Abstract
Fanconi anaemia (FA) and Bloom syndrome (BS) are autosomal recessive diseases characterised by chromosome fragility and cancer proneness. Here, we report that BLM and the FA pathway are activated in response to both crosslinked DNA and replication fork stall. We provide evidence that BLM and FANCD2 colocalise and co-immunoprecipitate following treatment with either DNA crosslinkers or agents inducing replication arrest. We also find that the FA core complex is necessary for BLM phosphorylation and assembly in nuclear foci in response to crosslinked DNA. Moreover, we show that knock-down of the MRE11 complex, whose function is also under the control of the FA core complex, enhances cellular and chromosomal sensitivity to DNA interstrand crosslinks in BS cells. These findings suggest the existence of a functional link between BLM and the FA pathway and that BLM and the MRE11 complex are in two separated branches of a pathway resulting in S-phase checkpoint activation, chromosome integrity and cell survival in response to crosslinked DNA.
Keywords: BLM helicase, DNA repair, Fanconi anaemia, replication fork arrest, S-phase checkpoint
Introduction
Fanconi anaemia (FA) is a rare autosomal recessive genetic condition associated with bone marrow failure, reduced fertility and predisposition to cancer, essentially myeloid leukaemia (Garcia-Higuera et al, 1999a; Ahmad et al, 2002). At the cellular level, FA is characterised by chromosome instability, hypersensitivity and altered mutability to agents inducing DNA interstrand crosslinks (ICLs) (Grompe and D'Andrea, 2001). Eight FANC genes (A, C, D1/BRCA2, D2, E, F, G/XRCC9 and L) have been cloned so far (Grompe and D'Andrea, 2001; Rosselli et al, 2003) and their products are thought to define a pathway involved in the control of genomic stability and checkpoint activation, the FA pathway (Garcia-Higuera et al, 1999a; D'Andrea and Grompe, 2003; Rosselli et al, 2003). Furthermore, six of the FANC proteins (A, C, E, F, G and L) form a nuclear complex (FA core complex) in response to DNA damage, and mutation in any of its component leads to inactivation of the complex (Garcia-Higuera et al, 1999b; Waisfisz et al, 1999; Medhurst et al, 2001) and loss of activation of the FA downstream effector FANCD2 (Grompe and D'Andrea, 2001).
Bloom syndrome (BS) is a rare autosomal recessive disorder characterised by reduced fertility, immunodeficiency and elevated predisposition to different types of cancer, including leukaemia (van Brabant et al, 2000). At the cellular level, BS is associated with chromosome instability, hypersensitivity to genotoxic agents and hypermutability (Hickson et al, 2001). The gene mutated in BS, BLM, encodes for a RecQ-class DNA helicase (Ellis et al, 1995). The cellular role of BLM is poorly defined, but a growing body of evidence indicates that it could be required for the maintenance of genome stability during DNA replication (Hickson et al, 2001).
Several observations suggest a functional interaction between BLM and the FA pathway. First, BS and FA patients share a partial overlapping phenotype. Second, BS and FA cells show a similar type of chromosomal instability, that is, chromosome interchanges and mainly quadriradial forms (van Brabant et al, 2000; Grompe and D'Andrea, 2001). Third, BLM is part of a supermolecular complex in which the key component is BRCA1, a functional partner of FANCD2 (Wang et al, 2000; Garcia-Higuera et al, 2001) and an interactor of FANCA (Folias et al, 2002). Fourth, both BLM and the FA core complex are necessary for the assembly of the MRE11 complex, another protein complex involved in genome stability, although in response to different genotoxic stresses (Franchitto and Pichierri, 2002a; Pichierri et al, 2002). Finally, BLM and FA core complex have been recently isolated as components of a common complex of unknown function (Meetei et al, 2003).
In this study, we investigated whether BLM and the FA pathway cooperate in the cellular response to agents inducing ICLs or replication fork arrest.
Our results show that in response to both ICLs or replication stresses, BLM and the FA pathway are activated and that BLM colocalises and co-immunoprecipitates with the monoubiquitinated isoform of FANCD2. Furthermore, our data suggest that a functional FA core complex is necessary for BLM phosphorylation specifically in response to ICLs and demonstrate that BLM and the MRE11 complex define two parallel branches, both under the control of the FA core complex, acting to ensure cell viability and chromosomal integrity in response to crosslinked DNA.
Results
Loss of the BLM or FA pathway results in hypersensitivity to crosslinked DNA and stalled replication forks
Sensitivity to DNA crosslinking agents such as mitomycin C (MMC) or photoactivated psoralens is characteristic of FA cells (Grompe and D'Andrea, 2001), whereas enhanced cell death in response to replication arrest has been associated to BS cells (Franchitto and Pichierri, 2002a; Davalos and Campisi, 2003). To determine the existence of a crosstalk between BLM and the FA pathway, we first examined cell survival of BS and FA cells to MMC, photoactivated 8-MOP or hydroxyurea (HU)-mediated replication arrest (Figure 1). Consistently to what reported earlier (Hook et al, 1984; Davalos and Campisi, 2003), we found that induction of crosslinked DNA by MMC resulted in a reduced cell survival in BS-derived cells (Figure 1A). Moreover, similar results were obtained after exposure to photoactivated 8-MOP (Figure 1B). Interestingly, ectopic expression of wild-type BLM restored normal sensitivity to ICL-inducing agent (Figure 1A and B). Although BS cell sensitivity was less pronounced than that of FA cells, these data demonstrate that a functional BLM is required to deal with crosslinked DNA. Conversely, FA cell lines showed a reduced survival to HU than normal or ectopically corrected counterpart cells (Figure 1C and D). Strikingly, lymphoblasts and fibroblasts from different FA complementation groups showed comparable levels of sensitivity to HU even if this sensitivity was intermediate between normal and BS cells. The sensitivity to HU observed in FA cells is unlikely attributable to some checkpoint response defect as described for BS cells (Heinrich et al, 1998; Ababou et al, 2002; Franchitto and Pichierri, 2002a). These observations suggest that stalled replication forks activate FA proteins.
Figure 1.
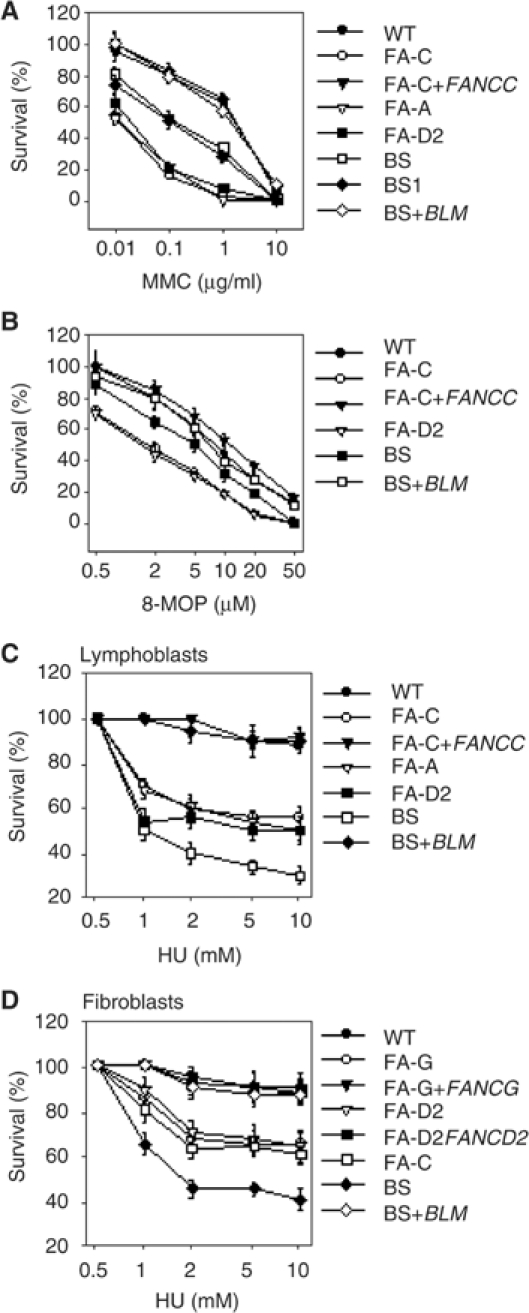
Loss of BLM or FA pathway results in enhanced cell death following crosslinked DNA and replication arrest. (A) MMC sensitivity of the indicated genotypes. (B) Photoactivated 8-MOP sensitivity of the indicated genotypes. (C, D) HU sensitivity of the indicated genotypes (see Materials and methods). The lymphoblasts used were GM3657 (WT), HSC536 (FA-C), HSC536 Corr (FA-C+FANCC), HSC72 (FA-A), EUFA143L (FA-G), GM16752 (FA-D2), GM3403 (BS1), GM3403BLM (BS1+BLM) and ZG (BS). The SV40-transformed fibroblasts used were MRC5 (WT), PD20 (FA-D2), PD20 315 (FA-D2+FANCD2), PD332 (FA-C), PD352 (FA-G), PD352+FANCG (FA-G+FANCG), GM8505 (BS) and GM8505+BLM (BS+BLM). Data are the mean±s.d. from three independent experiments. Data were analysed by χ2 test; the differences in sensitivity between wild-type and FA or BS cells were statistically significant (P<0.01, χ2 test).
