Abstract
We describe a new class of plant small heat stress proteins (sHsps) with dominant nuclear localization (Hsp17-CIII). The corresponding proteins in tomato, Arabidopsis, and rice are encoded by unique genes containing a short intron in the β4-encoding region of the α-crystallin domain (ACD). The strong nuclear localization results from a cluster of basic amino acid residues in the loop between β5 and β6 of the ACD. Using yeast 2-hybrid tests, analyses of native complexes of the sHsps, and immunofluorescence data, we demonstrate that, in contrast to earlier observations (Kirschner et al 2000), proteins of the sHsp classes CI, CII, and CIII interact with each other, thereby influencing oligomerization state and intracellular localization.
INTRODUCTION
The small heat stress proteins (sHsps) represent a ubiquitous family of stress proteins that range in size from 14 to 42 kDa. This diverse family of proteins, which includes the α-crystallin proteins of the vertebrate eye lens, is defined by a conserved C-terminal domain of approximately 90 amino acid residues, referred to as the α-crystallin domain (ACD), which is flanked by an N-terminal domain and a short C-terminal extension (Caspers et al 1995; Waters et al 1996; de Jong et al 1998; MacRae 2000; Scharf et al 2001; Haslbeck 2002; Narberhaus 2002). In contrast to the ACD, the N-terminal domains of sHsps show no sequence conservation, except between evolutionarily closely related members of the family.
Plants are characterized by an unusual complexity of sHsps, which evidently have evolved independently after the divergence of plants and animals (Waters et al 1996; de Jong et al 1998). In contrast to other organisms, plants are unique in expressing a multiplicity of cytosolic sHsps and, in addition, specific isoforms targeted to intracellular organelles. Based on the sequence comparison of sHsps in the Arabidopsis genome, there are at least 2 forms of sHsps in the nucleocytoplasmic compartment, referred to as class CI and class CII proteins (Vierling 1991; Scharf et al 2001). They share only ∼50% identity in the ACD and are estimated to have diverged over 400 million years ago (Waters and Vierling 1999). So far, 3 additional gene subfamilies encode mitochondrial (M), plastidial (P), and endoplasmic reticulum (ER)–localized sHsps, each with the appropriate organelle-targeting signals. Finally, the Arabidopsis genome encodes 7 sHsp-related proteins whose intracellular localization and role in the sHsp network remain to be elaborated (Scharf et al 2001).
An important feature of sHsps is the formation of homooligomeric complexes usually in the range of 200–750 kDa with 9 to >24 subunits. Frequently, the oligomeric complexes of the plant sHsps are in the range of 200–250 kDa, and they are probably built of 12 subunits (Chen et al 1994; Jinn et al 1995; Lee et al 1995, 1997; Ehrnsperger et al 1997, 1999; Helm et al 1997; Kirschner et al 2000; van Montfort et al 2001). The plant class CI and class CII sHsps form class-specific homooligomers (Lee et al 1995, 1997; Helm et al 1997; Kirschner et al 2000). However, detailed analyses of the oligomers of the class CI proteins showed that depending on the plant investigated (soybean, rice, pea, and mung bean), the proteins may actually be composed of 8 or more isoforms (Jinn et al 1995). This observation reflects the multiplicity of genes encoding class CI and class CII sHsps in plants.
Under heat stress conditions, a rapid reorganization of the sHsp oligomers is observed, which facilitates association with denatured proteins (Giese and Vierling 2002; Sobott et al 2002). Concomitantly, the assembly of large cytoplasmic multichaperone complexes built of 40-nm particles (heat stress granules [HSG]) is observed (Nover et al 1983; Neumann et al 1984). Although formed mainly of class CI and class CII sHsps, they include a number of other proteins (Nover et al 1989; Scharf et al 1998; Kirschner et al 2000).
In the following, we present experimental data supporting the existence of a third class of nucleocytoplasmic sHsps (class CIII) as defined earlier from sequence comparison (Scharf et al 2001). Because of a cluster of basic amino acid residues in the loop between β5 and β6 of the ACD (nuclear localization signal [NLS]), class CIII proteins are predominantly localized in the nucleus, but as a result of interaction with class CII sHsps, they can be recruited to the cytoplasmic chaperone complexes. Using expressed sequence tag (EST) database search, we identified clones encoding the corresponding proteins in different plants, including Arabidopsis, tomato, and rice.
MATERIALS AND METHODS
General materials and methods
The use of either tobacco (Nicotiana plumbaginifolia) or tomato (Lycopersicon esculentum) mesophyll protoplasts and Chinese Hamster ovary (CHO) cells for transient expression of plant Hsps and of antisera against sHsps classes CI and CII was described previously (Lyck et al 1997; Scharf et al 1998; Kirschner et al 2000; Mishra et al 2002). Polyclonal antibodies against LpHsp16.1-CIII were raised in guinea pig (Eurogentec, Seraing, Belgium) by immunization with purified GST-Hsp16.1-CIII fusion protein overexpressed in Escherichia coli BL21DE3. For immunodetection of 3HA-tagged proteins, the monoclonal HA antibody, clone 16B12 (Hiss Diagnostics, Freiburg, Germany), was used. Secondary antibodies conjugated with horseradish peroxidase or fluorescent dyes CY2 or CY3 were obtained from Sigma-Aldrich (Deisenhofen, Germany) or Dianova (Hamburg, Germany), respectively.
For 5′–random amplification of complementary deoxyribonucleic acid (cDNA) ends (RACE), we used a cDNA library generated from heat-stressed tomato cells (Bharti et al 2000) and the MARATHON RACE kit (Clontech Laboratories, Palo Alto, CA, USA). For polymerase chain reaction (PCR) amplification and cloning of a full-length cDNA of Hsp16.1-CIII, the adapter-specific primer AP1 was combined with the gene-specific primer Pr549R, which binds in the 3′–untranslated region of Hsp16.1-CIII cDNA. PCR fragments were cloned into pBluescript II+ (Table 1) by using introduced restriction sites NotI (primer AP1) and XbaI (Pr549R). The database accession number for the complete cDNA sequence is AF399821.
Table 1.
Vectors used for cloning and expression of sHsps
Nucleic acid analysis by reverse transcriptase (RT)–PCR and Southern and Northern hybridization and protein analysis by immunoblotting were performed as described previously (Mishra et al 2002).
The following primers were used for the generation of Hsp16.1-CIII–specific probes, for RT-PCR, and for diagnostic PCR reactions, as shown in Figure 1C: Pr520R, 5′-CTTAACATAAGGAGTAAAAGTGCC-3′; Pr547F, 5′-GAGCCAACTTCTCTTTCCAGAATC-3′; PR548R, 5′-CTCTTCCCGTTGCTTCGTATCACC-3′; Pr976F, 5′-CAATCTTGAAAATGAGCACTGTTG-3′; and Pr977R, 5′-AAAATTAATAATACTAAGTTAAATACAC-3′.
