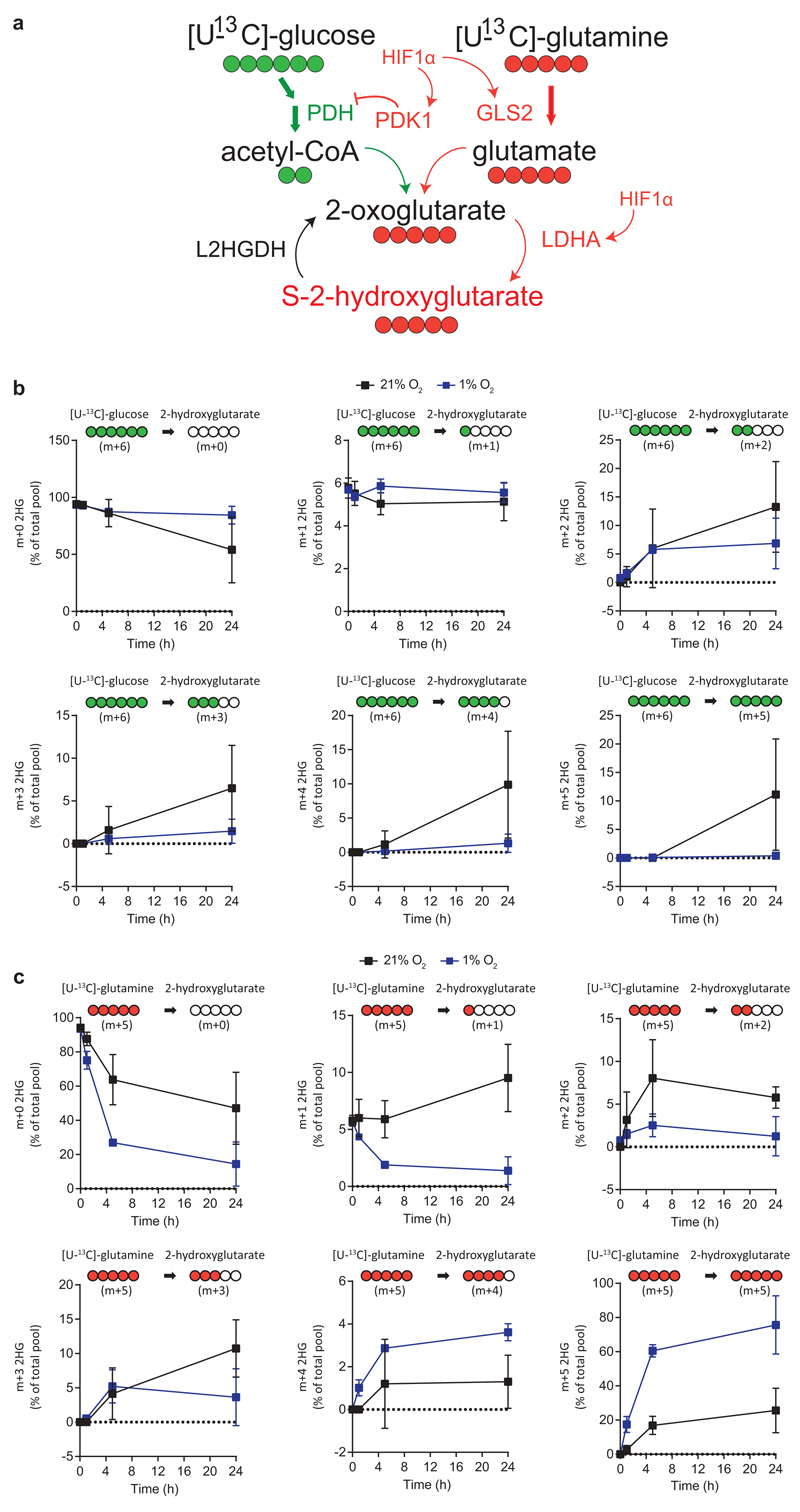
We next performed 13C-labelling experiments and in agreement with previous reports21,23, U-13C-glucose and U-13C-glutamine tracing indicates that glutamine is the major source of 2HG, (Fig. 2m, Extended Data Fig 4a-c). The m+5 isotopologue dominates, suggesting direct conversion of glutamine-derived 2-oxoglutarate to 2HG23,21. Furthermore, the glutamate pool increases in hypoxic (Extended Data Fig. 2p) and Vhl-/- CD8+ T-lymphocytes (Extended Data Fig. 1a) and depends on HIF-1α (Extended Data Fig. 2q) but not HIF-2α (Extended Data Fig. 2r). Inhibition of pyruvate dehydrogenase (Pdh) by pyruvate dehydrogenase kinases (Pdk), promotes glutaminolysis28–30. We reasoned that Pdk supports S-2HG production by diverting glutamine-derived 2-oxoglutarate to Ldha, and consistent with this, dichloroacetate (DCA) abrogates hypoxia-induced 2HG accumulation (Extended Data Fig. 2s-u). Pdk1 expression is impaired in hypoxic Hif1α-/- CD8+ T-lymphocytes (Extended Data Fig. 2j), as is phosphorylation of Pdh (Extended Data Fig. 2s), and re-expression of Pdk1 in this context increases S-2HG in hypoxia (Fig. 2l and Extended Data Fig. 2o). Inhibition of Pdk activity also impedes hypoxia-induced increases in the glutamate pool (Extended Data Fig. 2v). Hence, HIF-1α drives S-2HG production in hypoxic CD8+ T-lymphocytes via the Pdk-Pdh axis and Ldha induction (Extended Data Fig. 4a).
S-2HG alters CD8+ T-cell differentiation
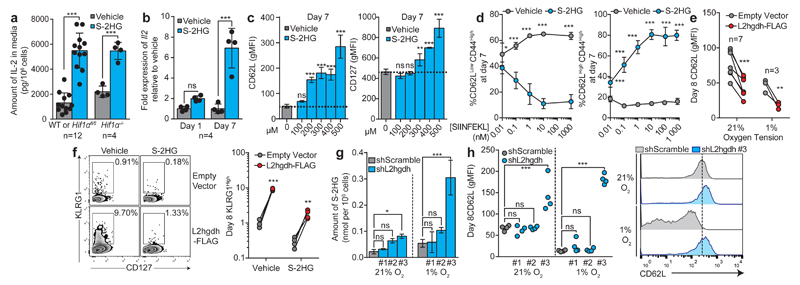
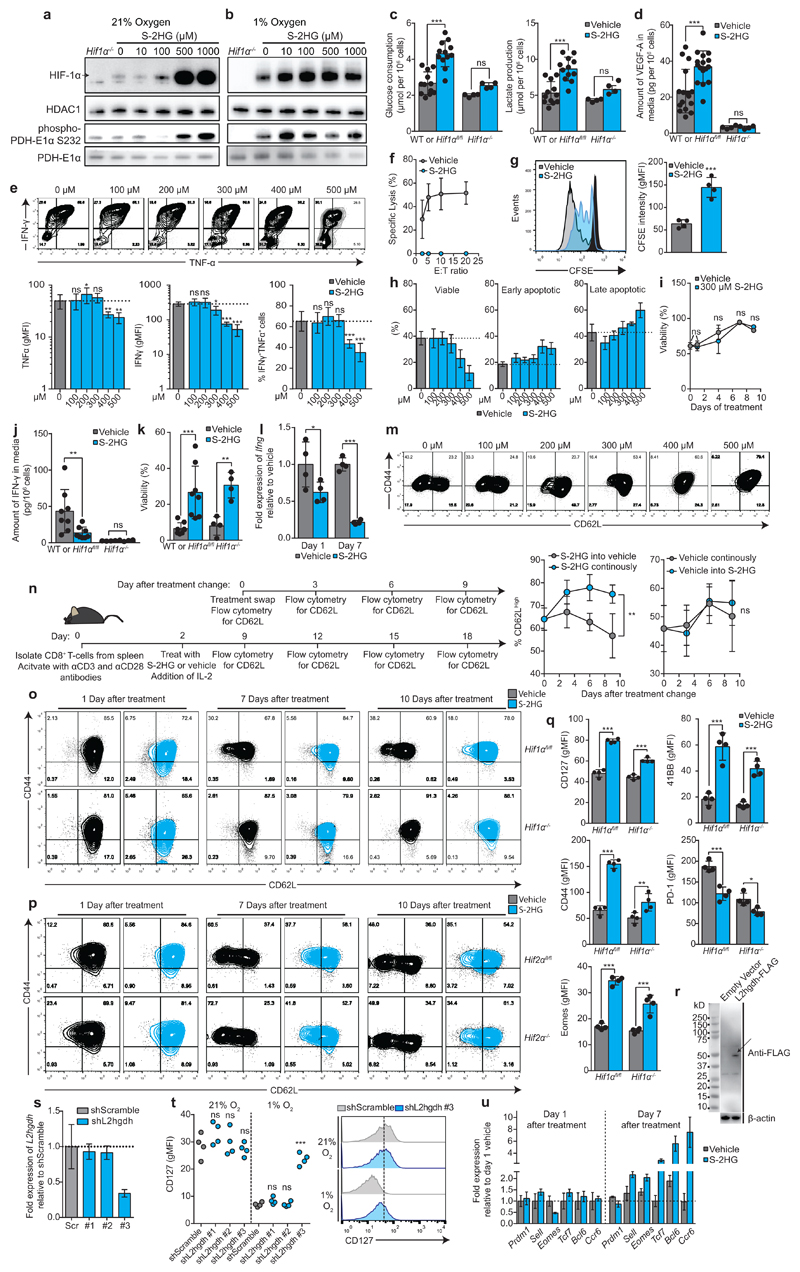
S-2HG inhibits 2-oxoglutarate-dependent dioxygenases31,32. Consistent with this, HIF-1α is stabilized in normoxic (Extended Data Fig. 5a) and hypoxic (Extended Data Fig. 5b) CD8+ T-lymphocytes by treatment with cell permeable S-2HG, suggesting that S-2HG augments HIF signalling in normoxia and hypoxia. Additionally, there is increased phosphorylation of PDH-E1α (Extended Data Fig. 5a and b), elevated glucose uptake, lactate secretion (Extended Data Fig. 5c) and VEGF-A production (Extended Data Fig. 5d), indicating HIF-1α-dependent effects. Since HIF-1α supports effector functions in CD8+ T-lymphocytes13,15, we reasoned that S-2HG promotes effector differentiation via HIF-1α. However, unexpectedly, there is suppression of effector cytokine production (Extended Data Fig. 5e) and decreased cytotoxicity (Extended Data Fig. 5f). Furthermore, S-2HG restrains cell expansion (Extended Data Fig. 5g) and there is a clear increase in apoptosis at doses > 300 μM (Extended Data Fig. 5h and i). Further characterization reveals decreased secretion of IFN-γ (Extended Data Fig. 5j), yet elevated production of IL-2 (Fig. 3a), with increased viability in the absence of IL-2 supplementation (Extended Data Fig. 5k). This possibly reflects an autocrine pro-survival effect. These effects are robustly mediated at the transcriptional level after prolonged treatment with S-2HG (Fig 3b. Extended Data Fig. 5l) and are independent of HIF-1α (Fig. 3a and Extended Data Fig 5j and k).