BLM and FANCD2 are both relocalised in response to crosslinked DNA and stalled replication forks
We sought to characterise the involvement of BLM in response to crosslinked DNA and the requirement of FA pathway in the resolution of replication arrest. To this purpose, on the one hand we analysed ICL-induced BLM assembly in nuclear foci and phosphorylation, two molecular events associated to its response to DNA damage (Ababou et al, 2000, 2002; Wang et al, 2000; Beamish et al, 2002; Franchitto and Pichierri, 2002a). On the other hand, we examined activation of FANCD2 by analysing its monoubiquitination, phosphorylation and relocalisation in nuclear foci in response to replication fork arrest.
Previous studies have shown that BLM relocalises in nuclear foci in response to HU or UVC (Wang et al, 2000; Franchitto and Pichierri, 2002a). Interestingly, we found that BLM accumulated in foci also in response to crosslinked DNA, although the increase with time is proportionally lower with respect to that observed after HU or 40 J/m2 UVC (Figure 2A and B and Supplementary Figure 1). Similarly, FANCD2 foci increased not only in response to crosslinked DNA and UVC-induced DNA damage (Garcia-Higuera et al, 2001), but also in response to HU (Figure 2A and B and Supplementary Figure 1). Besides being assembled in nuclear foci, BLM was phosphorylated in response to crosslinked DNA (Figure 2C). FANCD2 underwent monoubiquitination after HU- or UVC-mediated DNA damage, as well as following induction of crosslinks in DNA (Figure 2D). Moreover, both HU- and UVC-mediated DNA damage resulted in a reduction of the FANCD2 electrophoretic mobility that could be reverted by PPase treatment (Figure 2D), suggesting that both treatments determined phosphorylation of FANCD2 similarly to that observed in response to ICLs or γ-irradiation (Taniguchi et al, 2002b; Pichierri and Rosselli, 2004). Both BLM and FANCD2 accumulated in nuclear foci and underwent post-translational modification also following 20 J/m2 UVC, a dose resulting in cell survival comparable to that observed after 10 μM photoactivated 8-MOP (Supplementary Figure 1 and our unpublished results).
Figure 2.
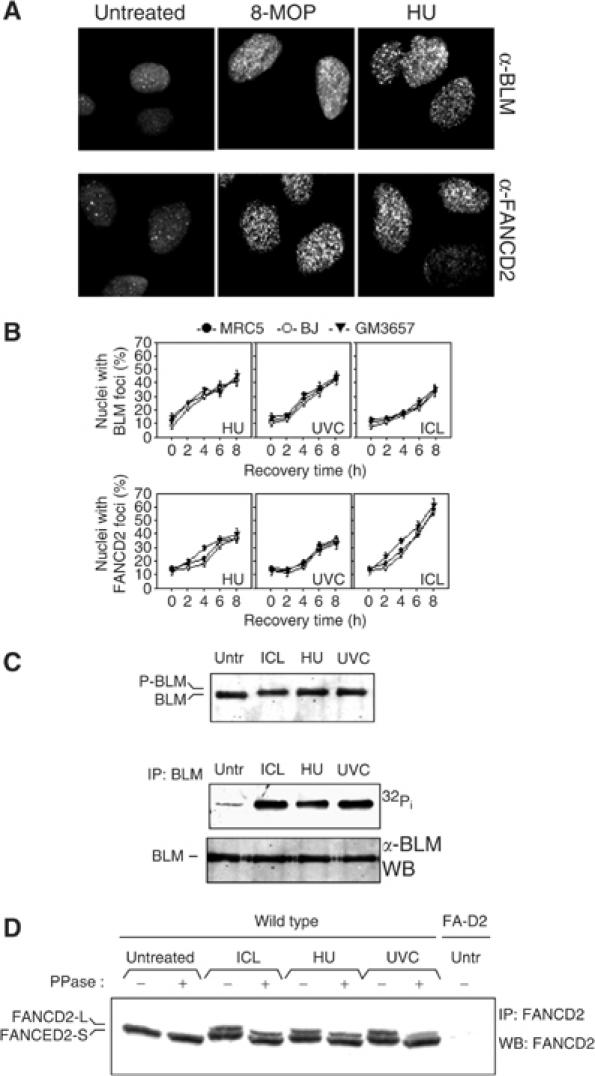
The BLM protein and FANCD2 respond similarly to DNA crosslink induction and replication fork arrest. (A) Representative images taken after 6 h of recovery from MRC5 fibroblasts. (B) ICL- and replication arrest-dependent assembly of BLM and FANCD2 nuclear foci. Wild-type fibroblasts (MRC5 and BJ) and lymphoblasts (GM3657) were exposed to photoactivated 8-MOP (ICL), HU or UVC light and analysed at the indicated time points. (C) BLM phosphorylation in response to crosslinked DNA and replication arrest. Upper blot: MRC5 fibroblasts were lysed 4 h after treatment with 10 μM photoactivated 8-MOP (ICL), 2 mM HU or 40 J/m2 UVC, and BLM phosphorylation was analysed by the shift in electrophoretic mobility as seen on 5% Tris-glycine gel. Lower blot: BLM phosphorylation was visualised by 32Pi metabolic labelling followed by autoradiography. The BLM band shift is not visible on the lower blot because of the higher percentage of acrylamide used for the SDS–PAGE. (D) FANCD2 monoubiquitination and phosphorylation in response to stalled replication fork. Cellular extracts prepared 4 h after exposure of wild-type cells (MRC5) to photoactivated 8-MOP (ICL), HU or UVC were analysed for the presence of the large isoform of FANCD2 (FANCD2-L) and for FANCD2 phosphorylation by Western blot analysis. Phosphorylation was assessed by observing the shift in the electrophoretic mobility of the FANCD2 doublet and confirmed by reversion of the band shift following PPase treatment.
These results indicate that both the FA pathway and BLM participate in the cellular response to crosslinked DNA and stalled replication forks.
BLM and FANCD2 colocalise and co-immunoprecipitate in response to crosslinked DNA and stalled replication forks
To examine the possibility that BLM and FANCD2 could act together in response to crosslinked DNA or stalled replication forks, we performed double immunofluorescent labelling in wild-type cells exposed to photoactivated 8-MOP or HU. We found that BLM and FANCD2 colocalised in a fraction of cells exhibiting both BLM and FANCD2 foci (Figure 3A and Supplementary Figure 2). BLM and FANCD2 foci colocalisation increased in a time-dependent manner, with the maximum between 6 and 8 h post-treatment (Supplementary Figure 2). In addition, BLM and FANCD2 were co-immunoprecipitated by anti-BLM antibodies in normal cells, either untreated or following induction of crosslinked DNA or replication arrest (Figure 3B). Interestingly, this association appeared to involve only a fraction of the FANCD2 protein and to be restricted to the monoubiquitinated isoform (Figure 3B). To further investigate whether BLM was able to preferentially associate with the monoubiquitinated FANCD2 isoform, we transfected MRC5 cells and FA-G fibroblasts, in which FANCD2 monoubiquitination is defective (Garcia-Higuera et al, 2001), with a cDNA encoding HA-tagged ubiquitin. Cellular exposure to HU or ICLs resulted in the appearance of an anti-HA immunoreactive band in BLM immunoprecipitates prepared from wild-type cells (Figure 3C). FANCD2 was not found in BLM immunoprecipitates prepared from FA-G cells (Figure 3C), demonstrating that BLM did not associate with the unmodified FANCD2 isoform.