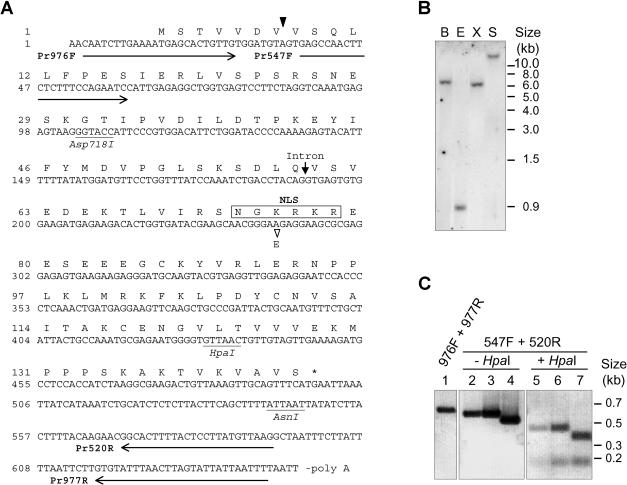
Fig 1.
Structure of tomato Hsp16.1-CIII. (A) The sequence represents the complementary deoxyribonucleic acid (cDNA) clone of LpHsp16.1-CIII including the 5′-random amplification of cDNA ends–derived extension and the open reading frame encoding amino acids 1 to 144. The arrowhead between codon 7 and 8 points to the 5′ end of the cDNA clone rescued by the yeast 2-hybrid screening. The position of the intron insertion at the genomic level is indicated by an arrow. The basic cluster of amino acid residues forming the putative nuclear localization signal, including the amino acid residue exchange K75E, is boxed. Arrows underlining the cDNA sequence correspond to the position and orientation of primers used for the polymerase chain reaction (PCR) analysis shown in (C). (B) Southern blot of genomic DNA from Lycopersicon peruvianum confirms the existence of a single Hsp16.1-CIII gene in tomato. B, BamHI; E, EcoRI; S, SalI; X, XbaI. (C) PCR analysis of genomic DNA (lanes 1 and 2), plasmid DNA containing an Hsp16.1-CIII cDNA clone including the 63-bp intron (MSA2, lanes 3 and 5), or the cDNA clone corresponding to the mature messenger ribonucleic acid of Hsp16.1-CIII (MSA3, lanes 4 and 7). The identity of the PCR products (lanes 2 to 4) was confirmed by restriction analysis using the diagnostic HpaI site (lanes 5–7). Numbers on top refer to primer pairs used for amplification of the corresponding PCR products (for primer sequences see Materials and Methods, and for their positioning in the Hsp16.1-CIII sequence see [A]). Size differences between fragments in lanes 3 and 4 or in lanes 6 and 7 correspond to the short intron of 63 bp
Expression plasmids for plant and mammalian cells
Standard procedures were used for cloning (Ausubel et al 1993; Sambrook and Russel 2001). PCR fragments for subcloning were amplified by using the Taq Plus Precision System (Stratagene, Amsterdam, Netherlands). Plant expression vectors were based on the pRT series of vectors (Töpfer et al 1988) and mammalian expression vectors on pcDNA3. For expression of Hsp16.1-CIII with N-terminal triple HA tag, pRT104 Neo was modified by insertion of a PCR-amplified 3HA fragment. The PCR was carried out on template plasmid pFA6a-3HA-kanMX6 (Longtine et al 1998) with forward primer Pr329F and reverse primer Pr330R (for details of primer sequences see below). The amplicon was cut with the appropriate restriction enzymes and ligated into NcoI-KpnI linearized pRT104. All vectors used in this study are summarized in Table 1.
The following primers were used to produce PCR fragments with appropriate restriction sites (underlined nucleotides) for subcloning (Table 1): Pr329F (NcoI), 5′-CTCTCTCTCCATGGTCTTTTACCCATACGATGTTCC-3′; Pr330R (KpnI), 5′-ATATATATGGTACCTGAGCAGCGTAATCTGGAACG-3′; Pr549R (XbaI), 5′-AATTAGCCTCTAGATAAGGAGTAAAAGTGCC-3′; Pr663F (EcoRI, NcoI), 5′-GGTAACAGAATTCACCATGGGCACTGTTG-3′; and Pr715F (XhoI), 5′-GGAGAGGACCCTCGAGGGCCACCATGG-3′.
For PCR-mediated mutagenesis K75E within the NLS of Hsp16.1-CIII, the forward primer Pr1006F (PvuI), 5′-GTGATACGATCGAACGGGGAGAGGAAG-3′, was used in combination with Pr549R. For complementation of the full-length open reading frame encoding the Hsp16.1-CIII K75E in frame to an N-terminal triple HA tag, the mutagenized PCR fragment was used in a second amplification step in combination with Pr715F on plasmid MS2 (Table 1) as template. The resulting DNA fragment was cut with the appropriate restriction sites and ligated into XhoI-XbaI linearized pRT104.
Yeast plasmids and 2-hybrid screening
The 2 μ vectors pADGal4 (Gal4p-AD amino acids 768 → 881, LEU2) and pBDGal4 (Gal4p-DBD amino acids 1 → 147, TRP1) were obtained from Stratagene. Generation of the tomato cDNA library in the yeast pADGal4 vector was described (Scharf et al 1998). Using pBDGal4×PsHsp18.1-CI as bait (Kirschner et al 2000), we obtained among the 11 positive clones 6 pAD vectors harboring LpHsp17-CI cDNAs and 1 containing a partial cDNA fragment encoding Hsp16.1-CIII (amino acids 8–144) as preys.
Two-hybrid interaction studies were performed by cotransformation of both 2-hybrid expression plasmids and selection of cotransformants on medium lacking leucine and tryptophan. The cotransformants were tested for histidine prototrophy.
Subcellular localization of proteins in protoplasts and CHO cells
For indirect immunofluorescence of protoplasts and CHO cells, we followed the procedures described by Scharf et al (1998) and Heerklotz et al (2001) respectively. For microscopic analysis, a Zeiss Axiophot microscope (Zeiss, Oberkochen, Germany) combined with an Olympus DP10 Photo System (Olympus, Hamburg, Germany) was used. Captured images were resized and combined using Photoshop 5.5 software (Adobe Systems, La Jolla, CA, USA). Confocal laser scan micrographs were captured using a Leica CLSM (Leica, Bensheim, Germany) and Imaris software (Bitplane, Zürich, Switzerland).