Figure 3. S-2HG alters phenotypic marker expression of CD8+ T-lymphocytes.
a/ Media IL-2 levels from S-2HG-treated cells.
b/ Expression of Il2 mRNA in S-2HG-treated cells.
c/ CD62L and CD127 expression on from S-2HG-treated OT-I cellsn=4 mice.
d/ % CD62LHighCD44Low and % CD62LLowCD44High cells with S-2HG treatment. n=4 mice.
e/CD62L expression on cells treated with S-2HG and transduced with empty or L2hgdh-FLAG over-epression vectors.
f/ Representative flow cytommetry plots of KLRG1 vs CD127 (n=4) on cells from e. Associated statistics are shown.
g/ S-2HG level in response to shRNA against L2hgdh (n=4).
h/ CD62L expression in response to L2hgdh knockdown (n=4). Representative flow cytometry histogram of CD62L surface levels is shown on the right.
Two-way ANOVA (a, b, e, f). One-way ANOVA (c, d, g, h). Error bars denote s.d. .Each dot in a, b, e, f, h, represents a mouse. ns=non-significant, *p<0.05, **p<0.01, ***p<0.001. gMFI=geometric mean fluorescence intensity.
We then characterized the phenotype of cells treated with a prolonged course of S-2HG. There is increased expression of CD62L (Fig. 3c and Extended Data Fig. 5m) and this is reversible upon withdrawal of treatment (Extended Data Fig. 5n).The effect does not occur when treating cells cultured in vehicle for 7 days (Extended Data Fig. 5n), demonstrating that S-2HG treatment of newly activated cells maintains this phenotypic marker. Importantly, CD62L downregulation does not occur when HIF-1α is absent15, masking the effect of S-2HG on CD62L following HIF-1α deletion (Extended Data Fig. 5o). HIF-2α is dispensable for CD62L maintenance in response to S-2HG (Extended Data Fig. 5p). With S-2HG, CD62L maintenance depends on the level of antigenic stimulation (Fig. 3d). Furthermore, S-2HG treated cells express more CD127 (Fig. 3c), CD44, 41BB, Eomes and less PD-1 in a HIF-1α-independent manner (Extended Data Fig. 5q).
To determine the role of endogenously produced S-2HG, over-expression of L2hgdh (Extended Data Fig. 5r), a dehydrogenase that oxidizes S-2HG, was performed. Over-expression of L2hgdh promotes the downregulation of CD62L in both 21% and 1% oxygen (Fig. 3e) indicating that endogenously produced S-2HG regulates CD62L expression. Furthermore, L2hgdh over-expression leads to an increase in the proportion of KLRG1High cells, which are decreased in the presence of exogenous S-2HG (Fig. 3f). Conversely, successful shRNA-mediated knockdown of L2hgdh by hairpin #3 (Extended Data Fig. 5s) increases endogenous S-2HG levels (Fig. 3g), especially in 1% oxygen, promoting maintenance of CD62L (Fig. 3h). Suppression of L2hgdh blocks loss of CD62L in response to low oxygen (Fig. 3h). The same effect is seen with CD127 in low oxygen (Extended Data Fig. 5t). These data demonstrate that L2hgdh activity regulates the expression of key phenotypic makers of CD8+ T-lymphocytes, by controlling endogenous S-2HG levels. Transcriptionally, S-2HG treatment increases expression of Eomes, Ccr6, Bcl-6, CD62L and Tcf-7, with repression of Prdm1, after 7 days (Extended Data Fig. 5u). This transcriptional program is similar to gene expression changes in memory CD8+ T-lymphocytes, suggesting that S-2HG treatment of CD8+ T-lymphocytes ex vivo may enhance long term persistence and survival in the context of adoptive cell transfer33.