Figure 3.
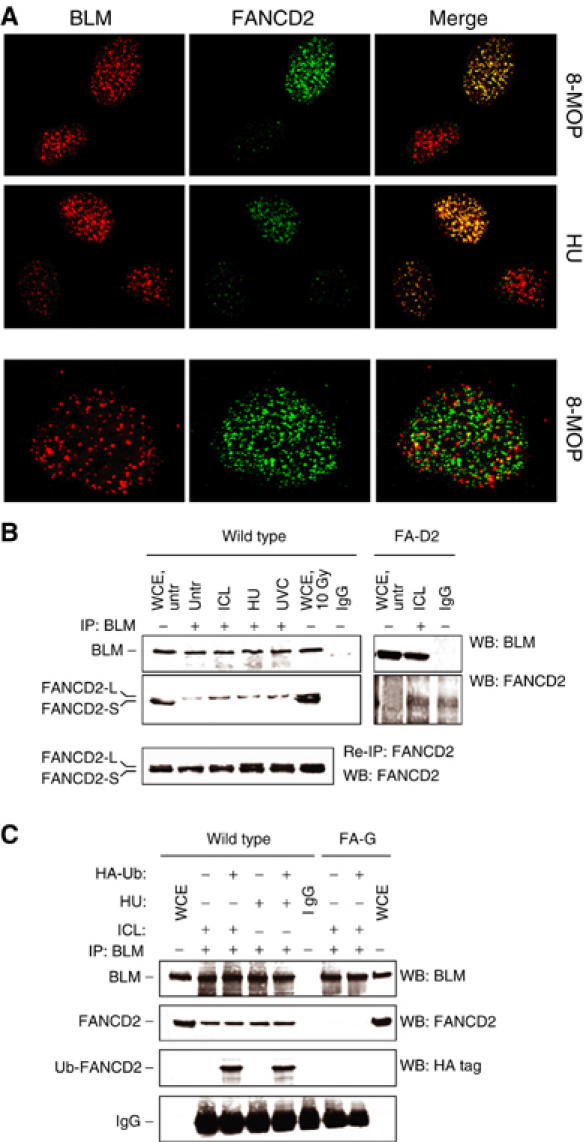
BLM and FANCD2 colocalise in response to crosslinked DNA and replication fork arrest. (A) ICL- and replication fork arrest-dependent BLM/FANCD2 colocalisation in wild-type cells. MRC5 cells (wild type) were exposed to photoactivated 8-MOP or UVC and immunofluorescence was carried out 6 h later. In the lower row is presented a nucleus in which BLM and FANCD2 foci are both present but not colocalising. (B) Co-IP of BLM and FANCD2. Cellular extracts prepared 6 h after exposure of wild-type or FA-D2 (PD20) cells to photoactivated 8-MOP (ICL), HU or UVC were immunoprecipitated and immunoprecipitates were analysed by Western blot. IgG: Extract prepared from untreated MRC5 cells immunoprecipitated with normal rabbit IgG. Lower panel: The supernatant from the first immunoprecipitation was used to re-immunoprecipitate FANCD2 and evaluate the fraction not bound to BLM. (C) Co-IP of BLM with the activated (i.e. ubiquitinated) form of FANCD2. MRC5 (wild type) and FA-G cells were transiently transfected with an empty vector or with a plasmid expressing HA-tagged ubiquitin and 36 h later exposed to photoactivated 8-MOP (ICL) or HU. Cellular extracts prepared 6 h after treatment were immunoprecipitated with a polyclonal antibody against BLM and immunoprecipitates were analysed by Western blot using anti-BLM, anti-FANCD2 and anti-HA-tag antibodies. IgG: Extract prepared from untreated MRC5 cells immunoprecipitated with normal rabbit IgG. Note that the FANCD2 doublet as well as the BLM band shift is not visible on the blots because of the percentage of acrylamide used for the SDS–PAGE (7% instead of 5%).
Taken together, these findings suggest that BLM is preferentially associated with the monoubiquitinated FANCD2 isoform.
FA complex is required for BLM phosphorylation following DNA crosslinks
Since BLM and the FA pathway might collaborate in response to both crosslinked DNA and replication arrest, we sought to determine whether the integrity of the FA pathway could affect BLM activation and vice versa. Therefore, on the one hand, we examined BLM relocalisation and phosphorylation in cells from different FA complementation groups exposed to photoactivated 8-MOP, UVC or HU. On the other hand, we analysed FANCD2 focalisation and phosphorylation, as well as FANCA, FANCG and FANCD2 chromatin shift in BS and BS complemented cells following ICLs or replication arrest.
We first looked at BLM activation in a FA genetic background. As shown in Figure 4A and B, BLM subnuclear relocalisation after ICLs was reduced in FA-C and FA-G cells, in which the FA core complex is absent (Waisfisz et al, 1999), but was normal in the absence of FANCD2. As it has been previously reported that BLM colocalises with phosphorylated histone H2AX (γ-H2AX) following replication arrest (Davalos and Campisi, 2003), we asked whether the reduction of BLM focalisation observed in FA core complex mutants was attributable to its inability to relocalise at γ-H2AX sites. BLM colocalisation with γ-H2AX was severely reduced in FA-G cells after ICLs but not after HU treatment (Supplementary Figure 3A and B), even though wild-type and FA-G cells showed a comparable induction of γ-H2AX foci (Rothfuss and Grompe, 2004) (Supplementary Figure 3C). Strikingly, BLM phosphorylation, a molecular event related to its response to DNA damage (Ababou et al, 2002; Beamish et al, 2002; Franchitto and Pichierri, 2002a; Davies et al, 2004), was abolished in FA-G and FA-C following ICL induction (Figure 4C and D). Re-introduction of the wild-type FANCC or FANCG in the corresponding FA cell strain, which is sufficient to restore the FA core complex functionality, recovered the ICL-dependent BLM phosphorylation (Figure 4D and data not shown). Interestingly, in FA core complex mutants, loss of BLM phosphorylation was observed over a range of recovery times after ICL induction (Figure 4E), excluding a kinetic effect. In vivo labelling experiments using [32P]orthophosphate further confirmed that BLM phosphorylation was dependent upon the FA core complex after ICLs (Figure 4F). In contrast, both the BLM assembly into nuclear foci and its phosphorylation were unaffected by mutations in the FA pathway after replication arrest or UVC radiation (Figure 4B–D and F).
Figure 4.
