Abstract
Many small lipophilic compounds in living cells can be modified by glycosylation. These processes can regulate the bioactivity of the compounds, their intracellular location and their metabolism. The glycosyltransferases involved in biotransformations of small molecules have been grouped into Family 1 of the 69 families that are classified on the basis of substrate recognition and sequence relatedness. In plants, these transfer reactions generally use UDP-glucose with acceptors that include hormones such as auxins and cytokinins, secondary metabolites such as flavonoids, and foreign compounds including herbicides and pesticides. In mammalian organisms, UDP-glucuronic acid is typically used in the transfer reactions to endogenous acceptors, such as steroid and thyroid hormones, bile acids and retinoids, and to xenobiotics, including nonsteroidal anti-inflammatory drugs and dietary metabolites. There is widespread interest in this class of enzyme since they are known to function both in the regulation of cellular homeostasis and in detoxification pathways. This review outlines current knowledge of these glycosyltransferases drawing on information gained from studies of plant and mammalian enzymes.
Keywords: biocatalysis, cellular homeostasis, detoxification, glycosyltransferases, regioselective glucosylation
Introduction
Glycosyltransferases transfer a sugar from an activated sugar donor to an acceptor molecule and as such are involved in the synthesis and modification of the multitude of glycoconjugates in existence in the biosphere. For a detailed analysis of glycosyltransferases and their classification into the 69 families known to exist, the reader is referred to the CAZy (carbohydrate-active enzymes) website (http://afmb.cnrs-mrs.fr/CAZY/). The classification system used depends both on the nature of substrates recognised by the enzymes and their sequence relatedness. Individual glycosyltransferases can therefore be classified either through biochemical studies to identify their substrates or through bioinformatic studies that reveal homology to genes encoding enzymes of known catalytic activity (Campbell et al, 1997; Coutinho et al, 2003).
To date, Family 1 contains >900 sequences with representatives in a wide range of prokaryotic and eukaryotic organisms. Substrates for these glycosyltransferases are small, lipophilic molecules in which single or multiple glycosylation can take place at –OH, –COOH, –NH2, –SH and C–C groups (Sandermann and Pflugmacher, 1998; Ikan, 1999; Radominska-Pandya et al, 1999; Vogt and Jones, 2000; Jones and Vogt, 2001). Within Family 1, there is a class of enzymes defined by the presence of a consensus sequence of 44 amino acids. This class is expanding rapidly since individuals can be easily recognised in genome sequencing projects. Currently, 48.3% of Family 1 contains this consensus (Coutinho and Henrissat, personal communication). Figure 1 illustrates the relatedness of the different glycosyltransferases described in this review.
Figure 1.
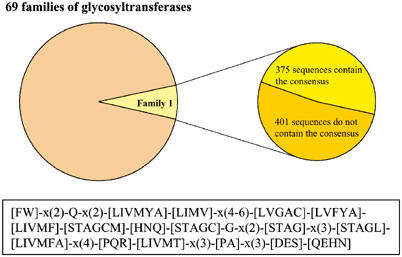
Family 1 constitute 7% of all glycosyltransferases that have been classified to date. This review focuses on the 48% of Family 1 containing the consensus sequence. The statistical data of Family 1 glycosyltransferases with and without the consensus are provided by Dr Pedro Coutinho and Professor Bernard Henrissat (AFMB-CNRS, France). The reader is referred to the CAZy (carbohydrate-active enzymes) website (http://afmb.cnrs-mrs.fr/CAZY/) for the up to date information. The 44-amino-acid consensus sequence defining the class of Family 1 glycosyltransferases described in this review is illustrated.
These enzymes have attracted very considerable interest because of their range of substrates, their potential role in developmental and metabolic homeostasis, and their function in detoxification processes of relevance to clinical and agricultural applications. The review will highlight some recent insights gained from studies of plant enzymes, in particular, those of Arabidopsis thaliana, and will discuss the findings in the wider context of related glycosyltransferases from other organisms.
Biomolecular studies of the enzymes
The consensus that defines the class of Family 1 enzymes discussed in this review was identified by Hundle et al (1992), when the sequence of a bacterial zeaxanthin glycosyltransferase was compared with those of a number of mammalian and plant enzymes. Hughes and Hughes (1994) first named the consensus as a ‘PSPG' motif in plant enzymes, and the entire class was formally classified by the Glycosyltransferase Nomenclature Committee (Mackenzie et al, 1997). In the completed genome of A. thaliana, there are 117 sequences containing the consensus, scattered across all of the five chromosomes (Li et al, 2001; Ross et al, 2001; Paquette et al, 2003). Based on their alignment, a number of the sequences are considered pseudogenes due to interruptions in their open-reading frames caused by nucleotide substitutions, insertions or deletions. In the human genome, 27 sequences have been identified (http://som.flinders.edu.au/FUSA/ClinPharm/UGT/; Bosio et al, 1996; Mackenzie et al, 1997; Tukey and Strassburg, 2000; Miners et al, 2004). The sequences are mapped only on two chromosomes (2 and 4) and can be further grouped into three subsets: UGT1 (consists of nine functional genes and four pseudogenes, located on chromosome 2), UGT2 (consists of eight functional genes and five pseudogenes, located on chromosome 4) and UGT8 (contains a single member, located on chromosome 4). Whereas >50% of the Arabidopsis genes do not have introns, all of the human glycosyltransferases are encoded by multiple exons. A particular feature of the human glycosyltransferases is alternative splicing. This occurs in the UGT1 subset and leads to variability in the N-terminal regions of the proteins (Gong et al, 2001).