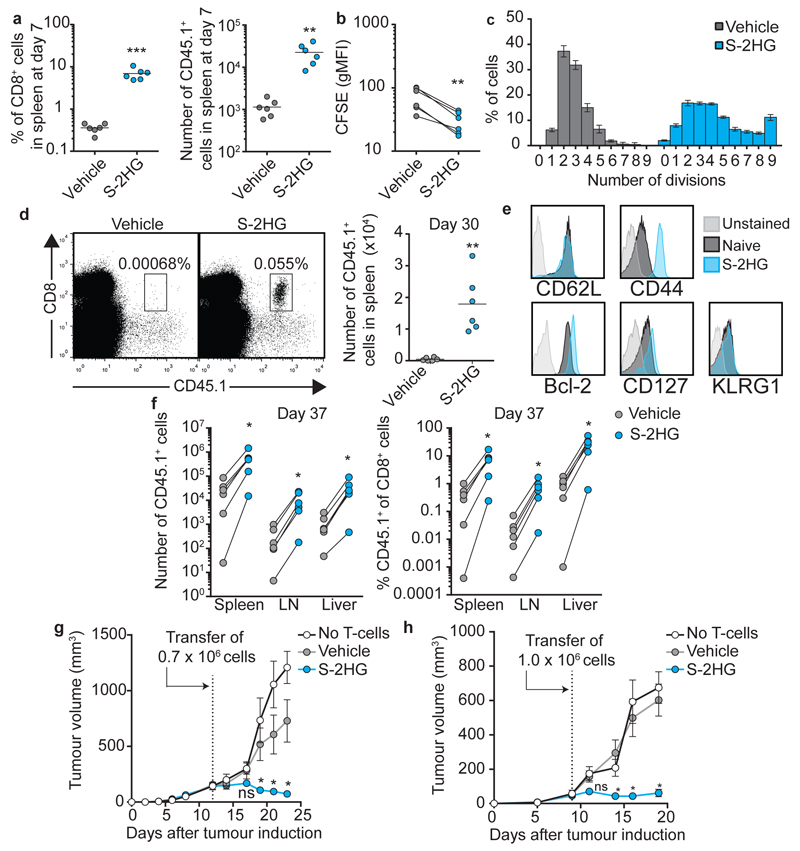
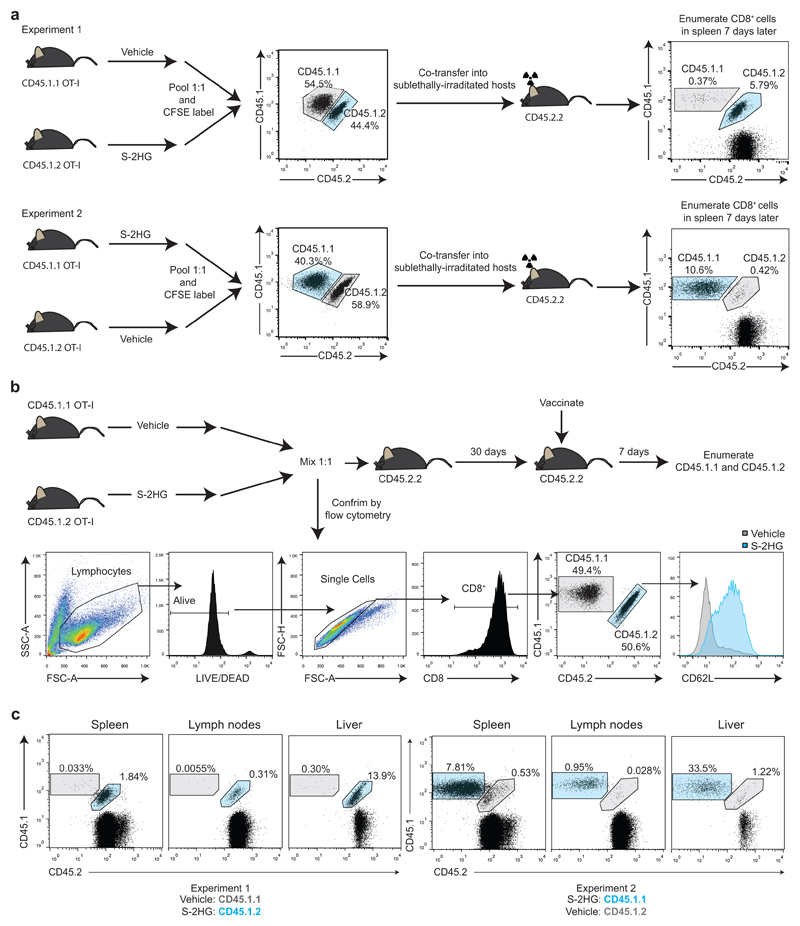
We thus co-transferred CFSE-labelled vehicle and S-2HG treated CD45.1.1 or CD45.1.2 OT-I CD8+ T-lymphocytes into lymphodepleted mice (Extended Data Fig. 6a) to assess their capacity for homeostatic proliferation34,35. S-2HG-treated cells display greater homeostatic proliferation (Fig. 4a, b), with more cells dividing >5 times (Fig. 4c).We then assessed the capacity of S-2HG treated cells to persist in lymphoreplete mice. Adoptively transferred CD45.1 OT-I CD8+ T-lymphocytes, pre-treated with S-2HG, show dramatically enhanced persistence 30 days after transfer (Fig. 4d), expressing elevated CD44, CD127 and Bcl-2 levels relative to naïve cells (Fig. 4e)36,37. In response to vaccination with SIINFEKL-loaded dendritic cells, S-2HG-treated OT-I CD8+ T-lymphocytes robustly recall (Fig. 4f and Extended Data Fig. 6b, c). Consistent with this, OT-I CD8+ T-lymphocytes, pre-treated with S-2HG are more proficient at controlling tumour growth in vivo in both lymphodepleted (Fig. 4g) and lymphoreplete (Fig. 4h) mice. These data demonstrate that S-2HG treatment ex vivo maintains cells in a state with increased proliferative and survival capacity, when transferred in vivo, that is otherwise decreased by effector differentiation.
Figure 4. S-2HG treatment promotes in vivo homeostatic renewal, persistence and anti-tumour capacity of transferred cells.
a/ Recovery of co-transferred CD45.1+ OT-I cells, from spleens (n=6).
b/In vivo CFSE levels in cells from a.
c/ % of cells in a that divided 0-9 times in vivo.
d/ Representative flow cytometry plots and associated statistics of recovered cells from spleens (n=6).
e/ Representative analysis of cells, in d, relative to naïve cells (n=6).
f/ Recovery of co-transferred cells, from spleens, lymphnodes and livers of vaccinated mice (n=6).
g/ Lymphodepleted mice bearing EG7-OVA tumours treated with no T-cells (n=7) or OT-I cells cultured with (n=6) or without (n=6) S-2HG. Error bars denote s.e.m.
h/ Lymphoreplete mice bearing EG7-OVA tumours treated with no T-cells (n=6) or OT-I cells cultured with (n=6) or without (n=6) S-2HG. Error bars denote s.e.m.
Paired t-test (a, b, f), unpaired t-test (d), one-way ANOVA (g, h). Error bars denote s.d. (not in g and h). Each dot in a, b, d and f represents a mouse. ns=non-significant, *p<0.05, **p<0.01, ***p<0.001. gMFI=geometric mean fluorescence intensity.
S-2HG alters methylation in CD8+ T-cells
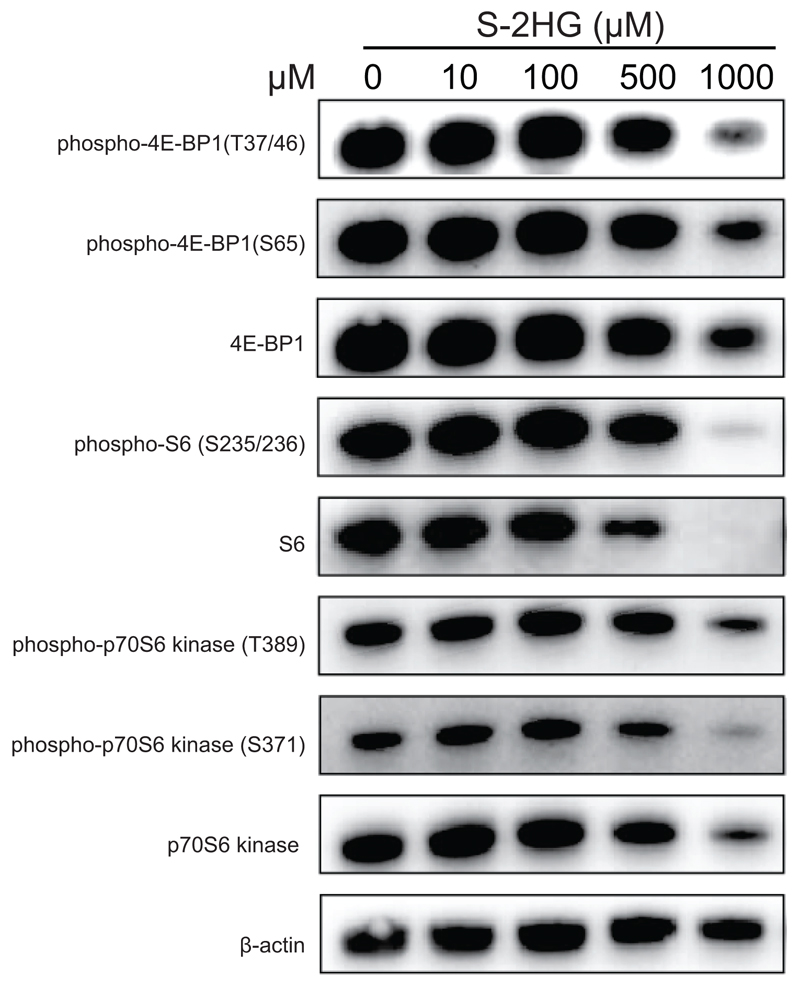
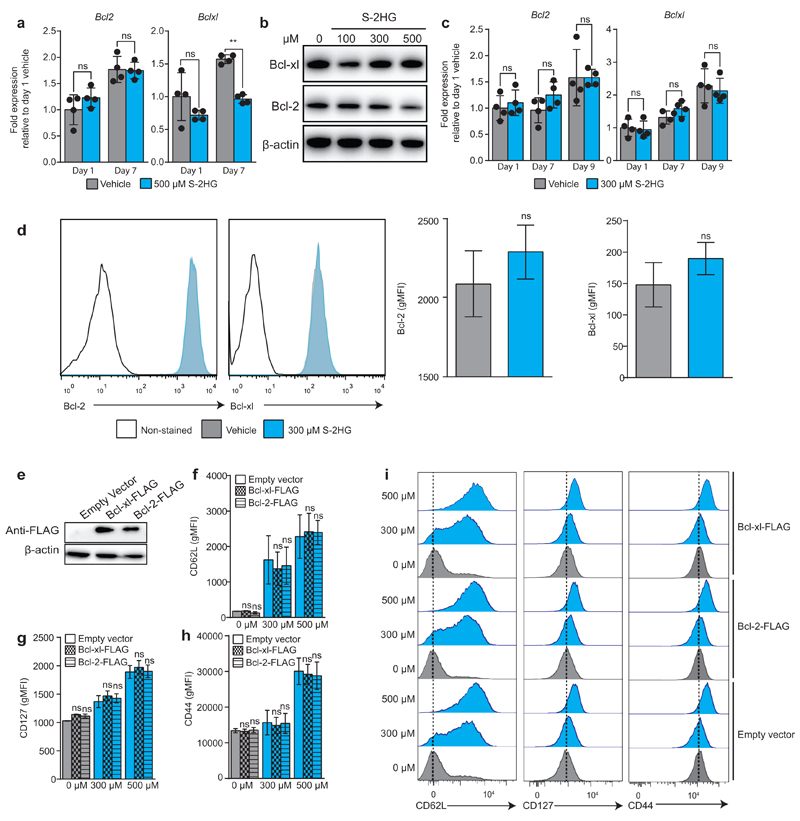
mTOR is a modifier of CD8+ T-lymphocyte differentiation; however, we do not observe mTOR inhibition36,38 at doses of S-2HG needed for this to occur (Extended Data Fig. 7). S-HG treatment ex vivo may be selecting cells that express higher levels of anti-apoptotic genes. Two critical anti-apoptotic genes implicated in CD8+ T-lymphocyte survival are Bcl-2 and Bcl-xl39,40. These genes are not induced by S-2HG treatment (Extended Data Fig. 8a-d). Moreover, over-expression of Bcl-2 or Bcl-xl (Extended Data Fig. 8e) does not influence the expression of CD62L, CD44 or CD127 in the presence or absence of S-2HG (Extended Data Fig. 8f-i), indicating that S-2HG is exerting these phenotypic changes independently of Bcl-2 or Bcl-xl.