Protein analysis of tomato cell cultures
Heat stress treatments of tomato cell suspension cultures were performed by incubation in a water bath at the indicated temperatures (Scharf et al 1998). Control samples were kept at 25°C. For protein extraction, harvested cells were ground in liquid nitrogen. One hundred milligrams of the frozen cell powder was transferred into microfuge tubes and sonified for 4 × 15 seconds with an interval of 15 seconds after each cycle (Sonopuls, Bandelin Electronics, Berlin, Germany) in 200 μL of NEB500 buffer containing 25 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.5, 500 mM NaCl, 5 mM MgCl2, 1 mM ethylenediamine–tetraacetic acid (EDTA), 10 mM NaF, 0.2% NP40, and 10% glycerol. For all buffers used for protein extractions in this study, Complete™ protease inhibitor cocktail tablets were added as recommended by the manufacturer (Roche Diagnostics, Mannheim, Germany). One volume of protein extract containing 30 μg of the protein was heated with 1 volume of 2× sodium dodecyl sulfate (SDS) sample buffer and separated on 14% SDS–polyacrylamide gels. For immunoblot analysis, proteins were transferred to 45-μm nitrocellulose membrane (PROTRAN BA85, Schleicher and Schuell, Dassel, Germany) and processed for chemiluminescence detection, according to the manufacturer's protocol (NEN, Life Science Products, Köln, Germany).
Native polyacrylamide gel electrophoresis
Protein extracts from protoplasts in nondenaturing sample buffer (50 mM Tris-HCl, pH 7.8, 25 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 14.2 mM β-mercaptoethanol, 5% glycerol, and proteinase inhibitors) were prepared and separated on native 3–20% polyacrylamide pore exclusion gels as described before (Kirschner et al 2000).
Gel filtration chromatography
After overexpression of indicated combinations of sHsps, samples of 500 000 protoplasts were lysed in 200 μL high-salt extraction buffer (50 mM Tris-HCl, pH 7.8, 500 mM NaCl, 25 mM KCl, 5 mM MgCl2, 30 mM EDTA, 0.5% NP40, 0.2% sarcosyl, 5% saccharose, 5% glycerol, 14.2 mM β-mercaptoethanol, and proteinase inhibitors). After centrifugation for 5 minutes at 10 000 × g at 4°C, 100 μL of the supernatant was injected on a Superdex 200 HR30/10 filtration column (Amersham Biosciences, Freiburg, Germany). Separation was performed at 4°C with elution buffer (20 mM Tris-HCl, pH 7.8, 500 mM NaCl, 0.5% NP40) at a flow rate of 0.4 mL/min. Fractions of 0.8 mL were collected, and after acetone precipitation, the distribution of sHsps was analyzed by SDS–polyacrylamide gel electrophoresis and immunoblot detection. Corresponding chemiluminescence signals were quantified by using the Image 1D software (Amersham Biosciences). The following molecular mass standards were used: thyroglobin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; lactate dehydrogenase, 140 kDa; and bovine serum albumin, 67 kDa.
RESULTS
Sequence and genomic structure
Using the yeast 2-hybrid screening with a tomato cDNA library in the prey and pea Hsp18.1-CI in the bait position, we recovered a cDNA fragment encoding a new type of sHsp distantly related to class CII. The cDNA clone, truncated at the 5′ end, was complemented by 5′-RACE (Fig 1A). The full-length cDNA encodes a basic protein of 144 amino acid residues with a pI of 8.45 (Hsp16.1-CIII). In contrast to the multiplicity of genes encoding the other 2 classes of cytosolic sHsps, the CIII genes in tomato and Arabidopsis are unique (see Southern blot analysis of tomato genomic DNA in Fig 1B). The PCR analyses with different primer combinations (Fig 1C) confirmed the existence of the Hsp16.1-CIII–encoding gene in the tomato genome, including the 5′ end added by the 5′-RACE and the short intron. A second cDNA clone was derived from the 5′-RACE, which contained additional 63 bp corresponding to the intron in the genomic sequence (Fig 1C).
Based on sequence comparison with other members of the classes CI and CII of cytosolic sHsps (Scharf et al 2001) and a number of properties outlined subsequently in this article, the tomato Hsp16.1 belongs to a new class (class CIII) of sHsps. The corresponding orthologs from other plants were identified from genomic sequences of Arabidopsis and rice or from the analysis of corresponding EST libraries (Discussion). The data from the 2 genomic sequences confirm the existence of a short intron found in the same position as that in the tomato Hsp16.1-CIII gene. Because of the considerable variation in the number of N-terminal amino acid residues preceding the ACD, the size of the proteins in this new class of plant sHsps ranges from 16.1 kDa for tomato to 19.1 kDa for the barley protein. Subsequently, we will collectively use the term Hsp17-CIII for sHsps belonging to this new class.
Expression of Hsp16.1-CIII is heat stress dependent
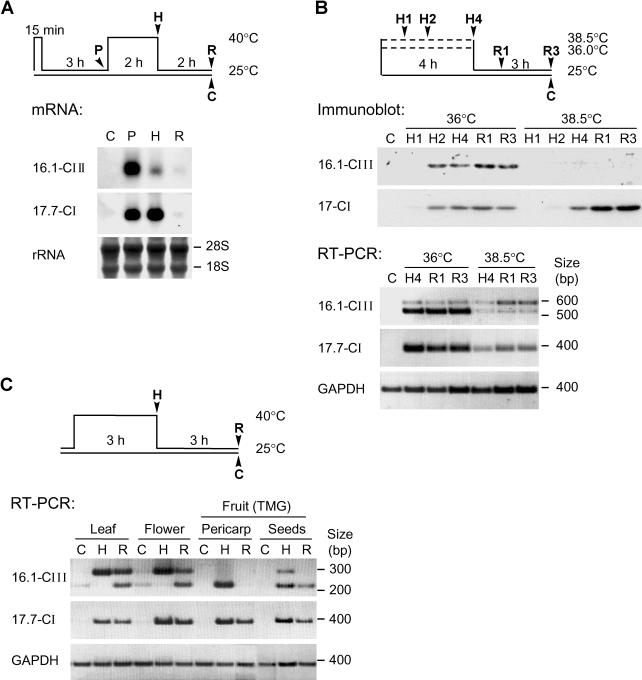
For expression analysis by Northern blots, ribonucleic acid (RNA) samples were prepared from tomato cell cultures subjected to the heat stress regimen indicated by the pictograph in Figure 2A. Hsp17-CIII expression was only observed after heat stress. For comparison, detection of Hsp17.7-CI messenger RNA (mRNA) was included. In contrast to the latter, Hsp16.1-CIII mRNA was mainly detected in preinduced cultures 3 hours after recovery from the inductive short heat stress at 40°C. Generation of a polyclonal antiserum against the tomato Hsp16.1-CIII allowed detection of the corresponding protein in immunoblots of protein extracts from tobacco protoplasts transformed with the Hsp16.1-CIII expression plasmids (data not shown). The new antiserum was used to analyze the expression of Hsp16.1-CIII in tomato cell cultures under different conditions of heat stress and recovery as defined in the pictograph in Figure 2B. In contrast to Hsp17-CI isoforms, which accumulate to high levels under all heat stress conditions, expression of Hsp16.1-CIII seems to be more temperature sensitive. Its accumulation is only observed in samples heat stressed at 36°C and in preinduced cultures (data not shown) but not in samples from cells heat stressed at 38.5°C. Evidently, deficiency in the splicing of the Hsp16.1-CIII pre-mRNA is responsible for these defects of Hsp16.1-CIII synthesis at higher temperatures (see results of RT-PCR analyses in Fig 2B).