None of the plant glycosyltransferases identified to date have a signal sequence, nor any clear membrane-spanning or targeting signals (Li et al, 2001). This suggests the enzymes function in the cytosol, although within that compartment the proteins may associate as peripheral components of the endomembrane system, as suggested by Winkel-Shirley (1999). In contrast, the human glycosyltransferases have a signal sequence involved in cotranslational translocation into the rough endoplasmic reticulum (RER), as well as a transmembrane-spanning domain and an ER retention signal (Sprong et al, 1998; reviewed, Radominska-Pandya et al, 1999). The enzymes therefore clearly function in the ER, necessitating transport of nucleotide-sugar from the cytosol into the ER lumen for the transfer reaction (reviewed, Hirschberg et al, 1998).
To date, only 13 crystal structures of enzymes from the 69 glycosyltransferase families have been solved. Of these, two structural groups can be identified, GT-A and GT-B (Ünligil and Rini, 2000; Bourne and Henrissat, 2001; Tarbouriech et al, 2001). GT-A structures consist of parallel β-strands flanked on either side by α-helices. In contrast, GT-B structures consist of two Rossmann-fold-like domains separated by a deep cleft (Figure 2A). While it is clear that the primary sequences of glycosyltransferases are significantly different from one another, Hu and Walker (2002) have suggested that their three-dimensional shapes may be similar and reflect either the GT-A or GT-B structures. This suggestion is supported by a recent study of Zhang et al (2003) in which 262 representative glycosyltransferase sequences from different families were analysed using a ‘fold recognition' strategy that revealed all of the sequences investigated adopted either the GT-A or GT-B structure.
Figure 2.
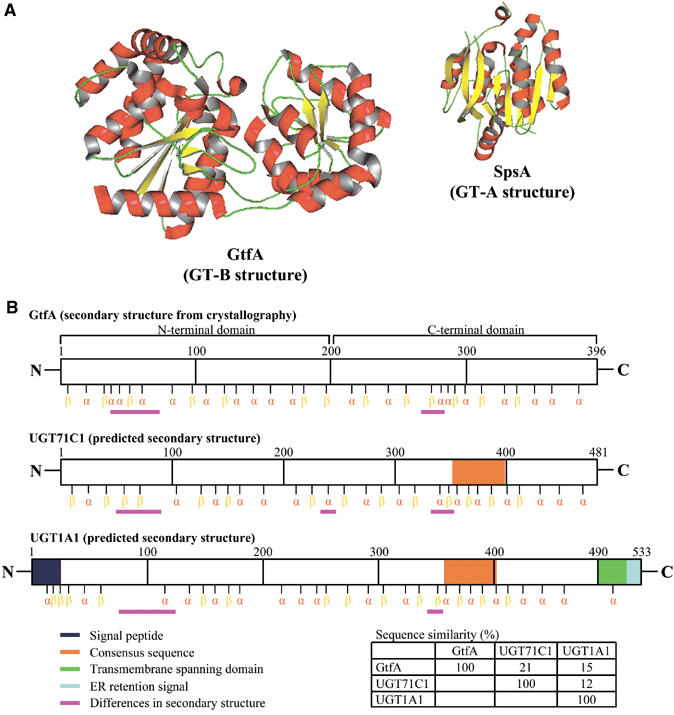
(A) Ribbon diagrams of two representative GT structures. A. orientalis GtfA (Family 1, PBD accession code 1PNV) and Bacillus subtilis SpsA (Family 2, PBD accession code 1H7L) were selected to illustrate the GT-B and GT-A structures, respectively. (B) Arabidopsis UGT71C1 and human UGT1A1 were chosen as the representative glycosyltransferases containing the consensus. Their secondary structures were predicted using a web-based programme (http://cubic.bioc.columbia.edu/predictprotein/) and were compared to the secondary structure of GtfA (without the consensus) gained from the crystallographic study.
In Family 1, two glycosyltransferases from the bacterium Amycolatopsis orientalis have been crystallised and shown to be GT-B structures (Mulichak et al, 2001, 2003). These enzymes, GtfA and GtfB, transfer sugar from NDP-sugar to the β-OH-Tyr6 and 4-OH-Phegly4 residues of vancomycin, respectively. The bacterial sequences are substantially different from those of plant and mammalian enzymes and are not classified in the same subset of Family 1, since they do not contain the 44-amino-acid consensus. Currently, no plant or mammalian glycosyltransferase has been crystallised. However, if the secondary structure of the bacterial enzyme GtfA is compared to a representative plant glycosyltransferase (UGT71C1) and a human glycosyltransferase (UGT1A1), a surprising similarity can be identified, particularly in the C-terminal region of the proteins (Figure 2B). It is possible therefore that the Family 1 class of enzymes described in this review will also adopt GT-B structures, containing the two Rossmann-fold-like domains.
Many of the aglycones recognised by human glycosyltransferases are also recognised by other enzymes and binding proteins. In some cases, the structures of these additional proteins are known and the amino acids involved in ligand binding have been identified. This has enabled a comparison to be made between sequences involved in aglycone glycosylation and other sequences that also recognise the same substrates. As reviewed by Radominska-Pandya et al (1999), conserved amino-acid residues in the N-terminal regions of the proteins have been identified that may be involved in substrate recognition. As yet, there are no similar studies on plant glycosyltransferases, but sequence alignment of the multigene family of enzymes in Arabidopsis has shown that there is much greater variability in the N-terminal regions of the proteins than in the C-terminal regions, which may reflect the diversity of aglycones recognised (Li et al, 2001).