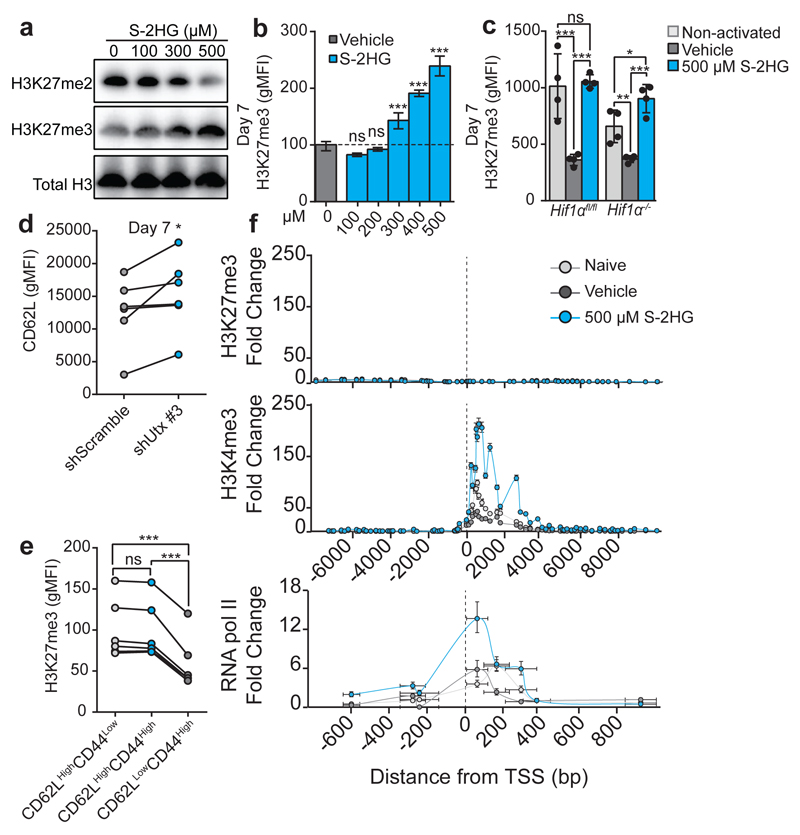
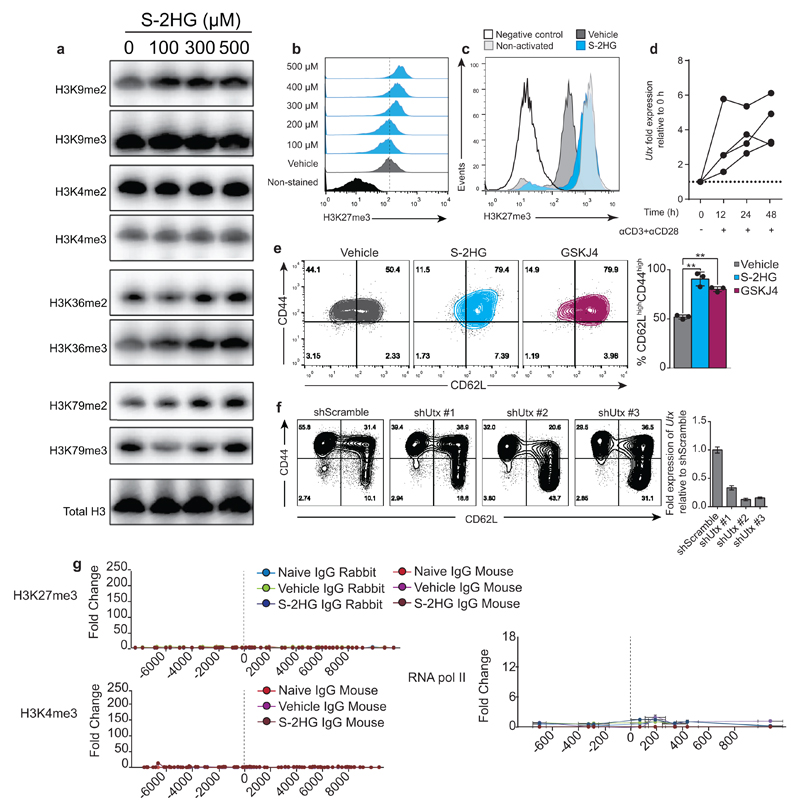
Inhibition of 2-oxoglutarate-dependent dioxygenases that demethylate histones (Jumonji C containing proteins) or oxidise 5-methylcytosine in DNA (Ten-eleven translocation (Tet) proteins) may mediate S-2HG’s effect 32,41–43. S-2HG alters global levels of various histone methylation marks (Extended Data Fig. 9a); in particular, di- and tri-methylation on H3K27 are reciprocally altered, indicating inhibition of H3K27me3 demethylation (Fig. 5a, b and Extended Data 9b). The H3K27me2/3 demethylase, Kdm6a (Utx), is an important regulator of thymocyte differentiation44; changes in H3K27me3 levels correlate with genes associated with regulation of CD8+ T-lymphocyte differentiation11. Global levels of H3K27me3 in CD8+ T-lymphocytes are reduced following activation, but remain high with S-2HG treatment in a HIF-1α-independent manner (Fig. 5c and Extended Data Fig. 9c). Utx is induced following TCR stimulation (Extended Data Fig. 9d) and inhibition of Utx reproduces the effect of S-2HG treatment on CD62L expression (Fig 5d and Extended Data Fig. 9e and f). In vivo, levels of H3K27me3 are highest in central memory (CD62LHighCD44High) and naïve (CD62LHighCD44Low) CD8+ T-lymphocytes, relative to effectors (CD62LLowCD44High) (Fig. 5e). To determine if histone methylation changes occur at the transcription start site (TSS) of CD62L with S-HG treatment, we performed ChIP-PCR for H3K27me3, H3K4me3 and RNA pol II, on naïve and activated CD8+ T-lymphocytes cultured with or without S-2HG (Fig. 5f and Extended Data Fig. 9g). We could find no enrichment for H3K27me3 at or around the TSS. However, naïve and S-2HG treated CD8+ T-lymphocytes have higher enrichment for H3K4me3 at the TSS that is reduced in vehicle-treated cells. Additionally, S-2HG treated CD8+ T-lymphocytes have markedly higher RNA pol II biding than both naïve and vehicle treated cells. Thus S-2HG is promoting CD62L transcription directly with enrichment of H3K4me3 at the TSS and indirectly via preservation of H3K27me3 elsewhere in the genome.
Figure 5. S-2HG alters global H3K27me3 in CD8+ T-lymphocytes.
a/Immunoblot analysis for H3K27me2 and H3K27me3 in cells treated with S-2HG.
b/ H3K27me3 in OT-I cells treated with S-2HG. n=6 mice.
c/ H3K27me3 abundance in Hif1αfl/fl (n=4) and Hif1αfl/fl dlckcre (n=4) cells treated with S-2HG.
d/ CD62L expression by cells with shUtx #3.
e/In vivo H3K27me3 levels in CD8+ populations. n=6 mice.
f/ ChIP-qPCR for H3K4me3, H3K27me and RNA pol II around the TSS for CD62L. A pool of n=6 mice was used. Error bars denote s.e.m.
One-way ANOVA (b, c). Error bars denote s.d. and each dot in c and e represents a mouse. ns=non-significant, *p<0.05, **p<0.01, ***p<0.001. gMFI=geometric mean fluorescence intensity. For immunoblot source images, see Supplementary Fig. 1.