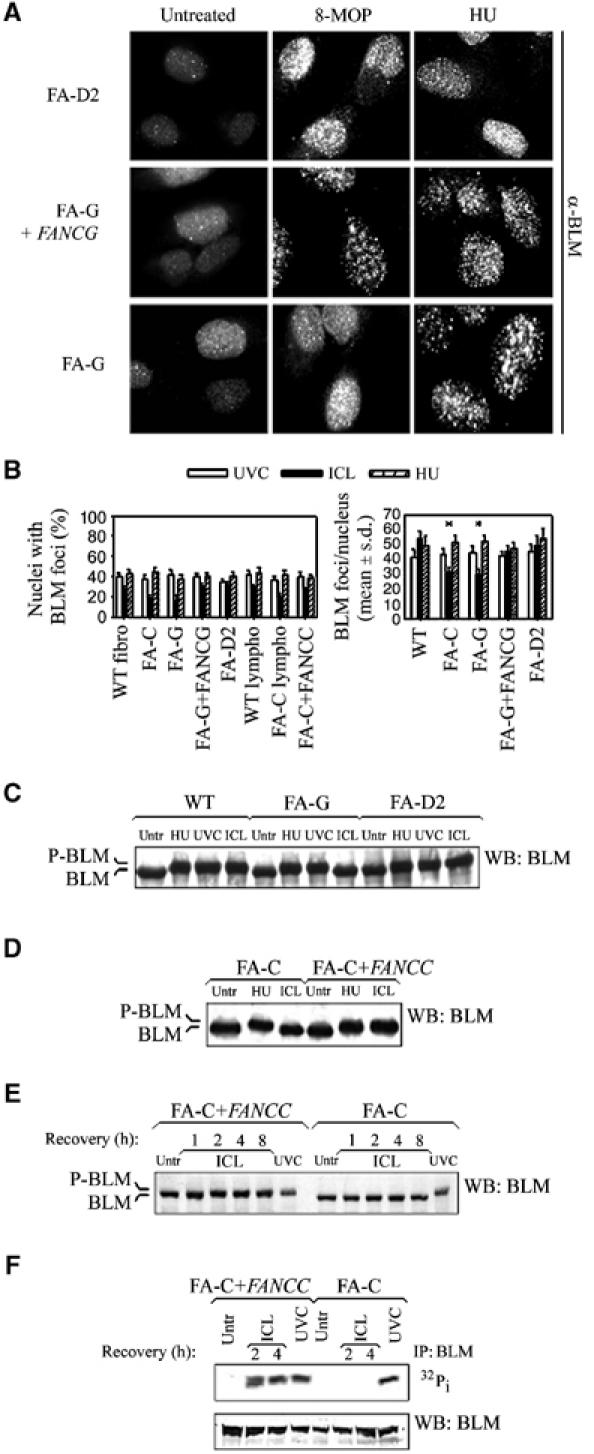
Integrity of the FA core complex is required for ICL-dependent BLM phosphorylation. (A) BLM relocalisation in FA cells following ICL and replication arrest. Wild-type (FA-G+FANCG), FA-G and FA-D2 SV40-immortalised fibroblasts were exposed to photoactivated 8-MOP (ICL) or to HU and immunofluorescence was carried out 6 h later. (B) Analysis of the BLM relocalisation in wild-type (WT), FA and phenotypically complemented cells. The analysis was carried out at 6 h post-treatment and similar results were obtained at 4 and 8 h. Data are the mean±s.d. from three independent experiments. *Statistically significant (P<0.01, χ2 test). (C) BLM phosphorylation assessed by Western blot following treatment with photoactivated 8-MOP (ICL), HU or UVC in wild-type and FA lymphoblasts. (D) Evaluation of BLM phosphorylation in FA-C cells. Cells were exposed to HU or photoactivated 8-MOP (ICL) and analysed for the presence of BLM phosphorylation by Western blot. (E) Analysis of BLM phosphorylation at different times after induction of ICL by 8-MOP. (F) Cells were exposed to UVC or photoactivated 8-MOP, and FANCD2 phosphorylation was visualised by 32Pi metabolic labelling followed by autoradiography. The FANCD2 band shift is not visible on the lower blot because of the higher percentage of acrylamide used in the SDS–PAGE (7% instead of 5%). Unless specified, BLM phosphorylation was evaluated as band shift 4 h after the indicated treatment.
Altogether, our results suggest that the FA core complex is specifically required for the phosphorylation of BLM in the presence of crosslinked DNA and for its efficient assembly into nuclear foci.
In converse experiments, FANCD2 relocalisation in nuclear foci following either ICLs or replication arrest was not affected by the absence of a functional BLM protein (Figure 5A and B). Similarly, the absence of BLM did not affect the post-translational modifications of FANCD2 in response to either crosslinked DNA or replication arrest (Figure 5C and D).
Figure 5.
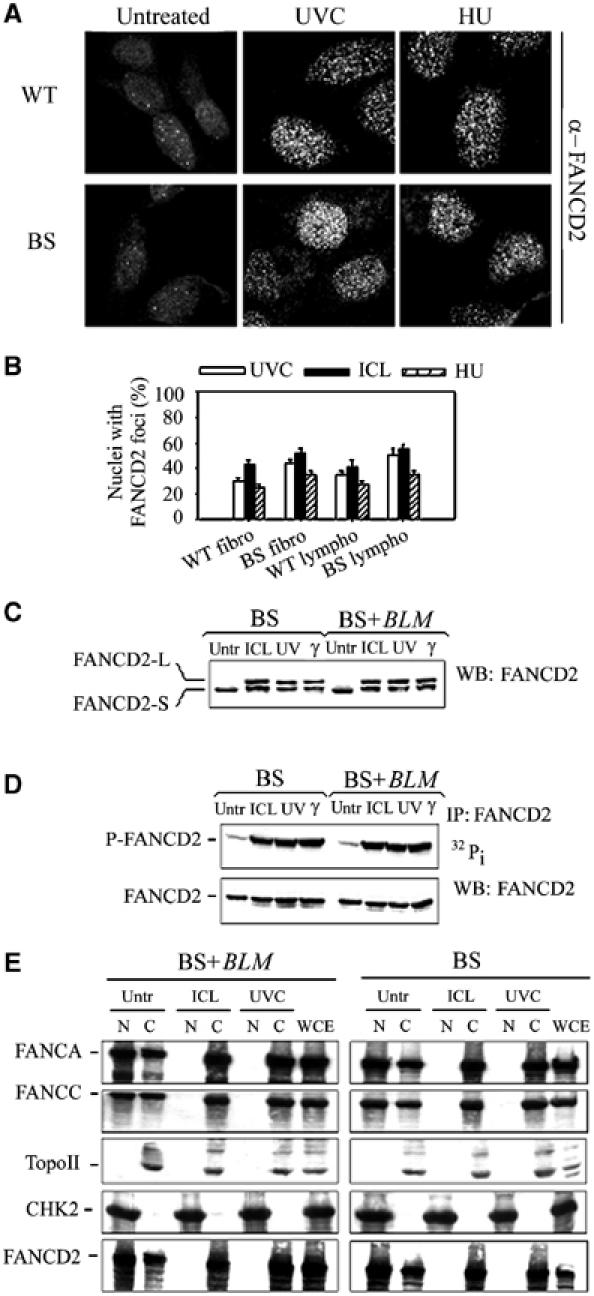
The FA core complex and FANCD2 migrate to chromatin in response to replication arrest in a BLM-independent manner. (A) FANCD2 relocalisation in BS cells following UVC or HU. Wild-type (WT) and BS hTert-immortalised fibroblasts were exposed to photoactivated 8-MOP (ICL) or to HU and immunofluorescence was carried out 6 h later. (B) Analysis of the FANCD2 relocalisation in wild-type (WT) and BS hTert-immortalised fibroblasts and EBV-transformed lymphoblasts. The analysis was carried out at 6 h post-treatment but consistent results were also obtained at other sampling times. Data are the mean±s.d. from three independent experiments. (C) FANCD2 monoubiquitination in BS lymphoblasts. Cellular extracts were prepared 4 h after exposure of cells to photoactivated 8-MOP (ICL) or UVC. A sample irradiated with 10 Gy of IR (γ) served as positive control. (D) FANCD2 phosphorylation analysed by 32Pi metabolic labelling. A cellular extract prepared from cells irradiated with 10 Gy of γ-rays was included as a positive control for FANCD2 phosphorylation. Note that the FANCD2 doublet is not visible on the blots because of the percentage of acrylamide used for SDS–PAGE (7.5% instead of 5%). (E) Cellular extracts prepared 6 h after exposure of BS and BLM-complemented lymphoblasts to photoactivated 8-MOP (ICL) or UVC were biochemically fractionated and the free nucleoplasmic (N) and the insoluble chromatin (C) fractions were sequentially analysed for the presence of FANC proteins. Similar results were obtained analysing FANCG distribution (data not shown). Immunoblotting using anti-topoisomerase II (TopoII) and anti-CHK2 antibodies served as control for the fractionation procedure and as loading control.