Fig 2.
Heat stress–dependent expression of Hsp16.1-CIII. (A) Tomato suspension culture cells were submitted to the indicated heat stress regime (see pictograph). Ribonucleic acid (RNA) samples were prepared after preinduction (P), heat stress (H), and recovery (R). The RNA was analyzed by Northern blotting using Hsp16.1-CIII– and Hsp17.7-CI–specific probes. Ribosomal RNA staining with methylene blue served as loading control. (B) Analysis of Hsp16.1-CIII expression in tomato suspension culture cells subjected to heat stress treatments at 36°C and 38.5°C (see pictograph on top). For total protein and RNA preparation, aliquots of cells were harvested as indicated, ie, samples H1, H2, and H4 at 1, 2, and 4 hours of heat stress, samples R1 and R3 at 1 and 3 hours of recovery, and sample C represents untreated control cells. The analysis of Hsp17-CI species in the immunoblot detection (above) and Hsp17.7-CI messenger RNA (mRNA) detection by reverse transcriptase–polymerase chain reaction (RT-PCR) (below) was used as positive control for the heat stress effect. The lack of Hsp16.1-CIII accumulation after heat stress treatment at 38.5°C is correlated to an increasing level of its unspliced pre-mRNA in the corresponding RT-PCR samples. For amplification of Hsp16.1-CIII transcripts, primers Pr547F and Pr520R were used (see Fig 1 and Materials and Methods). (C) RT-PCR analysis of Hsp16.1-CIII and Hsp17.7-CI expression in different tomato tissues under control (C), heat stress (H), and recovery (R) conditions. Total RNA was prepared from leaves, flowers, and pericarp and seeds of mature green fruits. In contrast to the RT-PCR analysis shown in part B, primer Pr520R was replaced by Pr548R, which binds complementarily to nucleotides 219–242 of the complementary deoxyribonucleic acid sequence (Fig 1A). As in part B, the glyceraldehyde-3-phosphate dehydrogenase signal was used as internal control for the RT-PCR reaction
Using RT-PCR, we analyzed different tissues of tomato to detect Hsp16.1-CIII– and Hsp17.7-CI–encoding mRNAs in comparison with the constitutive expressed glyceraldehyde-3-phosphate dehydrogenase mRNA as endogenous control (Fig 2C). As expected, mRNAs of the 2 heat stress–induced genes were only detected in the heat stress (H) and recovery (R) samples but not in the control (C) samples. As observed in cell cultures, splicing of Hsp16.1-CIII pre-mRNA was defective in the heat stress samples of leaves and flowers, but this splicing deficiency was not observed in fruit pericarp. However, under the given heat stress conditions, we were not able to detect a significant accumulation of Hsp16.1-CIII protein in any of these tissues. Compared with the sHsps of classes CI and CII, Hsp16.1-CIII evidently represents a minor Hsp.
Because of an NLS in the ACD, Hsp16.1-CIII is a nuclear protein
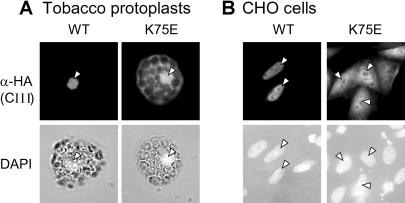
In contrast to the sHsps of classes CI and CII, the class CIII sHsps have an extended cluster of basic amino acid residues (NGKRKR) in the loop region between β5 and β6 of the ACD (Figs 1A and 7A). To investigate the intracellular localization of LpHsp16.1-CIII, we constructed plant and mammalian expression vectors encoding the protein with an N-terminal triple HA tag for immunodetection. After transformation of tobacco protoplasts, the protein was predominantly localized in the nucleus (Fig 3A). To test the particular role of the basic cluster as potential NLS, we created a mutant protein 3HA-Hsp16.1-CIII K75E by changing the basic cluster NGKRKR to NGERKR and investigated its intracellular localization in comparison with 3HA-Hsp16.1-CIII (Fig 3A). Clearly, a considerable part of the mutant protein remains now in the cytoplasm, indicating that the NGKRKR motif is responsible for the enhanced nuclear localization of this type of sHsp. Similar results were obtained when the 3HA-tagged Hsp16.1-CIII was expressed in CHO cells (Fig 3B). The tomato Hsp16.1-CIII protein was exclusively in the nucleus, and the K75E mutant protein was distributed between nucleus and cytoplasm. The advantage of the use of CHO cells for this type of investigation results from the fact that intracellular localization of plant proteins can be studied independently of any other intrinsic proteins that may influence the result (see below).
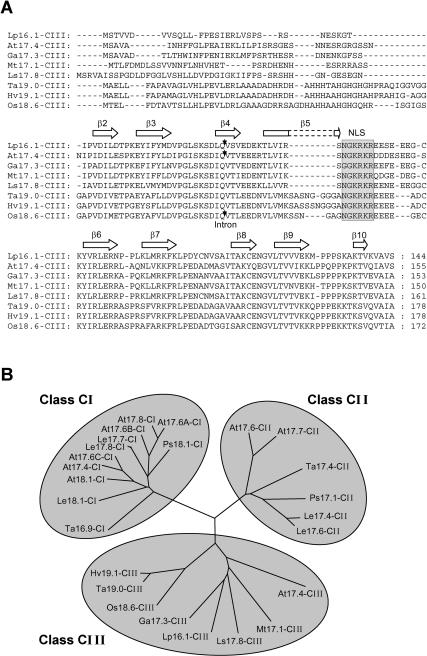
Fig 7.