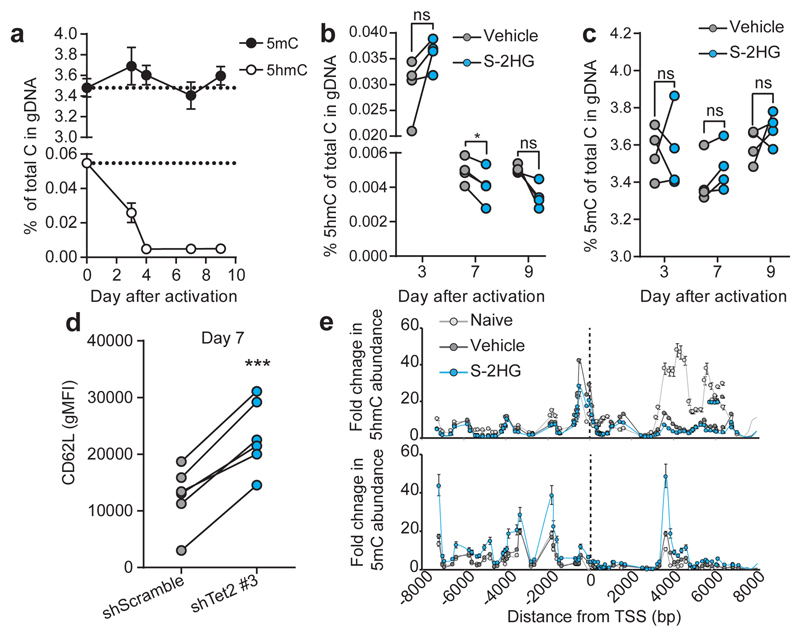
Total levels of 5-methylcytosine (5mC) in genomic DNA are largely unchanged by TCR triggering (Fig. 6a). However, total levels of 5-hydroxymethylcytosine (5hmC) decrease following TCR triggering (Fig. 6a). 5hmC removal in genomic DNA can occur via Tet-mediated oxidation (active) as well as DNA replication (passive)45,46. S-2HG treatment induces small changes in total 5hmC and 5mC in a time-dependent manner (Fig. 6b, c); at day 3, there are marginally higher levels of 5hmC (statistically non-significant) with no changes in 5mC (Fig. 6b, c). This is likely due to decreased proliferation in the presence of S-2HG (Extended Data Fig 5g). Following sustained treatment, at days 7 and 9, cells have less 5hmC and more 5mC (Fig. 6b, c). 5mC changes did not reach statistical significance at any time point; changes in 5hmC were statistically significant at day 7 only. Nonetheless, the changes at days 7 and 9 are consistent with inhibition of Tet proteins in the presence of sustained S-2HG treatment.
Figure 6. S-2HG alters global 5hmC and 5mC in DNA of CD8+ T-lymphocytes.
a/ 5hmC and 5mC in gDNA. n=4 mice.
b/ 5hmC in gDNA from cells treated with S-2HG. n=4 mice.
c/ 5mC in gDNA from cells treated with S-2HG-. n=4 mice.
d/ CD62L expression on CD8+ T-lymphocytes 7 days after transduction with shTet2 #3.
e/ DIP-qPCR for 5mC and 5hmC around the TSS for CD62L. A pool of n=6 mice was used. Error bars denote s.e.m.
One-way ANOVA (b and c). Error bars denote s.d. and each dot in b and c represents a mouse. ns= non-significant, *p<0.05, ***p<0.001. gMFI=geometric mean fluorescence intensity.
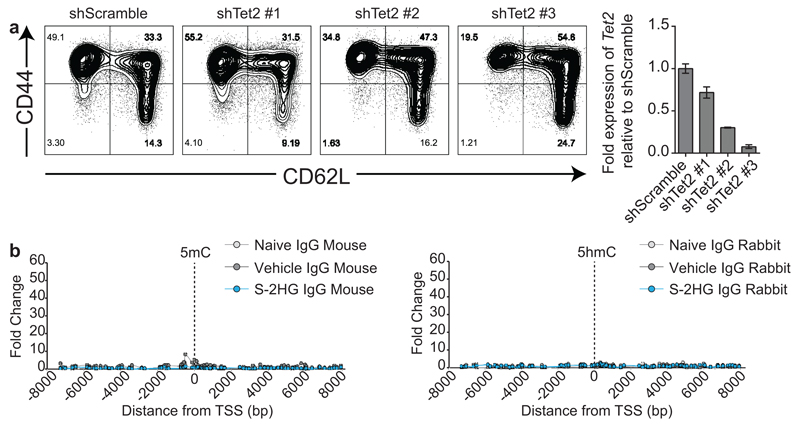
Tet2 regulates CD4+ T-lymphocyte function and is inhibited by 2HG47,48. Knockdown of Tet2 recapitulates the effect of S-2HG on CD62L, indicating that Tet2 also contributes to CD8+ T-lymphocyte effector differentiation (Fig. 6d and Extended Data Fig. 10a), implicating DNA demethylation as an added epigenetic modifier of CD8+ T-cell differentiation12. We performed both 5hmC and 5mC DIP-PCR around the TSS of CD62L (Fig. 6e and Extended Data Fig 10b). Naïve cells have the highest and lowest enrichment for 5hmC and 5mC respectively, while S-2HG treated cells display the opposite pattern. Vehicle treated cells have an intermediate level of these two marks.
Discussion
Our data support a model in which both Utx and Tet2 contribute to effector differentiation of CD8+ T-lymphocytes in vitro. The activity of these epigenetic modifiers is altered by S-2HG in a fashion that inhibits effector differentiation.
TCR-triggering induced loss of 5hmC in gDNA, whereas 5mC levels were relatively stable (Fig. 6a). 5hmC levels stabilized by 4 days after activation (Fig. 6a) and S-2HG treatment produced reciprocal changes in 5mC and 5hmC levels at later time points (Fig. 6b, c). We investigated 5mC and 5hmC presence at and around the TSS of CD62L (Fig. 6e). S-2HG treatment causes reciprocal 5mC increases and 5hmC decreases in this region consistent with inhibition of Tet proteins and transcriptional repression47, yet there is robust expression of CD62L with S-2HG treatment (Fig. 3c, d, e and h and Extended Data Fig. 5m-p). Inhibition of Tet2 maintains CD62L expression (Fig. 6d and Extended Data Figure 10a), indicating that S-2HG-induced DNA methylation changes elsewhere in the genome may indirectly promote CD62L expression.
We also investigated H3K4me3 and H3K27me3 patterns at the TSS of CD62L (Fig. 5f). S-2HG-treated and naïve CD8+ T-lymphocytes have high enrichment for the active H3K4me3 mark49 at the TSS that is lost in vehicle-treated cells. At least in S-2HG treated cells, this is accompanied by binding of RNA-pol II. Despite global increases in H3K27me3 with S-2HG (Fig. 5a-c), the lack of H3K27me3 at the TSS of CD62L is not surprising, as this mark is associated with repression49. Nevertheless, inhibition of Utx promotes CD62L maintenance (Fig. 5d and Extended Data Fig. 9e and f), indicating that H3K27me3 deposition at other genomic sites can indirectly promote CD62L expression. Due to the relationship between histone methylation and other marks such as histone acetylation, modulation of the latter may have similar effects to those seen with S-2HG treatment50.
Adoptively transferred cells treated with S-2HG ex vivo have an increased capacity to proliferate and persist in vivo, with enhanced anti-tumour efficacy (Fig. 4), demonstrating a new strategy to improve persistence of adoptive cell therapies for cancer. The data presented uncover a metabolic-epigenetic axis that controls aspects of T-cell fate. Factors regulating endogenous S-2HG levels, such as HIF signalling, TCR triggering, L2hdgh activity and potentially others, can alter the differentiation of CD8+ T-lymphocytes and thus shape the immune response.
Extended Data
Extended Data Figure 1. VHL-HIF-1α regulate central carbon metabolism and 2HG levels in CD8+ T-lymphocytes.
a/ Illustration of central carbon metabolism in CD8+ lymphocytes, including glycolysis and the tricarboxylic acid cycle, depicting relative levels of detected metabolites between Vhlfl/fl (n=5), Vhlfl/fldlckcre (n=5) and Hif1αfl/fl Vhlfl/fldlckcre (n=3) CD8+ T-lymphocyte groups.
b/ Glucose consumption and lactate production in Vhlfl/fl and Vhlfl/fldlckcre CD8+ T-lymphocytes 7 days after activation with αCD3 and αCD28 antibodies. n=4 mice per genotype from two independent experiments.
c/ Immunoblot analysis for HIF-1α and LaminB1, using nuclear extracts prepared from Vhlfl/fl and Vhl-/- CD8+ T-lymphocytes cultured in 21% oxygen.
d/ Rank of metabolite loadings in PC1 from PCA.
e/ Immunoblot analysis for HIF-1α, HIF-2α and β-tubulin, on whole cell extracts prepared from RCC4 and 786-O renal cancer cell lines, with and without expression of functional VHL.
f/ Deletion efficiency of Vhl in Vhlfl/fl MEFs following infection with Adeno-Cre virus, n=3 individual preparations. Accompanying immunoblot analysis for HIF-1α and β-tubulin, on whole cell extracts.
Two-tailed t-test (b), one-way ANOVA for multiple comparisons (a). Error bars denote s.d. and each dot represents an individual mouse in a and b. *p<0.05, **p<0.01, ***p<0.001. For immunoblot source images, see Supplementary Fig. 1.