Photoaffinity-labelling experiments have confirmed that residues in the consensus sequence of mammalian glycosyltransferases interact with UDP (reviewed, Radominska-Pandya et al, 1999). However, one study involving human UGT2B4 showed that there was also an interaction of the N-terminus with the sugar (Pillot et al, 1993). The role of this interaction in the catalytic mechanism will become clear when a structure for this enzyme class has been solved.
Glycosylation as a homeostatic mechanism of small molecule metabolism
In plants, enzymes of this class are known to recognise a great diversity of substrates including hormones, secondary metabolites and xenobiotics such as pesticides and herbicides (reviewed, Jones and Vogt, 2001; Ross et al, 2001). The sugar donor is generally UDP-glucose, although UDP-rhamnose, UDP-galactose and UDP-xylose have also been identified as activated sugars for the transfer reactions (Martin et al, 1999; Miller et al, 1999; Jones et al, 2003). There is considerable information available on the existence and diversity of glycosides, the effect of glycosylation on the activity of the acceptor molecules, and the consequences in relation to cellular homeostasis. For example, it is well documented that glycosylation alters the bioactivity of plant hormones. This has been reviewed for auxins, cytokinins, gibberellins and abscisic acid (reviewed, Kleczkowski and Schell, 1995). The reason that conjugation inactivates these hormones is unclear, but could in principle arise directly from a change in recognition by the hormone's receptor(s), or indirectly from events enabled by the glycosylation status. In this context, glycosylation is known to provide access to membrane-bound transporters. Glycosides and glucose esters of small molecules, including hormones, secondary metabolites and xenobiotics, have been shown to accumulate in the vacuolar lumen (reviewed, Coleman et al, 1997; Rea et al, 1998; Martinoia et al, 2002). Transporters for some of these compounds have been identified in the vacuolar membrane, and there is evidence to suggest that different mechanisms function for glucosides of endogenous metabolites compared to those of xenobiotics (Klein et al, 1996). Therefore, addition of a sugar residue onto an aglycone can lead both to a change in bioactivity and a change in its cellular location. If the compound is hydrophobic and can diffuse across lipid bilayers, glycosylation of the aglycone can in principle be used to contain the glycoside in a specific hydrophilic compartment—whether intracellular, such as the vacuole and endomembrane system, or extracellular, such as the cell wall matrix. In terms of control of flux in pathways of secondary metabolism, local concentrations of substrates and products in the cytosol can therefore be affected by glycosylation and removal from the compartment within which the reaction is occurring. In this way, as suggested by Hösel (1981), glycosylation and deglycosylation can be a regulatory mechanism altering levels of metabolites along a pathway through controlling exit from the cytosol or re-entry into the reaction mix.
In contrast to the diversity of sugar donors in plants, the mammalian UGT1 and UGT2 subset invariably use UDP-glucuronic acid. Known acceptors for these glucuronosyltransferases include endogenous substrates such as steroids, bilirubin and bile acids and exogenous xenobiotic substrates such as dietary flavonoids, and drugs such as morphine and naproxen. The reader is referred to recent reviews by Radominska-Pandya et al (1999), King et al (2000), Tukey and Strassburg (2000) and Miners et al (2004). UDP-glucuronosyltransferases represent the major class of enzymes involved in mammalian phase II detoxification pathways (reviewed, Bock, 2003). Glucuronides formed in mammalian cells are exported through multidrug resistance protein (MRP) transporters and are excreted from the body in the urine or bile (reviewed, König et al, 1999; Kruh and Belinsky, 2003). Unlike UGT1s and UGT2s, mammalian UGT8 transfers galactose from UDP-galactose to ceramide to form the glycosphingolipid galactosylceramide, which is the major component of the plasma membrane (reviewed, Kolter and Sandhoff, 1999).
Impact of changing the level of expression of plant glycosyltransferases
There is now a variety of genetic approaches available to investigate the action of the glycosyltransferases in the plant and how their catalytic activities may be related to physiological functions. These include the use of different promoters to upregulate the expression of glycosyltransferase genes such that enzyme activities are increased and, conversely, the use of an antisense strategy, T-DNA knockouts or gene silencing methods to downregulate expression of the endogenous gene(s). Phenotypic mutants have also been characterised and there is one example in which the defect has been surprisingly discovered in a glycosyltransferase gene (Quiel and Benders, 2003). As yet, these studies are limited in number and few general conclusions can be drawn, but examples of the effects observed will be briefly discussed.
Disturbance of hormonal homeostasis
Two studies have involved glycosyltransferases of plant hormones. The gene encoding ZOG1, a Phaseolus enzyme shown to O-glycosylate trans-zeatin in vitro, was overexpressed in tobacco callus. The only metabolite change observed involved a massive accumulation of the corresponding glucoside, and the callus required much higher levels of supplementary trans-zeatin for induction of shoot differentiation (Martin et al, 2001a). The second study involved UGT84B1, a glycosyltransferase recognising indol-3-acetic acid (IAA) in vitro (Jackson et al, 2001). The gene was overexpressed in Arabidopsis plants and the resulting transgenic plants displayed a phenotype similar to auxin deficiency (Figure 3) (Jackson et al, 2002). Interestingly, the loss of gravitropism by the transgenic root system could be recovered by applying an auxin analogue not glucosylated by UGT84B1 in vitro. While IAA glucose ester levels substantially accumulated in the transgenic plants, IAA also increased, suggesting a complex regulation of free hormone levels in planta.
Figure 3.