(A) Sequence comparison of class CIII sHsps from different plants and (B) phylogenetic relationship of nucleocytoplasmic sHsps. (A) Amino acid sequence alignment of LpHsp16.1-CIII with orthologous class CIII sHsps of Arabidopsis thaliana (At), Gossypium arboreum (Ga), Medicago truncatula (Mt), Lactuca sativa (Ls), Triticum aestivum (Ta), Hordeum vulgare (Hv), and Oryza sativa (Os) was done by using the Clustal X 1.8 software. Database accession numbers and number of corresponding expressed sequence tags (ESTs) are LpHsp16.1-CIII, AAK84869 (no EST); AtHsp17.4-CIII, AAD25777 (5 ESTs); GaHsp17.3-CIII, TC11872 (3 ESTs); MtHsp17.1-CIII, TC56172 (3 ESTs); LsHsp17.8-CIII, TC1271 (6 ESTs); TaHsp19.0-CIII, TC41583 (8 ESTs); HvHsp19.1-CIII, TC36830 (4 ESTs); and OsHsp18.6-CIII, OJ1311D08 (8 ESTs). Arrowheads point to the conserved position of introns in the genomic sequences from tomato (63 bp), Arabidopsis (97 bp), and rice (88 bp). Clusters of basic amino acid residues of the nuclear localization signal are shaded, and amino acid residues fitting the β-fold motifs in the α-crystallin domain (β2 to β9) and in the C-terminal extension (β10) are indicated by open arrows. (B) Based on Clustal alignment using the full-length amino acid sequence of the indicated proteins, a tree was drawn and visualized by the TreeView 32 software. The class CIII proteins are clearly clustered on a separate branch distinct from the other 2 classes. Database accession numbers for sequences of class CI and class CII sHsps are AAF79569 (AtHsp17.8-CI), AAD39328 (AtHsp17.6A-CI), AAC95188 (AtHsp17.6B-CI), CAA34208 (AtHsp17.6C-CI), CAA35182 (AtHsp17.4-CI), CAA35183 (AtHsp18.1-CI), CAA45039 (AtHsp17.6-CII), and CAA74399 (AtHsp17.7-CII) for A thaliana; CAA39603 (LeHsp17.8-CI), AAD30454 (LeHsp17.7-CI), LeTC101691 (LeHsp18.1-CI), AAC36312 (LeHsp17.4-CII), and AAC14577 (LeHsp17.6-CII) for Lycopersicon esculentum; AAA33672 (PsHsp18.1-CI) and AAA33670 (PsHsp17.1-CII) for Pisum sativum; and CAA31785 (TaHsp16.9-CI) and CAA41218 (TaHsp17.4-CII) for T aestivum. Accession numbers for class CIII sequences are the same as in (A).
Fig 3.
Immunofluorescence analysis of the intracellular distribution of LpHsp16.1-CIII and its nuclear localization signal (NLS) mutant K75E. Tobacco protoplasts (A) and Chinese Hamster ovary (CHO) cells (B) were used for transient expression of 3HA-tagged Hsp16.1-CIII in its wild type (WT) or NLS mutant form (K75E). As indicated on the left margin, detections were done with HA antiserum (α-HA) and with 4′,6-diamidino-2-phenylindole for nuclear staining. The corresponding nuclei of tobacco protoplasts and CHO cells with detectable Hsp16.1-CIII expression are indicated by arrowheads
Protein interaction and oligomeric state of Hsp16.1-CIII
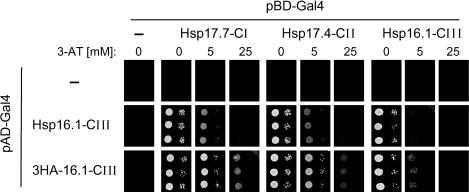
Identification of the tomato Hsp16.1-CIII clone from a yeast 2-hybrid screening using pea Hsp18.1-CI as bait indicated an unusual property of the class CIII protein. Generally, sHsps of classes CI and CII showed only class-specific interactions (Kirschner et al 2000), whereas Hsp16.1-CIII interacts with both members of the other 2 classes of cytosolic sHsps (Fig 4). These heterologous interactions, probably reflecting contacts through the dimer interfaces, are as strong as the homologous contacts with Hsp16.1-CIII in bait and prey positions. The results were similar when the tomato Hsp17.7-CI or Hsp17.4-CII in the bait positions were replaced by the corresponding pea proteins, ie, Hsp18.1-CI and Hsp17.1-CII, respectively (data not shown).
Fig 4.
Yeast 2-hybrid interaction test for different sHsps. As prey vectors, we used pADGal4 without insertion (negative control) and with Hsp16.1-CIII as well as its 3HA-tagged form as insertions. The bait vectors (pBDGal4) contained no insert (negative control) or the indicated complementary deoxyribonucleic acid fragments encoding LpHsp17.7-CI, LpHsp17.4-CII, and LpHsp16.1-CIII. Interaction of the fusion proteins was evaluated by growing the yeast strains in 1:100 dilutions on histidine-free media containing the indicated concentrations of 3-amino triazol (3-AT)
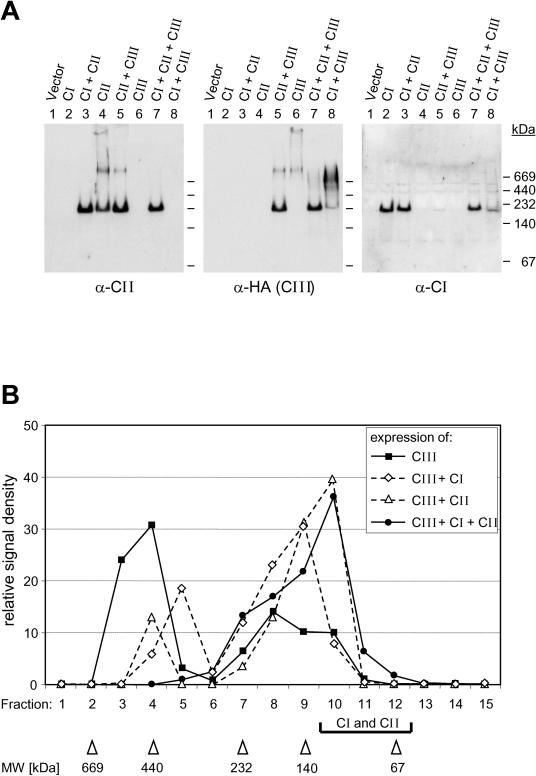
Because of the peculiarities of the test system based on the Gal4 fusion proteins, the yeast 2-hybrid system cannot reveal interactions of sHsps in their native oligomeric state, which is mainly dodecameric for proteins of classes CI and CII. To investigate interactions at this level of oligomerization, 3 types of experiments were performed. First, in pull-down assays with recombinant GST-Hsp16.1-CIII fusion protein and whole-cell extracts from tobacco protoplasts expressing either Hsp17-CI or Hsp17-CII in the native oligomeric state, we observed interactions of Hsp16.1-CIII with both sHsps (data not shown). Second, the interaction of the 3 types of sHsps was demonstrated by coexpression of HA-tagged Hsp16.1-CIII together with the indicated sHsps in tobacco protoplasts, followed by the analysis of the oligomeric complexes under nondenaturing conditions (Fig 5A). The 3 representatives used for this assay were PsHsp18.1-CI forming an oligomer of about 210 kDa (lane 2), LpHsp17.4-CII detected with 3 complexes (lane 4) of 220 kDa (dodecamer), >700 kDa, and a high–molecular weight complex at the sample loading site, as well as Hsp16.1-CIII (lane 6) with 2 native complexes, one of >700 kDa and the other a high–molecular weight complex on top. In the 3 types of dimeric coexpression mixtures, ie, CI plus CII (lane 3), CII plus CIII (lane 5) and CI plus CIII (lane 8), the migration of the native complexes changed (see especially the positions of the CII and CIII complexes in lanes 4 to 6), and in the trimeric mixture (lane 7), most of the CII and CIII as well as the CI protein were detected in a 220-kDa complex. These interactions between the 3 types of proteins were not heat stress dependent. Third, in agreement with these observations, the separation profile of 3HA-Hsp16.1-CIII on a gel filtration column changed markedly in the presence of coexpressed class CI and class CII Hsps (Fig 5B). Both class CI and class CII proteins contributed to the shift of Hsp16.1-CIII from high–molecular weight complexes (fractions 2 to 5) to much smaller complexes (fractions 7 to 12) with an elution profile similar to those of the native, homooligomeric complexes of class CI and class CII proteins (data not shown).