Extended Data Figure 2. HIF-1α-dependent metabolic alterations underlie S-2HG production in CD8+ T-lymphocytes.
a/ Example calculation of intracellular 2HG concentration.
b/ 1H-NMR analysis for 2HG from CD8+ T-lymphocytes cultured as in Fig. 2b.
c/ Deletion efficiency of Hif1α or Hif2α in CD8+ T-lymphocytes, isolated from Hif1αfl/fldlckcre or Hif2αfl/fldlckcre mice. n=4 mice.
d/ Total 2HG levels, normalized to viable cell count or protein content, in Hif1αfl/fl and Hif1αfl/fldlckcre CD8+ T-lymphocytes cultured as in Fig. 2b. n=4 mice per genotype.
e/ Total 2HG levels, normalized to viable cells or protein content, in Hif2αfl/fl and Hif2αfl/fldlckcre CD8+ T-lymphocytes cultured as in Fig. 2b. n=4 mice per genotype.
f/ Illustration outlining the workflow for metabolite extraction, deletion efficiency and viability experiments in Hif1αfl/fl, Hif1αfl/fldlckcre, Hif2αfl/fl and Hif2αfl/fldlckcre CD8+ T-lymphocytes. Also shown are viability measurements at day 4. n=4 mice per genotype.
g/ Total amount of 2HG in Hif1αfl/fl (n=6) and Hif1αfl/fldlckcre (n=7) CD8+ T-lymphocytes, at indicated times following activation. n≥4 mice per time point.
h/ Heat map indicating qPCR measurement of expression of enzymes involved in central carbon metabolism in CD8+ T-lymphocytes cultured as in Fig. 2b; n=4 mice per condition.
i/ LC-MS/MS quantification of total intracellular succinate, fumarate and malate levels in CD8+ T-lymphocytes isolated from C57BL/6J mice and cultured as in Fig. 2b; n=7 mice.
j/ Heat map indicating qPCR measurement, in Hif1αfl/fl (n=4) and Hif1αfl/fldlckcre (n=3) CD8+ T-lymphocytes growing in 1% oxygen, of expression of enzymes implicated in the hypoxic production of S-2HG.
k/ qPCR validation of shRNA-knockdowns in CD8+ T-lymphocytes isolated from C57BL/6J mice.
l/ LC-MS/MS quantification of S- and R-2HG in CD8+ T-lymphocytes isolated from C57BL/6J mice, with shRNA-mediated knockdown of Mdh1; n=4 pools of 4 mice per pool.
m/ LC-MS/MS quantification of S- and R-2HG in CD8+ T-lymphocytes isolated from C57BL/6J mice, with shRNA-mediated knockdown of Mdh2; n=4 pools of 4 mice per pool.
n/ LC-MS/MS quantification of R-2HG in CD8+ T-lymphocytes isolated from C57BL/6J mice, with shRNA-mediated knockdown of Ldha; n=4 pools of 4 mice per pool.
o/ Validation of Pdk1-FLAG and Ldha-FLAG expression in Hif1αfl/fldlckcre CD8+ T-lymphocytes by immunoblot analysis for FLAG.
p/ LC-MS/MS quantification of total intracellular glutamate levels in CD8+ T-lymphocytes cultured as in Fig. 2b; n=7 mice.
q-r/ LC-MS/MS quantification of total intracellular glutamate levels in Hif1αfl/fl, Hif1αfl/fldlckcre, Hif2αfl/fl and Hif2αfl/fldlckcre CD8+ T-lymphocytes cultured as in Fig. 2b; n=4 mice per genotype.
s/ Immunoblot of cytosolic fractions for phospho-PDH E1α (S232) and total PDH-E1α in CD8+ T-lymphocytes cultured in 1% oxygen in the presence of the indicated concentration of DCA for 48h.
t/ Total intracellular concentration of 2HG in CD8+ T-lymphocytes from C57BL/6J mice cultured as in Fig. 2b and treated with 5mM DCA for the latter 48h of culture; n=4 mice.
u/ Total intracellular amount of 2HG, nomralized to viable cell count or protein content in CD8+ T-lymphocytes from C57BL/6J mice cultured as in Fig. 2b and treated with 5mM DCA for the latter 48h of culture; n=4 mice.
v/ Total intracellular amount of glutamate in CD8+ T-lymphocytes from C57BL/6J mice cultured as in Fig. 2b and treated with 5mM DCA for the latter 48h of culture; n=4 mice.
Two-way ANOVA for grouped data (d, e, f, q, r, t, u, v). Paired t-test for matched comparisons (i, p), one-way ANOVA for multiple matched comparisons (g, l, m, n). Error bars denote s.d. and each dot represents an individual mouse in g, i and p. ns= non-significant, *p<0.05, **p<0.01, *** p<0.0001. Experiments were performed with indicated numbers of mice from multiple occasions. For immunoblot source images, see Supplementary Fig. 1.
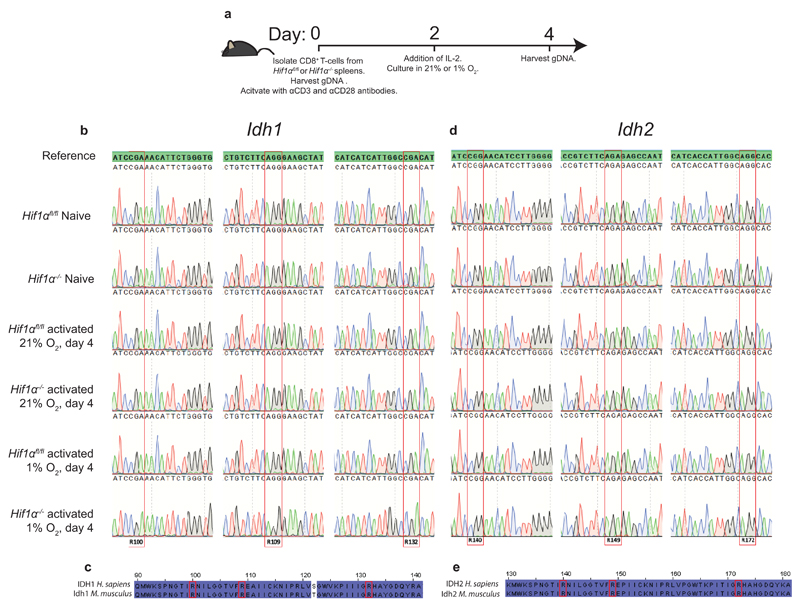
Extended Data Figure 3. Naïve and expanding primary CD8+ T-lymphocytes do not possess mutations in Idh1 or Idh2 that can explain the presence of high levels of 2HG.
a/ Illustration outlining the workflow for mutational analysis of Idh1 and Idh2
b/ Sanger sequencing chromatograms validating the presence of wild type Idh1 as compared to the C57BL/6J NCBI reference sequence.
c/ Alignment of mouse and human IDH1 protein indicating conservation of active site arginine residues.
d/ Sanger sequencing chromatograms validating the presence of wild type Idh2 as compared to the C57BL/6J NCBI reference sequence.
e/ Alignment of mouse and human IDH2 protein indicating conservation of active site arginine residues.
Extended Data Figure 4. Kinetics of 2HG labelling in 21% and 1% oxygen, by U-13C-glucose and U-13C-glutamine.
a/ Proposed mechanism by which HIF-1α controls S-2HG production in CD8+T-lymphocytes and 13C-labelling strategy using U-13C-glucose (m+6) and U-13C-glutamine (m+5) to label endogenous 2HG. Red and green represent pathways promoted and inhibited respectively by HIF-1α in hypoxia.
b/ Isotopologue distribution of 2HG (as a percentage of the total pool) in CD8+ T-lymphocytes, after labelling with U-13C-glucose for 1, 5 and 24 h in both 21% and 1% oxygen conditions; n=3 mice per time point. Error bars denote s.d.
c/ Isotopologue distribution of 2HG (as a percentage of the total pool) in CD8+ T-lymphocytes, after labelling with U-13C-glutamine for 1, 5 and 24 h in both 21% and 1% oxygen conditions; n=3 mice per time point. Error bars denote s.d.