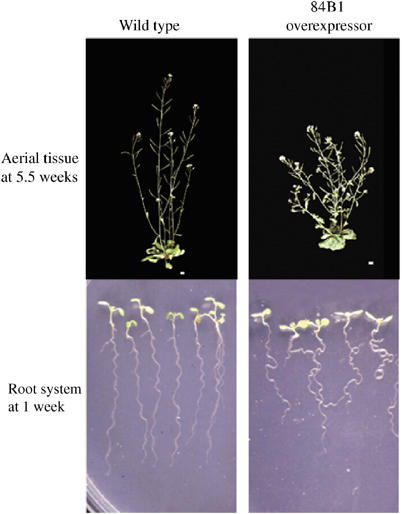
Phenotype of a transgenic line constitutively overexpressing an Arabidopsis gene UGT84B1 encoding a glycosyltransferase of the auxin indole-3-acetic acid. The aerial tissues of the transgenic Arabidopsis plant (upper panel) showed a higher degree of branching and shorter stature compared to wild type. When seedlings (lower panel) were grown vertically, the transgenic root system displayed a phenotype of impaired gravitropism.
Effects on secondary metabolism
In the above studies, overexpression of the glycosyltransferase genes led to a significant increase in their respective glucosides, but two other recent studies have shown that this is not a general consequence (Fukuchi-Mizutani et al, 2003; Lim et al, 2003b). These studies involved glycosyltransferases that glycosylate plant secondary metabolites. Under conditions in which high levels of recombinant enzyme activity in the transgenic plants were demonstrated, levels of product only marginally increased above wild type (Lim et al, 2003b).
Downregulation of TOGT, a tobacco glycosyltransferase recognising multiple phenolic substrates in vitro, led to a decreased accumulation of scopoletin glucoside in planta and, in parallel, a reduced resistance to Tobacco mosaic virus (Chong et al, 2002). Importantly, the study demonstrated that TOGT functions in planta during viral infection, although its precise role in the process was not defined. In another example of glycosyltransferase downregulation, T-DNA insertion lines for UGTs 73C6 and 78D1 were analysed. The Arabidopsis glycosyltransferases recognised quercetin and kaempferol in vitro; in the knockouts, lower levels of the glycosides were observed compared to wild type (Jones et al, 2003).
Detoxification of mycotoxins and xenobiotics
Several studies have investigated the potential of plant glycosyltransferases for detoxification applications. For example, screening of an Arabidopsis cDNA library in a yeast mutant hypersensitive to the mycotoxin deoxynivalenol revealed that colonies expressing a glycosyltransferase, UGT73C5, were protected against the effect of the mycotoxin. Overexpression of UGT73C5 in transgenic Arabidopsis led to mycotoxin resistance (Poppenberger et al, 2003). Glycosyltransferases capable of conjugating xenobiotics such as 2,4,5-trichlorophenol and 3,4-dichloroaniline in vitro have been identified (Loutre et al, 2003; Meßner et al, 2003), suggesting that transgenic plants overexpressing the respective glycosyltransferases may provide useful tools for phytoremediation. Interestingly, the enzymes that recognise the xenobiotics had previously been shown to glycosylate endogenous metabolites (Lim et al, 2001, 2002) suggesting that individual glycosyltransferases play multiple roles in the plant.
Utility of glycosyltransferases as regioselective biocatalysts
Chemical synthesis of glycosides is notoriously difficult when regioselectivity is a requirement. A classic example is the synthesis of glycosides of the important medicinal compound quercetin, which has five hydroxyl groups that can act as sugar acceptors (Aherne and O'Brien, 2002). To synthesise any single monoglycoside, four other hydroxyl groups must be protected (Bouktaib et al, 2002; Li et al, 2002). Thus, chemical synthesis of glycosides often involves multiple blocking–deblocking steps before any product can be obtained. In this context, glycosyltransferases potentially offer a simple means for the synthesis of regiospecific glycosides.
Many studies have demonstrated the regioselectivity of glycosylation by plant enzymes, as illustrated by the recent examples given in Table I that describe the substrates recognised by enzymes from different plant species and the glycosides formed. The availability of an entire multigene family of glycosyltransferases from Arabidopsis enabled a systematic investigation of regioselectivity since many activities towards a single substrate could be directly compared (Figures 4A and B). Thus, for example, 48 of the 107 Arabidopsis enzymes recognised esculetin in vitro but glucosylated the hydroxycoumarin in a regioselective manner at either the 6-OH or the 7-OH position (Lim et al, 2003a). In another example of regioselectivity, UGTs 74F1 and 74F2 are highly homologous, have evolved from a common ancestor, but UGT74F1 recognises the hydroxyl group on the benzene ring of salicylic acid, whereas UGT74F2 recognises the carboxyl group (Lim et al, 2002).
Table 1.