Fig 5.
Characterization of native complexes of class CI, class CII, and class CIII proteins. Tobacco protoplasts were transformed with the indicated combinations of expression plasmids encoding PsHsp18.1-CI (CI), LpHsp17.4-CII (CII), or 3HA-LpHsp16.1-CIII (CIII). After 18 hours at 25°C, native protein extracts were prepared and subjected for separation of sHsp complexes by nondenaturing polyacrylamide gel electrophoresis (A) or by gel filtration chromatography (B), as described in Materials and Methods. Part A shows the results of immunoblot detections on the same blot by sequential cycles of stripping and incubation with antisera specific for class CII (left panel), class CI (right panel), or the HA-tagged Hsp16.1-CIII (middle pannel). In part B, only the elution profiles for Hsp16.1-CIII are shown. Fractions corresponding to the elution of the majority of class CI and class CII proteins are marked by a bracket
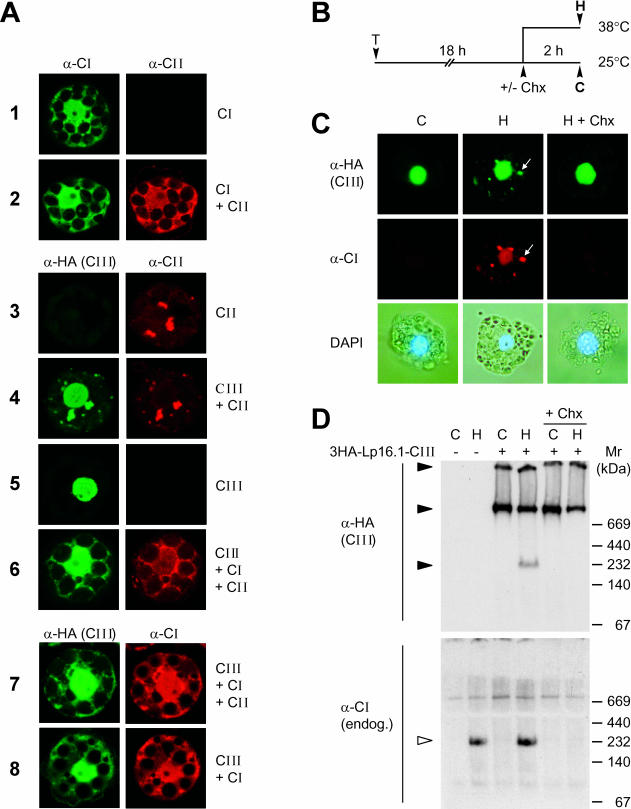
To prove whether the observed changes in the native states of sHsps expressed in different combinations could also result in changes of their intracellular localization, we complemented these studies using corresponding immunofluorescence data (Fig 6A). Hsp16.1-CIII alone was exclusively found in the nucleus (sample 5), but in the presence of Hsp17-CI (sample 8) or Hsp17-CII (sample 4), a portion of Hsp16.1-CIII colocalizes with them in the cytoplasm. The tendency for autoaggregation of tomato Hsp17.4-CII (sample 3) results in the formation of large cytosolic aggregates also incorporating Hsp16.1-CIII (sample 4). Class CI proteins were generally found as soluble proteins distributed between nucleus and cytoplasm (sample 1 and Kirschner et al 2000). Interestingly, they solubilize the class CII proteins in any combination tested here (samples 2 and 6 or 7). Control conditions were used for incubation of the protoplasts before processing for immunofluorescence. Under heat stress conditions, the aggregation tendency of CII proteins dominates the solubilizing effect of CI proteins, and considerable parts of all 3 proteins were found in cytoplasmic aggregates (data not shown).
Fig 6.
Expression and intracellular localization of Hsp16.1-CIII in mesophyll protoplasts. (A) Tomato protoplasts were transformed for transient expression of PsHsp18.1-CI (CI), LpHsp17.4-CII (CII), or 3HA-Hsp16.1-CIII (CIII) alone or for coexpression in combinations as indicated for each sample on the right margin. The specific primary antisera used for the immunofluorescence detection of the 3 types of sHsps are indicated on top of the corresponding group of images. (B) Tobacco protoplasts were transformed with plasmid encoding 3HA-LpHsp16.1-CIII. After 18 hours of transgene expression at 25°C, samples were subjected to 2 hours of heat stress at 38°C to induce the endogenous set of chaperones (see pictograph). Cycloheximide (Chx, 5 μg/mL) was added to aliquots of the samples before the heat stress treatment to prevent the formation of endogenous Hsps. (C) Immunofluorescence detection of class CI and class CIII proteins. Arrows point to colocalization of tomato Hsp16.1-CIII with endogenous tobacco Hsp17-CI in cytoplasmic HSG complexes. The lower panel presents pictures of the corresponding cells after nuclear staining with 4′,6-diamidino-2-phenylindole. (D) Analysis of 3HA-Hsp16.1-CIII complexes in nondenaturing gels by immunoblot detection with α-HA (upper part) and α-Hsp17-CI antisera (lower part). Samples corresponding to protoplast transformed with the expression plasmid for 3HA-Hsp16.1-CIII are indicated (+) on top of the gel. Oligomeric complexes are indicated by filled (3HA-Hsp16.1-CIII) and open arrowheads (Hsp17-CI)
To provide further indications for the interaction between the 3 types of sHsps in vivo, we studied the intracellular distribution in tobacco protoplasts expressing 3HA-Hsp16.1-CIII together with the endogenous set of Hsps formed after heat stress induction. Figure 6B shows the heat stress treatment of samples for analysis of the intracellular localization (Fig 6C) and the native complexes of Hsp16.1-CIII (Fig 6D). As shown before, Hsp16.1-CIII was exclusively found in the nuclei of protoplasts maintained at room temperature, but after 2 hours of heat stress at 38°C (pictograph, Fig 6B), part of the protein shifted to cytoplasmic aggregates, representing the HSG complexes (see colocalization of Hsp16.1-CIII with endogenous tobacco Hsp17-CI as indicator for HSG complexes formation, Fig 6C). If new synthesis of endogenous Hsps was inhibited by adding cycloheximide at the onset of the heat stress treatment, the redistribution of Hsp16.1-CIII was not observed, ie, recruitment to the cytoplasmic HSG complexes depends on the presence of endogenous class CI and class CII sHsps.