Extended Data Figure 5. S-2HG treatement promotes HIF-1α stability and alters the phenotypic and functional properties of CD8+ T-lymphocytes in a HIF-1α-independent manner.
a-b/ Immunoblot analysis of nuclear and cytosolic fractions, prepared from CD8+ T-lymphocytes cultured in (a) 21% and (b) 1% oxygen, for HIF-1α, HDAC1, phospho-PDH E1α (S232) and total PDH-E1α. Cells were activated for 48 h with αCD3+αCD28 antibodies and then expanded for a further 4 days in the presence of IL-2 followed by treatment with the indicated concentration of S-2HG for 16 hours. The arrow indicates HIF-1α protein.
c/ Glucose consumption and lactate production of C57BL/6J, Hif1αfl/fl (n=12) and Hif1αfl/fldlckcre (n=4) CD8+ T-lymphocytes treated with or without 500 μM S-2HG-octyl ester for 16 hours as in Extended Data Fig. 5a.
d/ VEGF-A production of wild type C57BL/6J, Hif1αfl/fl (n=16) and Hif1αfl/fldlckcre (n=4) CD8+ T-lymphocytes treated with or without 500 μM S-2HG-octyl ester for 16 hours as in Extended Data Fig. 5a.
e/ Representative flow cytometry plots of IFN-γ vs TNF-α in SIINFEKL re-stimulated OT-I CD8+ T-lymphocytes, as a function of increasing doses of S-2HG-octyl ester for 7 days. Associated quantification and statistics are shown in the graphs below.
f/ Specific killing of EG7-OVA cells by OT-I CD8+ T-lymphocytes. Total splenocytes were activated for 48 h with 1000 nM SIINFEKL and then expanded for a further 4 days in the presence of IL-2 followed by treatment with 500 μM S-2HG-octyl ester for 24 h. OT-I CD8+ T-lymphocytes were incubated with target and control cells for 4 hours; n=3 mice per condition.
g/ CFSE dilution assay, with associated statistics (n=4 mice per condition) at day 3 of CD8+ T-lymphocytes activated with αCD3+αCD28 antibodies and cultured with or without 500 μM S-2HG-octyl ester from day 0. Data are representative of 4 mice. Associated quantification and statistics are shown in the graph on the right.
h/ Viability and annexin V assay of CD8+ T-lymphocytes treated with increasing S-2HG doses for 4 days, n=4 mice.
i/ Viability of CD8+ T-lymphocytes cultured with 300 μM S-2HG-octyl ester for the indicated number of days, n=4 mice.
j/ Amount of IFN-γ protein in the media of wild type C57BL/6J, Hif1αfl/fl (n=8) and Hif1αfl/fl dlckcre (n=4) CD8+ T-lymphocytes treated for 24 h with or without 500 μM S-2HG-octyl ester.
k/Viability of Hif1αfl/fl (n=8) and Hif1αfl/fl dlckcre (n=4) OT-I CD8+ T-lymphocytes activated with 1000 nM SIINFEKL peptide and cultured for 7 days with or without 500 μM S-2HG-octyl ester in the absence of IL-2 supplementation from day 0.
l/ Expression of Ifng mRNA in CD8+ T-lymphocytes treated for either 24 h or 7 days with or without 500 μM S-2HG-octyl ester. n=4 mice per group.
m/ CD44 and CD62L surface expression on OT-I CD8+ T-lymphocytes treated with increasing doses of S-2HG for 7 days. Cells were activated with 1000 nM SIINFEKL peptide; n=3 mice. Gated on live, CD8+ cells.
n/ Illustration outlining the workflow for the experiment. Left panel: %CD62LHigh CD8+ T-lymphocytes, treated for 7 days with 500 μM S-2HG-octyl ester, followed by washout or maintenance of the compound and follow up every 3rd day, for 9 more days; n=4 mice. Right panel: %CD62LHigh CD8+ T-lymphocytes, treated for 7 days with vehicle, followed by addition of 500 μM S-2HG-octyl ester or vehicle and follow up every 3rd day, for 9 more days; n=4 mice. Gated on live, CD8+ cells.
o/ CD44 and CD62L surface expression on Hif1αfl/fl and Hif1αfl/fl dlckcre CD8+ T-lymphocytes treated with or without 500 μM S-2HG-octyl ester for 1, 7 and 10 days following treatment. Data are representative of 3 mice per genotype. Gated on live, CD8+ cells.
p/ CD44 and CD62L surface expression on Hif2αfl/fl and Hif2αfl/fl dlckcre CD8+ T-lymphocytes treated with or without 500 μM S-2HG-octyl ester for 1, 7 and 10 days following treatment. Data are representative of 2 mice per genotype. Gated on live, CD8+ cells.
q/ Flow cytometric characterisation of indicated phenotypic markers on Hif1αfl/fl (n=4) and Hif1αfl/fl dlckcre (n=4) CD8+ T-lymphocytes treated for 7 days with 500 μM S-2HG-octyl ester. Gated on live CD8+ cells.
r/ Validation of L2hgdh-FLAG expression in CD8+ T-lymphocytes from C57BL/6J mice by immunoblot analysis for FLAG. The arrow indicates L2hgdh-FLAG protein.
s/ qPCR validation of L2hgdh knockdowns in CD8+ T-lymphocytes isolated from C57BL/6J mice.
t/ CD127 surface expression in response to L2hgdh knockdown (n=4). Representative flow cytometry histogram of CD127 surface levels on transduced (GFP+) CD8+ T-lymphocytes in response to shScramble or shL2hgdh #3 in 21% or 1% oxygen is shown on the right.
u/qPCR quantification of Prdm1, Sell, Eomes, Tcf7, Bcl6 and Ccr6 expression in CD8+ T-lymphocytes treated for 1 or 7 days with or without 500 μM S-2HG-octyl ester .
Paired t-test for matched comparisons (g) and two-way ANOVA for grouped data (c, d, j, k, l, q). One-way ANOVA for multiple comparisons (i, n, t). Error bars denote s.d. and each dot in c, d, g, j, k, l, q, t represents an individual mouse. ns=non-significant, *p<0.05, **p<0.01, ***p<0.001. gMFI=geometric mean fluorescence intensity. Experiments were performed with indicated numbers of mice from multiple occasions. For immunoblot source images, see Supplementary Fig. 1.
Extended Data Figure 6. Ex vivo treatment of CD8+ T-lymphocytes with S-2HG promotes in vivo homeostatic proliferation and recall of adoptively transferred cells.
a/ Diagram outlining the homeostatic proliferation experiments in Figure 4a-c. Representative flow cytometry plots are shown for each pool before and after adoptive transfer. Flow cytometry plots show viable CD8+ cells.
b/Diagram outlining the recall experiments in Figure 4f.
c/ Representative flow cytometry plots of recalling CD45.1+ CD8+ T-lymphocytes in indicated organs on day 7 post vaccination (day 37 post transfer).
Extended Data Figure 7. S-2HG does not inhibit mTOR signalling at the doses necessary for the formation of memory-like CD8+ T-lymphocytes.
Immunoblot analysis on cytosolic extracts for mTOR signalling in CD8+ T-lymphocytes treated with the indicated doses of S-2HG for 24 h. For immunoblot source images, see Supplementary Fig. 1.
Extended Data Figure 8. S-2HG does not induce Bcl-2 or Bcl-xl that can explain the in vivo persistence of adoptively transferred CD8+ T-lymphocytes.
a/qPCR quantification of Bcl2 and Bclxl mRNA levels in response to 500 μM S-2HG-octyl ester treatment for either 1 or 7 days. n=4 mice
b/Immunnoblot analysis for Bcl-2 and Bcl-xl protein in response to increasing doses of S-2HG-octyl ester for 9 days.
c/qPCR quantification of Bcl2 and Bclxl mRNA levels in response to 300 μM S-2HG-octyl ester treatment for either 1, 7 or 9 days. n=4 mice
d/Representative flow cytometry histograms of Bcl-2 and Bcl-xl abundance in CD8+ T-lymphocytes treated with 300 μM S-2HG-octyl ester for 9 days. Quantification and associated statistics are shown in the graph on the right. n=3 mice.
e/Immunoblot analysis confirming the expression of Bcl-xl-FLAG and Bcl-2-FLAG in OT-I in CD8+ T-lymphocytes.
f-h/CD62L (f), CD127 (g) and CD44 (h) surface expression in OT-I CD8+ T-lymphocytes transduced with retrovirus expressing either Bcl-2-FLAG or Bcl-xl-FLAG and treated with the indicated concentration of S-2HG-octyl ester for 7 days. n=2 mice
i/ Representative flow cytometry histograms of CD62L, CD127 and CD44 surface expression in OT-I CD8+ T-lymphocytes transduced with retrovirus expressing either Bcl-2-FLAG or Bcl-xl-FLAG and treated with the indicated concentration of S-2HG-octyl ester for 7 days. The associated statistics of these flow cytometry data are shown in f, g and h. **P<0.01, ns= non-significant.
Paired t-test for matched comparisons (d) and two-way ANOVA for grouped data (a, b). One-way ANOVA of matched samples for multiple comparisons (c, f, g, h). Error bars denote s.d. and each dot in a and c represents an individual mouse. ns=non-significant, **p<0.01, gMFI=geometric mean fluorescence intensity. Experiments were performed with indicated numbers of mice from at least two occasions. For immunoblot source images, see Supplementary Fig. 1.
Extended Data Figure 9. S-2HG induces global histione H3 methylation changes in CD8+ T-lymphocytes.
a/ Immunoblot analysis on nuclear extracts for histone H3 methylation marks in activated CD8+ T-lymphocytes treated with the indicated doses of S-2HG for 7 days.
b/Representative flow cytometry histograms of H3K27me3 staining as a function of increasing S-2HG-octyl ester concentration.
c/ H3K27me3 staining in CD8+ T-lymphocytes treated with or without 500 μM S-2HG-octyl ester and stained with or without fluorophore conjugated C36B11 antibody.
d/ qPCR measurement for expression of Utx in unstimulated and stimulated CD8+ T-lymphocytes; n=4 mice. Expression for Utx is displayed for each mouse individually.
e/ Represetative flow cytomery plots of CD44 vs CD62L expression, with associated statistics, on activated CD8+ T-lymphocytes after 4 days of treatment with 500 μM S-2HG-octyl ester or 1 µM GSKJ4. Gated on live, CD8+ cells. n=3 mice.
f/ Representative flow cytometry plots of CD44 vs CD62L expression on CD8+ T-lymphocytes with shRNA-mediated knockdown of Utx, 7 days after trasnduction. Gated on live, CD8+GFP+ cells. Graph on right shows knockdown hairpin fidelity for Utx.
g/ IgG control ChIP-qPCR for H3K4me3, H3K27me and RNA pol II at and around the TSS for CD62L, in freshly isolated naïve or activated CD8+ T-lymphocytes treated with or without 500 μM S-2HG-octyl ester for 7 days. Each profile shows the fold change over the non-binding control primer. Each dot represents an individual primer pair. A pool of n=6 mice was used for each condition. Error bars denote s.e.m.