Recombinant plant glycosyltransferases reported recently (2001–2004)
| Substrate | Product | Organism | Reference |
|---|---|---|---|
| Hydroquinone | Arbutin | Rauvolfia | Arend et al (2001) |
| Indole-3-acetic acid | IAA glucose ester | Arabidopsis | Jackson et al (2001) |
| Phenylproponoids | Phenylpropanoid-4-O-Glc, glucose esters | Arabidopsis | Lim et al (2001) |
| Thiohydroximate | Glucosinolates | Brassica | Marillia et al (2001) |
| cis-Zeatin | cis-Zeatin-O-Glc | Zea | Martin et al (2001b) |
| Couramins | Coumarin-O-Glc | Nicotiana | Taguchi et al (2001) |
| Benzoxazinoids | Benzoxazinoid-O-Glc | Zea | von Rad et al (2001) |
| Benzoates | Benzoate-O-Glc, glucose esters | Arabidopsis | Lim et al (2002) |
| Betanidin | Betanidin-5-O-Glc, 6-O-Glc | Dorotheanthus | Vogt (2002) |
| Flavonoids | Flavonoid-3-O-Glc, 5-O-Glc | Petunia | Yamazaki et al (2002) |
| Flavonoids | Anthocyanin-3′-O-Glc | Gentiana | Fukuchi-Mizutani et al (2003) |
| Cyanohydrins | Dhurrin | Sorghum | Hansen et al (2003) |
| Flavonols | Flavonol-3-O-Rha; flavonol-3-O-Rha-7-O-Glc | Arabidopsis | Jones et al (2003) |
| Flavonoids | Flavonoid-3-O-Glc, 7-O-Glc, 4′-O-Glc | Allium | Kramer et al (2003) |
| Caffeic acid | Caffeic-3-O-Glc | Arabidopsis | Lim et al (2003a) |
| Coumarins | Coumarin-6-O-Glc, 7-O-Glc | Arabidopsis | Lim et al (2003b) |
| 3,4-Dichloroaniline | 3,4-Dichloroaniline-N-Glc | Arabidopsis | Loutre et al (2003) |
| Trichrolophenol | Trichrolophenol-O-Glc | Arabidopsis | Meßner et al (2003) |
| Deoxynivalenol | Deoxynivalenol-3-O-Glc | Arabidopsis | Poppenberger et al (2003) |
| Anthranilate | Anthranilate glucose ester | Arabidopsis | Quiel and Bender et al (2003) |
| Flavonoid/coumarin | Flavonol-7-O-Glc, coumarin-3-O-Glc | Nicotiana | Taguchi et al (2003) |
| cis-Zeatin | cis-Zeatin-O-Glc | Zea | Veach et al (2003) |
| Isoflavonoid | Formononetin-7-O-Glc | Glycyrrhiza | Nagashima et al (2004) |
| Quercetin | Quercetin-3-O-Glc, 7-O-Glc, 3′-O-Glc, 4′-O-Glc | Arabidopsis | Lim et al (2004) |
Figure 4.

(A) The sequences illustrated are those of the phylogenetic group E of Arabidopsis glycosyltransferases (Li et al, 2001). Regioselective glycosylation of esculetin falls into two distinct subsets in Group E, with the exception of UGTs 72B1 and 72B3, which suggests that a switching event in regioselectivity has occurred during evolution (Lim et al, 2003a). Sequences not analysed are labelled in black. (B) UGTs 74F1 and 74F2 are 82% identical at the amino-acid sequence level, but display different regioselectivity towards salicylic acid. (C) UrdGT1b and UrdGT1c are Streptomyces fradiae glycosyltransferases involved in the biosynthesis of the antibiotic urdamycin. Gene shuffling using the DNA sequences encoding these two glycosyltransferases enabled the generation of an engineered protein (cyan) with a catalytic activity different from the parental enzymes (Hoffmeister et al, 2002).
Studies such as these provide a good foundation for exploring the structural determinants involved in governing regioselectivity and substrate recognition and can also inform the design of novel biocatalysts for glycoside synthesis through directed evolution. In this context, Hoffmeister et al (2002) have recently reported the use of gene shuffling as a tool to generate mutants of urdamycin glycosyltransferase possessing novel catalytic activity (Figure 4C).
A limitation in the use of these enzymes as biocatalysts is the perceived requirement of activated sugars such as UDP-glucose, and several studies have addressed the potential for regeneration of the sugar-nucleotides (Heidlas et al, 1992; Ichikawa et al, 1992; Wong et al, 1992). Given that UDP-glucose is abundant in bacterial cells (Ross et al, 1991), the possibility of using the endogenous metabolite to synthesise glycosides in a whole-cell biocatalysis process has been explored (Arend et al, 2001; Lim et al, 2004). The success of this approach has been demonstrated for the synthesis of hydroquinone glucoside and a variety of quercetin glucosides. Furthermore, bacterial cells expressing different glycosyltransferases or a range of other enzymes involved in the substrate modification can be co-cultured in a fermenter to synthesise a diverse range of products using an in vivo combinatorial biochemistry (Willits et al, 2004).
Concluding remarks
The number of glycosyltransferases classified in this subset of Family 1 is likely to increase in the coming years through the increased availability of gene sequences from pro- and eukaryotic organisms. Studying these enzymes highlights a common problem in postgenomic science—how to relate gene sequence to cellular function. Catalytic activity can be screened in vitro, but the precise role of the glycosyltransferases in the cell continues to remain elusive. In plants, several tens of thousands of small molecule glycosides are known to exist, suggesting that the enzymes will recognise multiple substrates. The availability of the entire multigene family of enzymes in Arabidopsis has also shown for the first time that multiple enzymes of a plant are capable of recognising the same substrate. In addition, since glycosyltransferases are known to be involved in detoxification pathways in plant and mammalian cells, it is probable that the same enzymes will recognise both endogenous metabolites and xenobiotics. Genetic studies are now beginning to examine the metabolic and developmental consequences of silencing glycosyltransferases. In this context, it will be interesting to determine the effects of silencing individuals compared with silencing groups of sequences that share common substrates since the enzymes may well function in a network of complementary activities.
Acknowledgments
We thank Pedro Coutinho and Bernard Henrissat (AFMB-CNRS, France) for providing statistical information of Family 1 glycosyltransferases, and Gideon Davies and Jiaxing Yao (Department of Chemistry, University of York, York, UK) for help with preparation of the protein cartoons. The work described from the University of York was funded by Biotechnology and Biological Science Research Council (BBSRC) and the Garfield Weston Foundation.