Then, we investigated whether BLM functionality could affect the response of the FA core complex to DNA damage. It has been shown that the FA core complex associates specifically with chromatin following DNA crosslinking treatments (Qiao et al, 2001). Hence, we analysed chromatin localisation of FANCA and FANCC—two key components of the FA complex—and that of FANCD2, in response to crosslinked DNA or UVC-induced replication arrest by a biochemical fractionation technique followed by Western blot analysis of the free nucleoplasmic and the chromatin fraction (Mendez and Stillman, 2000). Both DNA crosslinks and UV irradiation induced association of the FA core complex proteins and FANCD2 with chromatin, as shown by the appearance of the FANC proteins in the chromatin fraction (Figure 5E). Consistently with normal FANCD2 activation, the absence of a functional BLM protein did not affect the ability of the FANC proteins to associate with chromatin, both in response to ICL treatment and replication fork arrest. As expected, the FANC proteins chromatin shift was absent in extracts from FA cells (data not shown).
Our results indicate that BLM is not required for the FA core complex translocation to chromatin in response to both crosslinked DNA and UVC-mediated replication arrest. Since BLM has no effect on FA proteins, whereas FA core complex is required for BLM phosphorylation, it is likely that FA core complex is an upstream regulator of BLM function in response to ICLs.
MRE11 complex and BLM act independently in response to crosslinked DNA
We have previously reported that the FA core complex is required for correct MRE11 complex relocalisation following ICL induction (Pichierri et al, 2002). Moreover, BLM phosphorylation appears necessary for the MRE11 assembly in nuclear foci after replication arrest (Franchitto and Pichierri, 2002a). Since we found that the integrity of the FA core complex is necessary for the observed DNA crosslink-dependent BLM phosphorylation (Figure 4), the question arises whether the MRE11 complex requires BLM to efficiently relocalise in response to crosslinked DNA. Consequently, MRE11 complex relocalisation was investigated in BS and BS-complemented cells by indirect immunofluorescence of MRE11.
MRE11 was found normally relocalised in response to ICLs also in the absence of a functional BLM, and NBS1 phosphorylation, a molecular event related to MRE11 complex functions, was also unaffected by loss of BLM (Supplementary Figure 4). On the contrary and as previously reported (Franchitto and Pichierri, 2002a), both MRE11 relocalisation and NBS1 phosphorylation were compromised in BS cells following replication arrest (Supplementary Figure 4).
These results suggest that the MRE11 complex and BLM are part of two independent branches in the response to crosslinked DNA, both controlled by the upstream FA core complex. To validate this hypothesis, we ablated MRE11 complex function by RNA interference (RNAi) in hTert-immortalised BS fibroblasts (Ouellette et al, 2000) and looked at cellular and chromosomal sensitivity, as well as at S-phase checkpoint activation in response to ICLs.
As shown in Supplementary Figure 5, transfection of siRNAs against MRE11 caused a significant reduction in the intracellular content of MRE11 protein and loss of MRE11 complex function as denoted by loss of NBS1 phosphorylation. Interestingly, the partial cellular (Figures 1A, 6A and B) and chromosomal (Figure 6C) sensitivities of the BS cells to ICLs were elevated to the same level as observed in FA cells by interference of the MRE11 expression. Strikingly and consistently with our published data (Pichierri and Rosselli, 2004), MRE11 RNAi in FA core complex mutants (FA-G) or FA-D2 cells did not increase the yield of ICL-induced chromosomal damage (Figure 6C).
Figure 6.
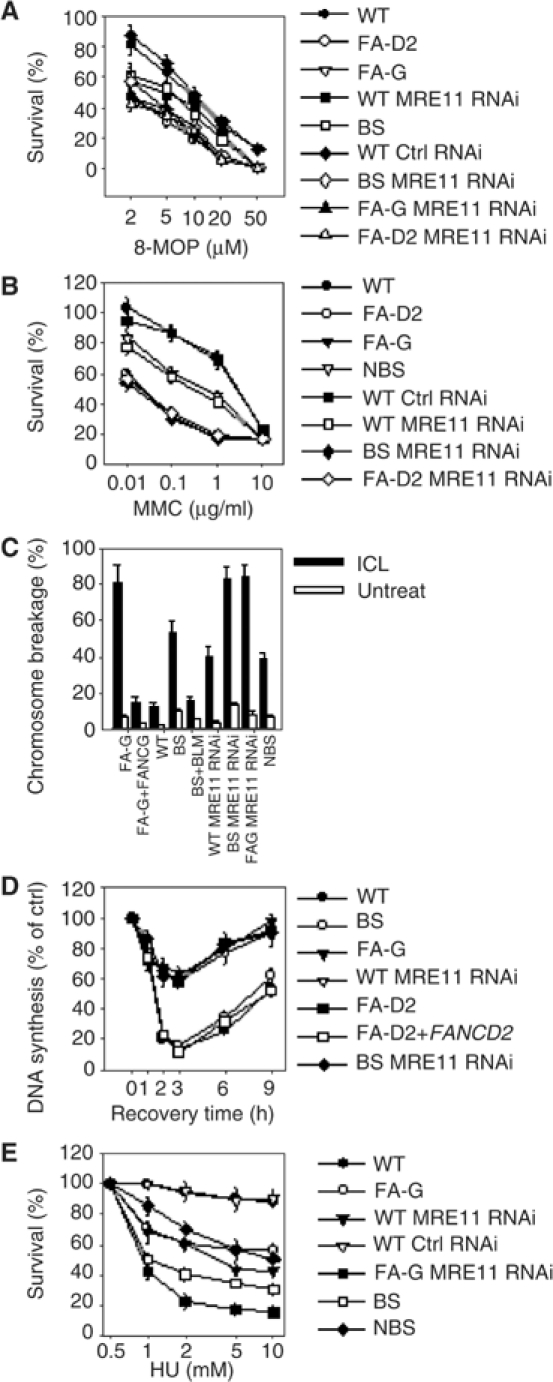
BLM and MRE11 contribute through two separate pathways to ensure cell viability and chromosome integrity following induction of crosslinked DNA. (A, B) ICL sensitivity of the indicated genotypes. Data are the mean±s.d. from three independent experiments. Results were found significant by the χ2 test (P<0.01). (C) Quantification of the photoactivated 8-MOP-induced chromosome breakage. Analysis was carried out as described in Materials and methods after exposure to 500 nM 8-MOP followed by 10 kJ/m2 UVA. Data are the mean±s.d. from three independent experiments. Results were significant as judged by the Student's t-test (P<0.01). (D) Analysis of the S-phase checkpoint activation following ICL induction. Replicative DNA synthesis was assessed at different time points after ICL induction with photoactivated 8-MOP. (E) Sensitivity to HU-induced replication arrest of the indicated genotypes. Data are the mean±s.d. from three independent experiments. Results were found significant by the χ2 test (P<0.01). In the case of siRNA-treated cells, exposure to genotoxins was always performed 72 h after transfection.
We have recently demonstrated that FA core complex proteins are required for efficient activation of the intra-S checkpoint following ICL induction (Pichierri and Rosselli, 2004). Since ICL-dependent BLM phosphorylation is under the genetic control of the FA core complex (Figure 4C–F), we analysed the activation of S-phase checkpoint in BS cells. As shown in Figure 6D, inhibition of DNA synthesis in response to ICLs was normal in BS cells. In contrast, DNA synthesis inhibition was affected in BS by MRE11 RNAi as observed in FA or NBS1 defective cells (Pichierri and Rosselli, 2004), in which formation of the MRE11 complex was hampered (Carney et al, 1998).
Our results indicate that BLM and the FA pathway act on two functionally separated pathways in response to replication fork arrest induced by HU, whereas functional crosstalks exist following ICL induction. Specifically, in response to HU-induced replication fork stall, BLM and MRE11 function in a common branch (Franchitto and Pichierri, 2002a), whereas FANC proteins work in a parallel pathway. To test this hypothesis we knocked down MRE11 expression in FA-G cells by RNAi and analysed cell survival after HU exposure. As expected, sensitivity of FA-G cells to HU was intermediate between that of wild type and BS (Figure 6E). However, inhibition of MRE11 expression in FA-G cells resulted in enhancement of HU-induced cell death even compared to that found in BS (Figure 6E).
Thus, our results suggest the existence of a branched pathway activated by replication fork arrest either by crosslinked DNA or by HU. Following HU exposure the FANC proteins and BLM operate in parallel arms, whereas after ICL induction BLM is placed downstream to the FA core complex. Functionality of both these branches is required to fully ensure cell survival, chromosome integrity and S-phase checkpoint activation following ICL induction.