One-way ANOVA for multiple matched comparisons (e). Each dot in c represents an individual mouse. Error bars denote s.d. **p<0.01. Experiments were performed with indicated numbers of mice from at least two occasions. For immunoblot source images, see Supplementary Fig. 1.
Extended Data Figure 10. S-2HG induces global changes in the content of 5hmC and 5mC in genomic DNA of CD8+ T-lymphocyte genomic DNA.
a/ Representative flow cytometry plots of CD44 vs CD6L expression on CD8+ T-lymphocytes with shRNA-mediated knockdown of Tet2, 7 days after trasnduction. Gated on live, CD8+GFP+ cells. Graph on right shows knockdown hairpin fidelity for Tet2.
b/ IgG control DIP-qPCR for 5mC and 5hmC at and around the TSS for CD62L, in freshly isolated naïve or activated CD8+ T-lymphocytes treated with or without 500 μM S-2HG for 7 days. Each profile shows the fold change over the non-binding control primer. Each dot represents an individual primer pair. A pool of n=6 mice was used for each condition. Error bars denote s.e.m.
Supplementary Material
Supplemental information is linked to the online version of the paper at www.nature.com/nature
Methods can be found in the Supplementary Information which is linked to the online version of the paper at www.nature.com/nature
Acknowledgements
We thank the Cambridge Institute PK Bioanalytics Core facility for MS measurements; Andy Cowburn, Alex Wood, Katharine Lodge and Edwin Chilvers for human PBMCs; Ben Jaggs for help with mouse work. P.A.T. was funded by CRUK, the MRC (1495954) and Wellcome Trust. J.R.G. was funded by CRUK. A.P. was funded by Marie-Curie IEF. A.T.P. was funded by UCSD NIH Grant (5T32GM007240-36). A.W.G. was funded by the NIH (A1096852, A1072117), Leukemia and Lymphoma Society, and Pew Scholars Fund. K.L.L., J.Y., G.S.C. and L.P. were funded by the Singapore National Research Foundation and Singapore Ministry of Education, the NMRC Clinician Scientist (NMRC/CIRG/1389/2014.) and the Swedish Research Council. R.S.J. and co-workers are funded by the Wellcome Trust (grant WT092738MA), the Swedish Cancer Foundation (Cancerfonden), and the Swedish Research Council (Vetenskapsrådet).
Footnotes
Author Contributions
P.A.T. and A.P. carried out metabolic analyses and ex vivo and in vivo T cell experiments, analysed the data, and wrote the manuscript. A.T.P., A.D., A.W.G. and R.S.J. carried out the initial metabolome survey. D.M, P.V., J.S., A.A., and C.E.E. aided in experimental execution and analysis. K.L.L. G.S.C. J. Y. and L.P. carried out ChIP and DIP experiments. J.R.G, K.L.L., L.P., and A.W.G. designed experiments and analysed data. R.S.J. designed experiments, analysed data, wrote the manuscript and administered the project.
This work is dedicated to the memory of Lorenz Poellinger.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no conflict of interest.
References
- 1.Ho PC, et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang CH, et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oestreich KJ, et al. Bcl-6 directly represses the gene program of the glycolysis pathway. Nat Immunol. 2014;15:957–964. doi: 10.1038/ni.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annual review of immunology. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Sullivan D, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crompton JG, et al. Lineage relationship of CD8 T cell subsets is revealed by progressive changes in the epigenetic landscape. Cell Mol Immunol. 2015 doi: 10.1038/cmi.2015.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harland KL, et al. Epigenetic plasticity of Cd8a locus during CD8(+) T-cell development and effector differentiation and reprogramming. Nat Commun. 2014;5:3547. doi: 10.1038/ncomms4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araki Y, et al. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30:912–925. doi: 10.1016/j.immuni.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russ BE, et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity. 2014;41:853–865. doi: 10.1016/j.immuni.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palazon A, Goldrath AW, Nizet V, Johnson RS. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41:518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol. 2013;14:1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finlay DK, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clever D, et al. Oxygen Sensing by T Cells Establishes an Immunologically Tolerant Metastatic Niche. Cell. 2016;166:1117–1131 e1114. doi: 10.1016/j.cell.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghesquiere B, Wong BW, Kuchnio A, Carmeliet P. Metabolism of stromal and immune cells in health and disease. Nature. 2014;511:167–176. doi: 10.1038/nature13312. [DOI] [PubMed] [Google Scholar]
- 18.Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saha SK, et al. Mutant IDH inhibits HNF-4alpha to block hepatocyte differentiation and promote biliary cancer. Nature. 2014;513:110–114. doi: 10.1038/nature13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Intlekofer AM, et al. Hypoxia Induces Production of L-2-Hydroxyglutarate. Cell metabolism. 2015;22:304–311. doi: 10.1016/j.cmet.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oldham WM, Clish CB, Yang Y, Loscalzo J. Hypoxia-Mediated Increases in l-2-hydroxyglutarate Coordinate the Metabolic Response to Reductive Stress. Cell metabolism. 2015;22:291–303. doi: 10.1016/j.cmet.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullen AR, et al. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7:1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel KP, et al. Diagnostic testing for IDH1 and IDH2 variants in acute myeloid leukemia an algorithmic approach using high-resolution melting curve analysis. J Mol Diagn. 2011;13:678–686. doi: 10.1016/j.jmoldx.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang DJ, et al. Selective expression of the Cre recombinase in late-stage thymocytes using the distal promoter of the Lck gene. Journal of immunology. 2005;174:6725–6731. doi: 10.4049/jimmunol.174.11.6725. [DOI] [PubMed] [Google Scholar]
- 27.Gibson KM, et al. Stable-isotope dilution analysis of D- and L-2-hydroxyglutaric acid: application to the detection and prenatal diagnosis of D- and L-2-hydroxyglutaric acidemias. Pediatr Res. 1993;34:277–280. doi: 10.1203/00006450-199309000-00007. [DOI] [PubMed] [Google Scholar]
- 28.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fendt SM, et al. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat Commun. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajagopalan KN, et al. Metabolic plasticity maintains proliferation in pyruvate dehydrogenase deficient cells. Cancer Metab. 2015;3:7. doi: 10.1186/s40170-015-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koivunen P, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arsenio J, et al. Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nat Immunol. 2014;15:365–372. doi: 10.1038/ni.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldrath AW, et al. Cytokine requirements for acute and Basal homeostatic proliferation of naive and memory CD8+ T cells. J Exp Med. 2002;195:1515–1522. doi: 10.1084/jem.20020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murali-Krishna K, et al. Persistence of memory CD8 T cells in MHC class I-deficient mice. Science. 1999;286:1377–1381. doi: 10.1126/science.286.5443.1377. [DOI] [PubMed] [Google Scholar]
- 36.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grayson JM, Zajac AJ, Altman JD, Ahmed R. Cutting edge: increased expression of Bcl-2 in antigen-specific memory CD8+ T cells. Journal of immunology. 2000;164:3950–3954. doi: 10.4049/jimmunol.164.8.3950. [DOI] [PubMed] [Google Scholar]
- 38.Fu X, et al. 2-Hydroxyglutarate Inhibits ATP Synthase and mTOR Signaling. Cell metabolism. 2015 doi: 10.1016/j.cmet.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurtulus S, et al. Bcl-2 allows effector and memory CD8+ T cells to tolerate higher expression of Bim. Journal of immunology. 2011;186:5729–5737. doi: 10.4049/jimmunol.1100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song A, Tang X, Harms KM, Croft M. OX40 and Bcl-xL promote the persistence of CD8 T cells to recall tumor-associated antigen. Journal of immunology. 2005;175:3534–3541. doi: 10.4049/jimmunol.175.6.3534. [DOI] [PubMed] [Google Scholar]
- 41.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manna S, et al. Histone H3 Lysine 27 demethylases Jmjd3 and Utx are required for T-cell differentiation. Nat Commun. 2015;6:8152. doi: 10.1038/ncomms9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachman M, et al. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nature chemistry. 2014;6:1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ichiyama K, et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015;42:613–626. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Zhang F, et al. Epigenetic manipulation restores functions of defective CD8(+) T cells from chronic viral infection. Mol Ther. 2014;22:1698–1706. doi: 10.1038/mt.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.