References
- Aherne SA, O'Brien NM (2002) Dietary flavonols: chemistry, food content, and metabolism. Nutrition 18: 75–81 [DOI] [PubMed] [Google Scholar]
- Arend J, Warzecha H, Hefner T, Stockigt J (2001) Utilizing genetically engineered bacteria to produce plant-specific glucosides. Biotechnol Bioeng 76: 126–131 [DOI] [PubMed] [Google Scholar]
- Bock KW (2003) Vertebrate UDP-glucuronosyltransferases: functional and evolutionary aspects. Biochem Pharmacol 66: 691–696 [DOI] [PubMed] [Google Scholar]
- Bosio A, Binczek E, Lebeau MM, Fernald AA, Stoffel W (1996) The human gene CGT encoding the UDP-galactose ceramide galactosyl transferase (cerebroside synthase): cloning, characterization, and assignment to human chromosome 4, band q26. Genomics 34: 69–75 [DOI] [PubMed] [Google Scholar]
- Bouktaib M, Atmani A, Rolando C (2002) Regio- and stereoselective synthesis of the major metabolite of quercetin, quercetin-3-O-β-D-glucuronide. Tetrahedron Lett 43: 6263–6266 [Google Scholar]
- Bourne Y, Henrissat B (2001) Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr Opin Struct Biol 11: 593–600 [DOI] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B (1997) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J 326: 929–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Baltz R, Schmitt C, Beffa R, Fritig B, Saindrenan P (2002) Downregulation of a pathogen-responsive tobacco UDP-Glc:phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell 14: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JOD, Blake-Kalff MMA, Davies TGE (1997) Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci 2: 144–151 [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317 [DOI] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M, Okuhara H, Fukui Y, Nakao M, Katsumoto Y, Yonekura-Sakakibara K, Kusumi T, Hase T, Tanaka Y (2003) Biochemical and molecular characterization of a novel UDP-glucose:anthoyanin 3′-O-glucosyltransferase, a key enzyme for blue anthocyanin biosynthesis, from gentian. Plant Physiol 132: 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q-H, Cho JW, Huang T, Potter C, Gholam N, Basu NK, Kubota S, Carvalho S, Pennington MW, Owens IS, Popescu NC (2001) Thirteen UDPglucuronosyltransferase genes are encoded at the human UGT1 gene complex locus. Pharmacogenetics 11: 357–368 [DOI] [PubMed] [Google Scholar]
- Hansen KS, Kristensen C, Tattersall DB, Jones PR, Olsen CE, Bak S, Møller BL (2003) The in vitro substrate regiospecificity of recombinant UGT85B1, the cyanohydrin glucosyltransferase from Sorghum bicolour. Phytochemistry 64: 143–151 [DOI] [PubMed] [Google Scholar]
- Heidlas JE, Lees WJ, Whitesides GM (1992) Practical enzyme-based syntheses of uridine 5′-diphosphogalactose and uridine 5′-diphospho-N-acetylgalactosamine on a gram scale. J Org Chem 57: 152–157 [Google Scholar]
- Hirschberg CB, Robbins PW, Abeijon C (1998) Transporters of nucleotide sugar, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem 67: 49–69 [DOI] [PubMed] [Google Scholar]
- Hoffmeister D, Wilkinson B, Foster G, Sidebottom PJ, Ichinose K, Bechtold A (2002) Engineered urdamycin glycosyltransferases are broadened and altered in substrate specificity. Chem Biol 9: 287–295 [DOI] [PubMed] [Google Scholar]
- Hösel W (1981) Glycosylation and glycosidases. In The Biochemistry of Plant, Stumpf PK, Conn EE (eds) pp 725–753. New York: Acedemic Press [Google Scholar]
- Hu Y, Walker S (2002) Remarkable structural similarities between diverse glycosyltransferases. Chem Biol 9: 1287–1296 [DOI] [PubMed] [Google Scholar]
- Hughes J, Hughes MA (1994) Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Sequence 5: 41–49 [DOI] [PubMed] [Google Scholar]
- Hundle BS, O'Brien DA, Alberti M, Beyer P, Hearst JE (1992) Functional expression of zeaxanthin glucosyltransferase from Erwinia herbicola and a proposed uridine-diphosphate binding site. Proc Natl Acad Sci USA 89: 9321–9325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa Y, Lin YC, Dumas DP, Shen GJ, Garcia-Junceda E, Willams MA, Bayer R, Ketcham C, Walker LE, Paulson JC, Wong CH (1992) Chemical-enzymatic synthesis and conformational analysis of sialyl Lewis x and derivatives. J Am Chem Soc 114: 9283–9298 [Google Scholar]
- Ikan R (1999) Naturally Occurring Glycosides. Chichester (UK): Wiley [Google Scholar]
- Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J, Sandberg G, Bowles DJ (2002) Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterisation of transgenic lines. Plant J 32: 573–583 [DOI] [PubMed] [Google Scholar]
- Jackson RG, Lim E-K, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ (2001) Identification and biochemical characterisation of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem 276: 4350–4356 [DOI] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima J-I, Schäffner A, Saito K (2003) UGT73C6 and UGT78D1—glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278: 43910–43918 [DOI] [PubMed] [Google Scholar]
- Jones P, Vogt T (2001) Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213: 164–174 [DOI] [PubMed] [Google Scholar]
- King CD, Rios GR, Green MD, Tephly TR (2000) UDP-glucuronosyltransferases. Curr Drug Metab 1: 143–161 [DOI] [PubMed] [Google Scholar]