Discussion
We have shown that BLM and the FA pathway are activated in response to both crosslinked DNA and replication fork stall. Moreover, we have provided evidence that BLM and FANCD2 colocalise and co-immunoprecipitate following treatment with either DNA crosslinkers or agents inducing replication arrest. Finally, we have demonstrated that BLM and the MRE11 complex are functionally linked to the FA pathway and act as two independent components of the DNA damage response to crosslinked DNA, both downstream to the FA core complex.
HU- and UVC-induced DNA adducts are able to stall replication forks, although by different mechanisms (Hyrien, 2000; Osborn et al, 2002). ICLs are also strong inhibitors of the replication fork progression, and their repair is thought to rely, in mammalian cells, mainly on a replication-dependent pathway (Akkari et al, 2000; De Silva et al, 2000; Wang et al, 2001). Thus, the stall of replication machinery might be the common basis to the activation of the BLM and the FA pathway in response to crosslinked DNA, HU or UVC exposure. Indeed, the BLM helicase is clearly involved in the response to stalled replication forks, either participating directly in the resolution of DNA structures arising at forks or during the homologous recombination (HR)-mediated restart of replication (Hickson, 2003). The reported connection between BLM and FA proteins argues for a direct implication of the FA pathway in the recovery of the arrested replication fork. Several observations support this possibility: the FA core complex is shifted to chromatin (i.e. activated) in response to ICLs (Qiao et al, 2001 and this study), but also following replication arrest; FANCD2 is found phosphorylated during unperturbed S-phase progression (Taniguchi et al, 2002a) and FANCD2 phosphorylation in response to UVC-mediated fork stall is dependent on ATR (P Pichierri and F Rosselli unpublished observations), the key kinase in the response to replication stresses (Abraham, 2001).
After HU- or UVC-induced replication arrest, BLM relocalisation and phosphorylation are normal in FA cells and, conversely, FANCD2 is correctly assembled in nuclear foci and phosphorylated in BS cells. This implicates that, despite the BLM/FANCD2 colocalisation, the two proteins act independently against replication stress caused by HU or UVC exposure. On the contrary, following ICL-mediated replication fork stall, phosphorylation of BLM is impaired in FA core complex mutant cells. This observation establishes a functional role for the previously reported FA core complex/BLM interaction (Meetei et al, 2003).
Finally, we have previously reported that MRE11 complex nuclear relocalisation is dependent upon a functional FA core complex following DNA crosslinks, but not after HU treatment (Pichierri et al, 2002). Here, we show that, in response to ICLs, MRE11 focalisation and NBS1 phosphorylation are correctly executed in BS cells and both the FA-MRE11 pathway and the BLM helicase are required for cell survival and chromosome integrity. On the contrary, only the FA-MRE11 branch is required for efficient intra-S checkpoint activation in response to crosslinked DNA. Indeed, BS cells normally arrest DNA replication in the presence of crosslinked DNA, and incorrect checkpoint activation is observed only following MRE11 knock-down.
These observations indicate that to cope with ICLs, BLM and MRE11 complex need to be coordinately activated by the FA core complex. Altogether, our results allow us to depict a model showing the cooperation between BLM and the FA pathways in the response to replication stresses, such as ICLs, UVC or HU (Figure 7).
Figure 7.
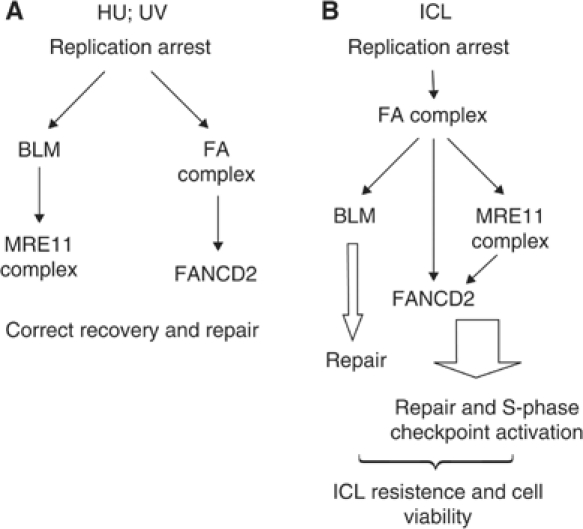
Model showing the cooperation of the BLM and the FA pathways in the response to replication arrest induced by either DNA crosslinks or UVC and HU exposure. In response to HU or UVC (A), stall of the replication machinery triggers activation of both the BLM–MRE11 and the FA branches. Possibly, the BLM–MRE11 branch is required for direct resolution of the replication arrest, whereas the FA pathway might be required for the subsequent recombinational-based replication rescue. FANCD2–BLM interaction might be functional at this step. In response to ICL-dependent replication fork arrest (B), the upstream FA core complex is activated to trigger the FA-MRE11 and the BLM branches of the pathway. Both the branches might be involved in the correct execution of the recombination-based rescue of replication, ensuring cell viability and chromosomal integrity.
In eukaryotic cells, replication arrest, as induced by HU- or UVC-mediated DNA damage, can be overcome by different mechanisms (Osborn et al, 2002). First, by processing the stalled forks in order to eliminate DNA structures or lesions that halted fork progression, and second by the induction of double-strand breaks (DSBs) at stalled forks to create the substrate for HR-driven replication restart. BLM and the MRE11 complex could cooperate in the first pathway, thus avoiding formation of DSBs at the forks, as already proposed (D'Amours and Jackson, 2002; Franchitto and Pichierri, 2002b). In contrast, the FA pathway might be involved in the latter process. Supporting this hypothesis, FANCD2 colocalises with RAD51 and BRCA1, both components of the HR pathway (Garcia-Higuera et al, 2001; Taniguchi et al, 2002a). Thus, loss of function of the BLM–MRE11 pathway could force the cell to use alternative mechanisms to restore replication, as that defined by the FANC proteins (i.e. HR). Consistently, inhibition of the BLM–MRE11 pathway in FA cells results in enhancement of cell death and chromosomal damage (Figure 6 and our unpublished results). The colocalisation and co-immunoprecipitation (Co-IP) of BLM and FANCD2 in response to stalled forks are not necessarily in contrast with the hypothesised existence of two parallel branches. In fact, BLM has been thought to act also during HR, in virtue of its ability to bind and process Holliday junctions and interact with RAD51 (Wu et al, 2001; Yang et al, 2002). So, the reported BLM/FANCD2 association could be functional during HR, but not during the earlier processing of stalled forks. Consistently, BLM/FANCD2 colocalisation is found only in a portion of cells containing BLM nuclear foci.
After ICL-induced DNA damage (see Figure 7B), the FA core complex could be required to process the crosslinked DNA in order to facilitate loading of other proteins required in the DNA repair/replication recovery process. The FA core complex-coordinate, recruitment/activation of the BLM helicase and the MRE11 complex could allow correct handling of DSBs at stalled replication forks in order to trigger HR-mediated replication rescue and facilitate activation of the S-phase checkpoint (D'Amours and Jackson, 2002). In fact, in contrast to the HU- and UVC-dependent replication arrest, which can be recovered also through DSB-independent mechanisms, the hurdle represented by an ICL can be only eliminated by creation of DSBs at the forks followed by HR (Grompe and D'Andrea, 2001). The observed BLM/FA pathway interaction could be required for the correct execution of HR. Consistently, loss of BLM results in an ICL sensitivity that is lower than that caused by mutations affecting the FANC proteins or the MRE11 complex (Nakanishi et al, 2002 and this study). Indeed, the MRE11 complex function appears essential, together with that of the FA core complex, to FANCD2 activation after DNA damage (Nakanishi et al, 2002). BLM may be required to ensure fidelity of the HR process, avoiding hyper-recombination, which could trigger chromosomal rearrangements and cell death (Yamagata et al, 1998; Pichierri et al, 2001; Saintigny et al, 2002).
In conclusion, our study establishes a functional link between the RecQ helicase BLM and the FA pathway in the recovery from replication arrest either induced by HU and UVC or crosslinked DNA. Our findings also demonstrate that BLM and the MRE11 complex–FANCD2 pathway act separately under the control of the FA core complex to ensure cell survival and genome integrity following ICL induction to DNA.