We used protein extracts from these protoplasts to analyze the expression levels and oligomeric states of the proteins (Fig 6D). In contrast to the class CI sHsps with predominant oligomeric state of dodecamers with 230 kDa (see open arrowhead for the position of the tobacco Hsp17-CI complex), the 3HA-Hsp16.1-CIII forms much larger complexes (>700 kDa). Interestingly, when sufficient amounts of the endogenous sHsps were present after 2 hours of heat stress (Fig 6D, sample H), part of the Hsp16.1-CIII was detected together with the Hsp17-CI band at 230 kDa (3HA immunoreactive material at 230 kDa in Fig 6D, upper panel). This evidently corresponds to the formation of heterooligomeric complexes shown in the coexpression experiments (Fig 5).
DISCUSSION
Using pea Hsp18.1-CI as bait and a tomato cDNA library, we isolated a new type of nucleocytoplasmic sHsp (Hsp16.1-CIII). Members of the class CIII sHsps are discriminated from the other 2 cytoplasmic classes, classes CI and CII, by a number of properties.
The class CIII–encoding genes, at least in rice, tomato, and Arabidopsis, are singular and contain short introns in the region encoding the β4 strand of the ACD. Based on their phylogenetic relationships, the known class CIII sHsps of monocots and dicots are clearly separated from the members of the other 2 classes (Fig 7; Scharf et al 2001).
An important marker motif of class CIII sHsps is the highly conserved cluster of basic amino acid residues in the extended loop between β5 and β6 of the ACD with consensus motif (73)NGKRKR (Fig 7A). In the other 2 classes of tomato sHsps, the corresponding sequences are (85)SGERNV for LpHsp17.7-CI and (84)SGERKR for LpHsp17.4-CII, ie, a glutamic acid residue replaces the lysine residue in Hsp16.1-CIII and the related proteins of other plants. The function of this basic cluster as NLS was supported by mutating the lysine residue (Hsp16.1-CIII, K75E) and demonstrating that the cytoplasmic distribution of the mutant protein was enhanced (Fig 3).
The tomato Hsp16.1-CIII forms oligomeric complexes in the range of 1 MDa, ie, much larger than the dodecamers of 200–230 kDa usually found for other plant sHsps. However, if coexpressed in tobacco protoplasts, some of the Hsp16.1-CIII cross-reactive material associated with the dodecameric complexes of CI and CII proteins (Fig 5). In addition, a significant recruitment of Hsp16.1-CIII into cytosolic chaperone complexes was observed under conditions providing sufficient amounts of Hsp17-CII. This was achieved either by coexpression with LpHsp17.4-CII or by heat stress induction of the endogenous set of tobacco sHsps, leading to the formation of HSG complexes (Nover et al 1989). In support of our earlier observations (Kirschner et al 2000), class CII proteins apparently represent the components of the cytosolic chaperone complexes essential for the recruitment of other sHsps.
Similar to the other plant sHsps analyzed so far, expression of Hsp17-CIII is heat stress dependent in Arabidopsis (data not shown) and tomato (Fig 2). However, the efficient splicing of the intron seems to be inhibited at higher temperatures and results in an accumulation of unspliced pre-mRNA. This effect is especially pronounced in leaves and flowers (Fig 2). In tomato cell culture, the accumulation of Hsp16.1-CIII protein was only observed under mild heat stress conditions, ie, either after direct temperature shift to 36°C or after preinduction by a short heat stress treatment at 40°C but not when directly shifted to 38.5°C (Fig 2). In this respect, ie, the temperature sensitivity of splicing, Hsp16.1-CIII reminds one of the Drosophila Hsp82, which is likewise encoded by an intron-containing gene (Yost and Lindquist 1986).
Particularly striking are our observations on the special role of the class CIII proteins in the network of nucleocytoplasmic sHsps. Tests with the yeast 2-hybrid system demonstrated that in contrast to the class-specific interaction of CI and CII sHsps, the dimer interface of CIII proteins is evidently compatible also with the interfaces of CI and CII proteins (Fig 4). This mutual compatibility of interactions was also observed at the level of larger oligomers when representatives of the 3 classes were coexpressed in tobacco protoplasts. This is documented by the analyses of native complexes and the immunofluorescence data on the intracellular localization.
The investigations on protein interactions between the sHsps in the nucleocytoplasmic compartment (classes CI, CII, and CIII) revealed interesting new insights. Evidently, each of the 3 classes of sHsps contributes with specific properties to the functional integrity of the sHsp network in plants.
As shown by Kirschner et al (2000), class CII proteins are indispensible for the heat stress–induced recruitment of class CI sHsps into HSG and HSG-like complexes. The strong tendency of class CII proteins to autoaggregate could be a prerequisite for this specific property (Port M et al, personal communication), and in case of tomato Hsp17.4-CII, the formation of cytoplasmic complexes and corecruitment of Hsp16.1-CIII were observed even under nonstress conditions.
In contrast to this, class CI proteins have a solubilizing effect on class CII proteins. The ratio between complex-bound and soluble amounts depends on the balance between the 2 types of sHsps and the temperature. In the natural set of sHsps induced by heat stress treatment of cell cultures or leaves, the class CI proteins always prevail and help to keep sHsps in a soluble state under nonstress conditions, eg, during the recovery period. Aggregation accompanied by HSG formation is only observed at elevated temperatures, and the reversibility may be critically dependent on the ratio of Hsp17-CI and Hsp17-CII.
Although probably expressed at much lower levels than the other 2 classes of cytoplasmic sHsps, class CIII proteins may be considered as mediators between them. Whether this could influence the coordinated incorporation of the sHsps into cytoplasmic HSG complexes remains to be investigated. Considering the tendency for heterooligomerization under nonstress conditions, it is also reasonable to assume that the NLS could contribute not only to the nuclear localization of the CIII proteins but also to an enhanced nuclear localization of CI and CII proteins. Nuclear localization of sHsps of classes CI and CII is documented in Figure 6A and was reported earlier by Wollgiehn et al (1994).