- Kleczkowski K, Schell F (1995) Phytohormone conjugates: nature and function. Crit Rev Plant Sci 14: 283–298 [Google Scholar]
- Klein M, Weissenböck G, Dufaud A, Gaillard C, Kreuz K, Martinoia E (1996) Different energization mechanisms drive the vacuolar uptake of a flavonoid glucoside and a herbicide glucoside. J Biol Chem 271: 29666–29671 [DOI] [PubMed] [Google Scholar]
- Kolter T, Sandhoff K (1999) Sphingolipids—their metabolic pathways and the pathobiochemistry of neurodegenerative diseases. Angew Chem Int Ed 38: 1532–1568 [DOI] [PubMed] [Google Scholar]
- König J, Nies AT, Cui Y, Leier I, Keppler D (1999) Conjugate export pumps of the multidrug resistance protein (MRP) family: localization, substrate specificity, and MRP2-mediated drug resistance. Biochem Biophys Acta 1461: 377–394 [DOI] [PubMed] [Google Scholar]
- Kramer CM, Prata RTN, Willits MG, De Luca V, Steffens JC, Graser G (2003) Cloning and regiospecificity studies of two flavonoid glucosyltransferases from Allium cepa. Phytochemistry 64: 1069–1076 [DOI] [PubMed] [Google Scholar]
- Kruh GD, Belinsky MG (2003) The MRP family of drug efflux pumps. Oncogene 22: 7537–7552 [DOI] [PubMed] [Google Scholar]
- Li M, Han X, Yu B (2002) Synthesis of quercetin 3-O-(2″-galloyl)-α-L-arabinopyranoside. Tetrahedron Lett 43: 9467–9470 [Google Scholar]
- Li Y, Baldauf S, Lim E-K, Bowles DJ (2001) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Lim E-K, Ashford DA, Hou B, Jackson RG, Bowles DJ (2004) Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotech Bioeng, (in press) [DOI] [PubMed] [Google Scholar]
- Lim E-K, Baldauf S, Li Y, Elias L, Worrall D, Spencer SP, Jackson RG, Taguchi G, Ross J, Bowles DJ (2003a) Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 13: 139–145 [DOI] [PubMed] [Google Scholar]
- Lim E-K, Doucet CJ, Elias L, Worrall D, Li Y, Ross J, Bowles DJ (2002) Activity of the group 1 glycosyltransferases of Arabidopsis towards salicylic acid, para-hydroxybenzoic acid and other benzoates. J Biol Chem 277: 586–592 [DOI] [PubMed] [Google Scholar]
- Lim E-K, Higgins GS, Li Y, Bowles DJ (2003b) Regioselectivity of glucosylation of caffeic acid by a UDP-glucose:glucosyltransferase is maintained in planta. Biochem J 373: 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E-K, Li Y, Parr A, Jackson R, Ashford DA, Bowles DJ (2001) Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J Biol Chem 276: 4344–4349 [DOI] [PubMed] [Google Scholar]
- Loutre C, Dixon DP, Brazier M, Slater M, Cole DJ, Edwards R (2003) Isolation of a glucosyltransferase from Arabidopsis thaliana active in the metabolism of the persistent pollutant 3,4-dichloroaniline. Plant J 34: 485–493 [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Owen IS, Burchell B, Bock KW, Bairoch A, Bélanger A, Fournel-Gigleux S, Green M, Hum DW, Iyanagi T, Lancet D, Louisot P, Magdalou J, Chowdhury JR, Ritter JK, Schachter H, Tephly TR, Tipton KF, Nebert DW (1997) The UDP glycosyltransferases gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7: 255–269 [DOI] [PubMed] [Google Scholar]
- Marillia EF, MacPherson JM, Tsang EWT, van Audenhove K, Keller WA, GrootWassink JWD (2001) Molecular cloning of a Brassica napus thiohydroximate S-glucosyltransferase gene and its expression in Escherichia coli. Physiol Plant 113: 176–184 [DOI] [PubMed] [Google Scholar]
- Martin RC, Mok DWS, Smets R, van Onckelen HA, Mok MC (2001a) Development of transgenic tobacco harbouring a zeatin O-glucosyltransferase gene from Phaseolus. In vitro Cell Dev Biol Plant 37: 354–360 [Google Scholar]
- Martin RC, Mok MC, Habben JE, Mok DWS (2001b) A maize cytokinin gene encoding an O-glucosyltransferase specific to cis-zeatin. Proc Natl Acad Sci USA 98: 5922–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RC, Mok MC, Mok DWS (1999) A gene encoding the cytokinin enzyme zeatin O-xylosyltransferase of Phaseolus vulgaris. Plant Physiol 120: 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu Ü, Müller-Röber B, Schulz B (2002) Multifunctionality of plant ABC transporters—more than just detoxifiers. Planta 214: 345–355 [DOI] [PubMed] [Google Scholar]
- Meßner B, Thulke O, Schäffner AR (2003) Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta 217: 138–146 [DOI] [PubMed] [Google Scholar]
- Miller KD, Guyon V, Evans JNS, Shuttleworth WA, Taylor LP (1999) Purification, cloning, and heterologous expression of a catalytically efficient flavonol 3-O-galactosyltransferase expressed in the male gametophyte of Petunia hybrida. J Biol Chem 274: 34011–34019 [DOI] [PubMed] [Google Scholar]
- Miners JO, Smith PA, Sorich MJ, Mckinnon RA, Mackenzie PI (2004) Predicting human drug glucuronidation parameters: application of in vitro and in silico modelling approaches. Annu Rev Pharmacol Toxicol 44: 1–25 [DOI] [PubMed] [Google Scholar]
- Mulichak AM, Losey HC, Lu W, Wawrzak Z, Walsh CT, Garavito RM (2003) Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc Natl Acad Sci USA 100: 9238–9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulichak AM, Losey HC, Walsh CT, Garavito RM (2001) Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure 9: 547–557 [DOI] [PubMed] [Google Scholar]