Materials and methods
Cell lines
The SV40-transformed FA fibroblasts group C (PD332), G (PD352) and D2 (PD20) along with the FA-D2 complemented cell line (PD20 315) were from the Fanconi anaemia fund cell repository (Portland, OR) and were handled as described (Waisfisz et al, 1999). The phenotypically reverted SV40-transformed PD352 (FA-G) strain has been already described (Pichierri et al, 2002). The EBV-transformed lymphoblastoid cell lines (LCLs) from normal, BS and FA patients, and their genetically reverted counterparts were grown as described (Ridet et al, 1997; Pichierri et al, 2002). The hTert-immortalised wild-type (BJ) and BS primary fibroblasts were handled as described (Ouellette et al, 2000).
Immunological reagents
The antibodies used in this study were rabbit polyclonal anti-BLM (AbCam Ltd, diluted 1:200 for immunofluorescence and 1:3000 for Western blotting), mouse monoclonal anti-FANCD2 (Santa Cruz Biotech, diluted 1:200), mouse monoclonal anti-MRE11 (GenTex, diluted 1:500 for immunofluorescence and 1:1000 for Western blotting), rabbit polyclonal anti-CHK2 (Santa Cruz Biotech, diluted 1:200), mouse monoclonal anti-γ-H2AX (Upstate, diluted 1:100), rabbit polyclonal anti-NBS1 (AbCam Ltd, diluted 1:5000), mouse monoclonal anti-topoisomerase II (Oncogene Research, diluted 1:200), mouse monoclonal anti-HA (Roche Biochemicals, diluted 1:2000), goat polyclonal anti-FANCA (Santa Cruz Biotech, diluted 1:300), goat polyclonal anti-FANCC (Santa Cruz Biotech, diluted 1:200) and rabbit polyclonal anti-FANCG (Waisfisz et al, 1999; a gift of Dr M Hoatlin, diluted 1:500).
Treatments and ICL-resistant DNA synthesis assay
ICL induction by activate psoralens was achieved exposing cells to 10 μM 8-metoxypsoralen (8-MOP) for 20 min followed by 10 kJ/m2 of UVA. Replication arrest was induced by exposure to 2 mM HU for 3 h or 40 J/m2 UVC. The UVC dose used in this study completely saturates nucleotide excision repair resulting in massive blockage of replication fork progression (Pichierri et al, 2003). For evaluation of cell survival, cells were exposed to different doses of 8-MOP, mitomycin-C (MMC) or HU (2 h pulse treatment) and after 36 h tested for the percentage of viable cells with respect to a mock-treated control (Pichierri et al, 2002). The analysis of the ICL-induced chromosomal damage was carried out on metaphase cells as described (Yang et al, 2001) 24 h after ICL induction by treatment with 2 μM 8-MOP followed by 10 kJ/m2 of UVA. When RNAi-treated cells were used, treatments were performed 72 h post-transfection with siRNAs. A minimum of 200 metaphase cells were scored for each experimental point and the results were analysed with the χ2 test. Evaluation of the DNA synthesis after ICL induction was performed as described (Xu et al, 2001).
Immunofluorescence
We carried out immunofluorescence microscopy and colocalisation studies on cells grown on coverslips. Cells were handled and fixed as described (Pichierri et al, 2002). To better investigate relocalisation and colocalisation of BLM and FANCD2 proteins, we analysed only the fraction of the proteins tightly chromatin-bound, which is thought to actually participate in the different DNA transactions, by the use of an in situ fractionation method as already reported (Mirzoeva and Petrini, 2003). Images were acquired as grey-scale files using the Metaview software and then processed using Adobe Photoshop. For each time point, at least 200 nuclei were examined and foci scored at a × 100 magnification. Only nuclei showing >10 bright foci were considered as positive. A nucleus was considered positive for colocalisation when at least two-thirds of the foci present colocalisation as observed in the merged image by the appearance of yellow spots. Parallel samples incubated with either the appropriate normal serum or only with the secondary antibody confirmed that the observed fluorescence pattern was not attributable to artifacts.
Immunoprecipitation and Western blot analysis
In Co-IP experiments, 2.5 × 107 cells for experimental point were used. A 2 mg of lysate prepared in Co-IP buffer (1% Triton X-100, 0.5% Na-doxycholate, 150 mM NaCl, 2.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 20 mM Tris–HCl pH 8.0) supplemented with protease and phosphatase inhibitors (10 μg/ml aprotinin, 10 μg/ml PMSF, 10 μg/ml leupeptin, 25 mM β-glycerophosphate, 1 mM Na-orthovanadate, 1 mM NaF) was precleared with protein A/G Sepharose beads and then incubated overnight at 4°C with rabbit polyclonal anti-FANCD2 (6 μg; AbCam Ltd) or with rabbit polyclonal anti-BLM (5 μg; Santa Cruz Biotech Ltd) and tumbled with 30 μl of protein A/G Sepharose beads for 2 h at 4°C. After extensive washing in Co-IP buffer, proteins were eluted by boiling treatment in 2 × electrophoresis sample buffer and subjected to SDS–PAGE followed by immunoblotting. For treatments with phosphatase, immunoprecipitates were resuspended in phosphatase buffer and incubated in the presence or absence of 300 U λ-phosphatase (NEB) for 1 h. Ponceau red staining of the blots was used to assess equal loading and transfer. When indicated, the WCE represents 20% of the input.
Chromatin fractionation
Analysis of the distribution of FA proteins in the chromatin and nucleoplasmic fraction was carried out by a standard protocol of chromatin fractionation (Mendez and Stillman, 2000) using 1.5 × 107 cells. As control for the proper purification of the chromatin fraction, we stripped and re-probed blots for topoisomerase II, which is a protein exclusively found in the chromatin fraction (Champoux, 2001), and for CHK2, which is a chromatin-unbound nuclear protein (Lukas et al, 2003).
32P in vivo labelling
In vivo labelling experiments used exponentially growing BS and BS corrected (BS+BLM) lymphoblasts were performed as described (Pichierri and Rosselli, 2004). After 30 min incubation at 37°C, cells were either mock-treated or exposed to 8-MOP, UVC, HU or γ-rays. The immunoprecipitated proteins were resolved by SDS–PAGE, analysed by autoradiography and Western blotting.
Inhibition of MRE11 expression by RNAi
MRE11 expression was knocked down by transfection with a mix of three siRNAs directed against the following sequences of the MRE11 mRNA: CCTGCCTCGAGTTATTAAG; CTGCGAGTGGACTATAGTG; GATGCCATTGAGGAATTAG. Each siRNA duplex (Dharmacon) was resuspended in RNase-free water at 60 μM concentration and aliquots of each were mixed to give a working concentration of 20 μM. Transfection was carried out using Oligofectamine reagent (Invitrogen) according to the manufacturer's protocol for adherent cells. As a control, an siRNA duplex directed against GFP was used. All the experiments were carried out for 72 h after transfection when maximal inhibition of MRE11 expression was observed as analysed by Western blot and immunofluorescence.
Supplementary Material
Supplementary Figures
Acknowledgments
We thank Drs A Sarasin, M Bignami and M Crescenzi for their critical reading of the manuscript and for fruitful discussions. We thank Drs WE Wrigth and JW Shay (University of Texas, Southwestern Medical Center, Dallas, TX, USA) for kindly providing the wild-type and the BS hTert-immortalised primary fibroblasts. We also thank Dr BA Cox (Fanconi Anemia Cell Repository) for kindly providing the FA-D2 SV40 fibroblasts. We are indebted to Dr D Bohmann for the plasmid encoding the HA-tagged ubiquitin. We thank the three anonymous reviewers who contributed to improve this work. This work was realised with the support of Association pour la Recherche sur le Cancer (PP and FR), Electricite de France (FR) and MIUR/Università della Tuscia (PP).