The data presented in this article demonstrate the special properties of a new class (class CIII) of nucleocytoplasmic sHsps capable of interaction with members of the other 2 cytoplasmic classes of sHsps. Although the expression levels of Hsp16.1-CIII in heat-stressed tomato tissues are clearly lower than those of class CI and class CII proteins, this does not preclude a role in the network of sHsps, eg, as a component of heterooligomeric complexes, as shown in Figure 5. Although it is premature to speculate about the situation in other plants, it is interesting to notice that, in contrast to tomato, multiple ESTs were identified in all plant EST databases investigated by us (see legend of Fig 7). This indicate, that in these cases the orthologous genes are expressed in a heat stress–independent manner. RNA interference experiments with transient or permanent knockout of the expression of the Hsp17-CIII gene may help to clarify its specific role in the sHsp network including possible functions in plant development.
Acknowledgments
We thank Daniela Bublak and Gisela Englich for excellent technical assistance and Sascha Gernhard for experimental contributions to the analysis of Hsp16.1-CIII expression in tomato cell cultures. We thank Markus Fauth and Kapil Bharti for their suggestions during performing the experimental work and Lutz Nover for many helpful discussions and comments during the preparation of the manuscript. The work was supported by grants to K.-D. S. from the Deutsche Forschungsgemeinschaft (SCHA 577/6-1) and by the Fonds der Chemischen Industrie.
REFERENCES
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, and Struhl K. (ed). 1993 Current Protocols in Molecular Biology. John Wiley and Sons, Inc, Indianapolis, IN, USA. [Google Scholar]
- Bharti K, Schmidt E, Lyck R, Bublak D, Scharf KD. Isolation and characterization of HsfA3, a new heat stress transcription factor of Lycopersicon peruvianum. Plant J. 2000;22:355–365. doi: 10.1046/j.1365-313x.2000.00746.x. [DOI] [PubMed] [Google Scholar]
- Caspers GJ, Leunissen JAM, de Jong WW. The expanding small heat-shock protein family, and structure predictions of the conserved α-crystallin domain. J Mol Evol. 1995;40:238–248. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- Chen Q, Osteryoung K, Vierling E. A 21-kDa chloroplast heat shock protein assembles into high molecular weight complexes in vivo and in organelle. J Biol Chem. 1994;269:13216–13223. [PubMed] [Google Scholar]
- de Jong WW, Caspers GJ, Leunissen JAM. Genealogy of the α-crystallin-small heat-stress superfamily. Int J Biol Macromol. 1998;22:151–162. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnsperger M, Lilie H, Gaestel M, Buchner J. The dynamics of Hsp25 quarternary structure. J Biol Chem. 1999;274:14867–14874. doi: 10.1074/jbc.274.21.14867. [DOI] [PubMed] [Google Scholar]
- Giese KC, Vierling E. Changes in oligomerization are essential for the chaperone activity of a small heat shock protein in vivo and in vitro. J Biol Chem. 2002;277:46310–46318. doi: 10.1074/jbc.M208926200. [DOI] [PubMed] [Google Scholar]
- Haslbeck M. sHsps and their role in the chaperone network. Cell Mol Life Sci. 2002;59:1649–1657. doi: 10.1007/PL00012492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz D, Döring P, Bonzelius F, Winkelhaus S, Nover L. The balance of nuclear import and export determines the intracellular distribution of tomato heat stress transcription factor HsfA2. Mol Cell Biol. 2001;21:1759–1768. doi: 10.1128/MCB.21.5.1759-1768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm KW, Lee GJ, Vierling E. Expression and native structure of cytosolic class II small heat-shock proteins. Plant Physiol. 1997;114:1477–1485. doi: 10.1104/pp.114.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn TL, Chen YM, Lin CY. Characterization and physiological function of class I low-molecular-mass heat-shock protein complex in soybean. Plant Physiol. 1995;108:693–701. doi: 10.1104/pp.108.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Winkelhaus S, Thierfelder J, Nover L. Transient expression and heat stress induced aggregation of endogenous and heterologous small heat stress proteins in tobacco protoplasts. Plant J. 2000;24:397–412. doi: 10.1046/j.1365-313x.2000.00887.x. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Pokala N, Vierling E. Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem. 1995;270:10432–10438. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lyck R, Harmening U, Höhfeld I, Treuter E, Scharf KD, Nover L. Intracellular distribution and identification of the nuclear localization signals of two plant heat-stress transcription factors. Planta. 1997;202:117–125. doi: 10.1007/s004250050110. [DOI] [PubMed] [Google Scholar]
- MacRae TH. Structure and function of small heat shock/alpha-crystallin proteins: established concepts and emerging ideas. Cell Mol Life Sci. 2000;57:899–913. doi: 10.1007/PL00000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002;16:1555–1567. doi: 10.1101/gad.228802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F. α-Crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev. 2002;66:64–93. doi: 10.1128/MMBR.66.1.64-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann D, Scharf KD, Nover L. Heat shock induced changes of plant cell ultrastructure and autoradiographic localization of heat shock proteins. Eur J Cell Biol. 1984;34:254–264. [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol. 1983;3:1648–1655. doi: 10.1128/mcb.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW 2001 Molecular Cloning: A Laboratory Manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Scharf KD, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol. 1998;18:2240–2251. doi: 10.1128/mcb.18.4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (Acd proteins) Cell Stress Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobott F, Benesch JLP, Vierling E, Robinson CV. Subunit exchange of multimeric protein complexes—real-time monitoring of subunit exchange between small heat shock proteins by using electrospray mass spectrometry. J Biol Chem. 2002;277:38921–38929. doi: 10.1074/jbc.M206060200. [DOI] [PubMed] [Google Scholar]
- Töpfer R, Schell J, Steinbiss HH. Versatile cloning vectors for transient gene expression and direct gene transfer in plant cells. Nucleic Acids Res. 1988;16:8725. doi: 10.1093/nar/16.17.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfort RLM, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eucaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- Vierling E. The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:579–620. [Google Scholar]
- Waters ER, Lee GJ, Vierling E. Evolution structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. [Google Scholar]
- Waters ER, Vierling E. The diversification of plant cytosolic small heat shock proteins preceded the divergence of mosses. Mol Biol Evol. 1999;16:127–139. doi: 10.1093/oxfordjournals.molbev.a026033. [DOI] [PubMed] [Google Scholar]
- Wollgiehn R, Neumann D, zur Nieden U, Müsch A, Scharf KD, Nover L. Intracellular distribution of small heat stress proteins in cultured cells of Lycopersicon peruvianum. J Plant Physiol. 1994;144:491–499. [Google Scholar]
- Yost HJ, Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986;45:185–193. doi: 10.1016/0092-8674(86)90382-x. [DOI] [PubMed] [Google Scholar]