- Nagashima S, Inagaki R, Kubo A, Hirotani M, Yoshikawa T (2004) cDNA cloning and expression of isoflavonoid-specific glucosyltransferase from Glycyrrhiza echinata cell-suspension cultures. Planta 218: 456–459 [DOI] [PubMed] [Google Scholar]
- Paquette S, Møller BL, Bak S (2003) On the origin of family 1 plant glycosyltransferases. Phytochemistry 62: 399–413 [DOI] [PubMed] [Google Scholar]
- Pillot T, Ouzzine M, Fournel-Gigleux S, Lafaurie C, Tebbi D, Treat S, Radominska A, Lester R, Siest G, Magdalou J (1993) Determination of the human liver UDP-glucuronosyltransferase 2B4 domains involved in the binding of UDP-glucuronic acid using photoaffinity labeling of fusion proteins. Biochem Biophys Res Commun 197: 785–791 [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Berthiller F, Lucyshyn D, Sieberer T, Schuhmacher R, Krska R, Kuchler K, Glössl J, Luschnig C, Adam G (2003) Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J Biol Chem 278: 47905–47914 [DOI] [PubMed] [Google Scholar]
- Quiel JA, Bender J (2003) Glucose conjugation of anthranilate by the Arabidopsis UGT74F2 glucosyltransferase is required for tryptophan mutant blue fluorescence. J Biol Chem 278: 6275–6281 [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A, Czernik PJ, Little JM, Battaglia E, Mackenzie PI (1999) Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab Rev 31: 817–899 [DOI] [PubMed] [Google Scholar]
- Rea PA, Li Z-S, Lu Y-P, Drozdowicz YM (1998) From vacuole GS-X pumps to multispecific ABC transporters. Annu Rev Plant Physiol Plant Mol Biol 49: 727–760 [DOI] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim E-K, Bowles DJ (2001) Higher plant glycosyltransferases. Genome Biol 2: 3004.1–3004.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Mayer R, Benziman M (1991) Cellulose biosynthesis and function in bacteria. Microbiol Rev 55: 35–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandermann H, Pflugmacher S (1998) Taxonomic distribution of plant glucosyltransferases acting on xenobiotics. Phytochemistry 49: 507–511 [DOI] [PubMed] [Google Scholar]
- Sprong H, Kruithof B, Leijendekker R, Slot JW, van Meer G, van der Sluijs P (1998) UDP-galactose:ceramide galactosyltransferase is a class I integral membrane protein of the endoplasmic reticulum. J Biol Chem 273: 25880–25888 [DOI] [PubMed] [Google Scholar]
- Taguchi G, Ubukata T, Hayashida N, Yamamoto H, Okazaki M (2003) Cloning and characterization of a glucosyltransferase that reacts on 7-hydroxyl group of flavonol and 3-hydroxyl group of coumarin from tobacco cells. Arch Biochem Biophys 420: 95–102 [DOI] [PubMed] [Google Scholar]
- Taguchi G, Yazawa T, Hayashida N, Okazaki M (2001) Molecular cloning and heterologous expression of novel glucosyltransferases from tobacco cultured cells that have broad substrate specificity and are induced by salicylic acid and auxin. Eur J Biochem 268: 1–10 [DOI] [PubMed] [Google Scholar]
- Tarbouriech N, Charnock SJ, Davies GJ (2001) Three-dimensional structures of the Mn and Mg dTDP complexes of the family GT-2 glycosyltransferase SpsA: a comparison with related NDP-sugar glycosyltransferases. J Mol Biol 314: 655–661 [DOI] [PubMed] [Google Scholar]
- Tukey RH, Strassburg CP (2000) Human UDP-glucuronosyltransferases: metabolism, expression and disease. Annu Rev Pharmacol Toxicol 40: 581–616 [DOI] [PubMed] [Google Scholar]
- Ünligil UM, Rini JM (2000) Glycosyltransferase structure and mechanism. Curr Opin Struct Biol 10: 510–517 [DOI] [PubMed] [Google Scholar]
- Veach YK, Martin RC, Mok DWS, Malbeck J, Vankova R, Mok MC (2003) O-glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol 131: 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T (2002) Substrate specificity and sequence analysis define a polyphyletic origin of betanidin 5- and 6-O-glucosyltransferase from Dorotheanthus bellidiformis. Planta 214: 492–495 [DOI] [PubMed] [Google Scholar]
- Vogt T, Jones P (2000) Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci 5: 380–386 [DOI] [PubMed] [Google Scholar]
- von Rad U, Hüttl R, Lottspeich F, Gierl A, Frey M (2001) Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J 28: 633–642 [DOI] [PubMed] [Google Scholar]
- Willits MG, Giovanni M, Prata RTN, Kramer CM, Luca VD, Steffens JC, Graser G (2004) Bio-fermentation of modified flavonoids: an example of in vivo diversification of secondary metabolites. Phytochemistry 65: 31–41 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (1999) Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant 107: 142–149 [Google Scholar]
- Wong CH, Wang R, Ichikawa Y (1992) Regeneration of sugar nucleotide for enzymatic oligosaccharide synthesis: use of Gal-1-phosphate uridyltransferase in the regeneration of UDP-galactose, UDP-2-deoxygalactose, and UDP-galactosamine. J Org Chem 57: 4343–4344 [Google Scholar]
- Yamazaki M, Yamagishi E, Gong ZZ, Fukuchi-Mizutani M, Fukui Y, Tanaka Y, Kusumi T, Yamaguchi M, Saito K (2002) Two flavonoid glucosyltransferases from Petunia hybrida: molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol Biol 48: 401–411 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kochhar S, Grigorov M (2003) Exploring the sequence-structure protein landscape in the glycosyltransferase family. Protein Sci 12: 2291–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]