References
- Ababou M, Dumaire V, Lecluse Y, Amor-Gueret M (2002) Bloom's syndrome protein response to ultraviolet-C radiation and hydroxyurea-mediated DNA synthesis inhibition. Oncogene 21: 2079–2088 [DOI] [PubMed] [Google Scholar]
- Ababou M, Dutertre S, Lecluse Y, Onclercq R, Chatton B, Amor-Gueret M (2000) ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene 19: 5955–5963 [DOI] [PubMed] [Google Scholar]
- Abraham RT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Ahmad SI, Hanaoka F, Kirk SH (2002) Molecular biology of Fanconi anaemia—an old problem, a new insight. BioEssays 24: 439–448 [DOI] [PubMed] [Google Scholar]
- Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M (2000) DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol 20: 8283–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, Ellis N, Kondo N, Lavin MF (2002) Functional link between BLM defective in Bloom's syndrome and the ataxia-telangiectasia-mutated protein, ATM. J Biol Chem 277: 30515–30523 [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR III, Hays L, Morgan WF, Petrini JH (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93: 477–486 [DOI] [PubMed] [Google Scholar]
- Champoux JJ (2001) DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70: 369–413 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP (2002) The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol 3: 317–327 [DOI] [PubMed] [Google Scholar]
- D'Andrea AD, Grompe M (2003) The Fanconi anaemia/BRCA pathway. Nat Rev Cancer 3: 23–34 [DOI] [PubMed] [Google Scholar]
- Davalos AR, Campisi J (2003) Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J Cell Biol 162: 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID (2004) Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol 24: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA (2000) Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol 20: 7980–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J (1995) The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83: 655–666 [DOI] [PubMed] [Google Scholar]
- Folias A, Matkovic M, Bruun D, Reid S, Hejna J, Grompe M, D'Andrea A, Moses R (2002) BRCA1 interacts directly with the Fanconi anemia protein FANCA. Hum Mol Genet 11: 2591–2597 [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pichierri P (2002a) Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J Cell Biol 157: 19–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A, Pichierri P (2002b) Protecting genomic integrity during DNA replication: correlation between Werner's and Bloom's syndrome gene products and the MRE11 complex. Hum Mol Genet 11: 2447–2453 [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Kuang Y, D'Andrea AD (1999a) The molecular and cellular biology of Fanconi anemia. Curr Opin Hematol 6: 83–88 [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Kuang Y, Naf D, Wasik J, D'Andrea AD (1999b) Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol Cell Biol 19: 4866–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD (2001) Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7: 249–262 [DOI] [PubMed] [Google Scholar]
- Grompe M, D'Andrea A (2001) Fanconi anemia and DNA repair. Hum Mol Genet 10: 2253–2259 [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Hoatlin ME, Zigler AJ, Silvey KV, Bakke AC, Keeble WW, Zhi Y, Reifsteck CA, Grompe M, Brown MG, Magenis RE, Olson SB, Bagby GC (1998) DNA cross-linker-induced G2/M arrest in group C Fanconi anemia lymphoblasts reflects normal checkpoint function. Blood 91: 275–287 [PubMed] [Google Scholar]
- Hickson ID (2003) RecQ helicases: caretakers of the genome. Nat Rev Cancer 3: 169–178 [DOI] [PubMed] [Google Scholar]
- Hickson ID, Davies SL, Li JL, Levitt NC, Mohaghegh P, North PS, Wu L (2001) Role of the Bloom's syndrome helicase in maintenance of genome stability. Biochem Soc Trans 29: 201–204 [DOI] [PubMed] [Google Scholar]
- Hook GJ, Kwok E, Heddle JA (1984) Sensitivity of Bloom syndrome fibroblasts to mitomycin C. Mutat Res 131: 223–230 [DOI] [PubMed] [Google Scholar]
- Hyrien O (2000) Mechanisms and consequences of replication fork arrest. Biochimie 82: 5–17 [DOI] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J (2003) Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5: 255–260 [DOI] [PubMed] [Google Scholar]
- Medhurst AL, Huber PA, Waisfisz Q, de Winter JP, Mathew CG (2001) Direct interactions of the five known Fanconi anaemia proteins suggest a common functional pathway. Hum Mol Genet 10: 423–429 [DOI] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W (2003) A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol 23: 3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH (2003) DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol Cancer Res 1: 207–218 [PubMed] [Google Scholar]
- Nakanishi K, Taniguchi T, Ranganathan V, New HV, Moreau LA, Stotsky M, Mathew CG, Kastan MB, Weaver DT, D'Andrea AD (2002) Interaction of FANCD2 and NBS1 in the DNA damage response. Nat Cell Biol 4: 913–920 [DOI] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ, Zou L (2002) Checking on the fork: the DNA-replication stress–response pathway. Trends Cell Biol 12: 509–516 [DOI] [PubMed] [Google Scholar]
- Ouellette MM, McDaniel LD, Wright WE, Shay JW, Schultz RA (2000) The establishment of telomerase-immortalized cell lines representing human chromosome instability syndromes. Hum Mol Genet 9: 403–411 [DOI] [PubMed] [Google Scholar]
- Pichierri P, Averbeck D, Rosselli F (2002) DNA cross-link-dependent RAD50/MRE11/NBS1 subnuclear assembly requires the Fanconi anemia C protein. Hum Mol Genet 11: 2531–2546 [DOI] [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Mosesso P, Palitti F (2001) Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol Biol Cell 12: 2412–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F (2004) The DNA crosslink-induced S-phase checkpoint depends on ATR–CHK1 and ATR–NBS1–FANCD2 pathways. EMBO J 23: 1178–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F, Franchitto A (2003) Werner's syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene 22: 1491–1500 [DOI] [PubMed] [Google Scholar]
- Qiao F, Moss A, Kupfer GM (2001) Fanconi anemia proteins localize to chromatin and the nuclear matrix in a DNA damage- and cell cycle-regulated manner. J Biol Chem 276: 23391–23396 [DOI] [PubMed] [Google Scholar]
- Ridet A, Guillouf C, Duchaud E, Cundari E, Fiore M, Moustacchi E, Rosselli F (1997) Deregulated apoptosis is a hallmark of the Fanconi anemia syndrome. Cancer Res 57: 1722–1730 [PubMed] [Google Scholar]
- Rosselli F, Briot D, Pichierri P (2003) The Fanconi anemia pathway and the DNA interstrand cross-links repair. Biochimie 85: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Rothfuss A, Grompe M (2004) Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol Cell Biol 24: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ Jr (2002) Homologous recombination resolution defect in Werner syndrome. Mol Cell Biol 22: 6971–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD (2002a) S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100: 2414–2420 [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D'Andrea AD (2002b) Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109: 459–472 [DOI] [PubMed] [Google Scholar]
- van Brabant AJ, Stan R, Ellis NA (2000) DNA helicases, genomic instability, and human genetic disease. Annu Rev Genomics Hum Genet 1: 409–459 [DOI] [PubMed] [Google Scholar]
- Waisfisz Q, de Winter JP, Kruyt FA, de Groot J, van der Weel L, Dijkmans LM, Zhi Y, Arwert F, Scheper RJ, Youssoufian H, Hoatlin ME, Joenje H (1999) A physical complex of the Fanconi anemia proteins FANCG/XRCC9 and FANCA. Proc Natl Acad Sci USA 96: 10320–10325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L (2001) Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol Cell Biol 21: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 14: 927–939 [PMC free article] [PubMed] [Google Scholar]
- Wu L, Davies SL, Levitt NC, Hickson ID (2001) Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem 276: 19375–19381 [DOI] [PubMed] [Google Scholar]
- Xu B, Kim S, Kastan MB (2001) Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol Cell Biol 21: 3445–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H (1998) Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc Natl Acad Sci USA 95: 8733–8738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Zhang R, Wang XW, Spillare EA, Linke SP, Subramanian D, Griffith JD, Li JL, Hickson ID, Shen JC, Loeb LA, Mazur SJ, Appella E, Brosh RM Jr, Karmakar P, Bohr VA, Harris CC (2002) The processing of Holliday junctions by BLM and WRN helicases is regulated by p53. J Biol Chem 277: 31980–31987 [DOI] [PubMed] [Google Scholar]
- Yang Y, Kuang Y, De Oca RM, Hays T, Moreau L, Lu N, Seed B, D'Andrea AD (2001) Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood 98: 3435–3440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


