Abstract
The final irreversible step in the duplication and dissemination of eukaryotic genomes takes place when sister chromatid pairs split and separate in anaphase. This is triggered by the protease separase that cleaves the Scc1 subunit of ‘cohesin', the protein complex responsible for holding sister chromatids together in metaphase. Only part of cellular cohesin is bound to chromosomes in metaphase, and it is unclear whether and how separase specifically targets this fraction for cleavage. We established an assay to compare cleavage of chromatin-bound versus soluble budding yeast cohesin. Scc1 in chromosomal cohesin is significantly preferred by separase over Scc1 in soluble cohesin. The difference is most likely due to preferential phosphorylation of chromatin-bound Scc1 by Polo-like kinase. Site-directed mutagenesis of 10 Polo phosphorylation sites in Scc1 slowed cleavage of chromatin-bound cohesin, and hyperphosphorylation of soluble Scc1 by Polo overexpression accelerated its cleavage to levels of chromosomal cohesin. Polo is bound to chromosomes independently of cohesin's presence, providing a possible explanation for chromosome-specific cohesin modification and targeting of separase cleavage.
Keywords: chromosome segregation, cohesin cleavage, Polo-like kinase, Scc1, separase
Introduction
Accurate chromosome segregation is crucial for the faithful inheritance of genomes during growth and proliferation of eukaryotic organisms. Failures in this process lead to aneuploidy, a state of missing, or supernumerous chromosomes, which is the cause of developmental abnormalities, for example, Down's syndrome, and is associated with malignant tumour development (reviewed in Jallepalli and Lengauer, 2001).
Sister chromatids of each chromosome remain connected with each other after their synthesis by a chromosomal protein complex called cohesin. This identifies the pairs of sister chromatids for bipolar alignment on the mitotic spindle. In metaphase, cohesin counteracts the pulling force that the spindle exerts on sister chromatids. The separation of sister chromatids at the metaphase to anaphase transition is triggered when the Scc1 subunit of cohesin is proteolytically cleaved by a conserved protease called separase (Uhlmann et al, 2000). Scc1 is cleaved at two specific separase recognition sites, leading to opening of the closed proteinaceous ring structure of the cohesin complex, that may have held sister chromatids together by topological embrace (for reviews, see Nasmyth, 2001; Haering and Nasmyth, 2003; Uhlmann, 2003).
Cohesin cleavage is a tightly regulated event. Separase is kept inactive for most of the cell cycle by an inhibitory protein, securin. Only when all chromosomes have reached bipolar attachment on the mitotic spindle, the anaphase promoting complex (APC) is activated to ubiquitylate securin and prime it for degradation by the proteasome (reviewed in Yanagida, 2000; Uhlmann, 2001). Budding yeast cells lacking securin cannot delay anaphase onset in response to faulty chromosome attachment (Yamamoto et al, 1996). Under unchallenged conditions, however, budding yeast and vertebrate securin are not essential but in their absence chromosome segregation is inefficient (Ciosk et al, 1998; Mei et al, 2001). This stems from securin's second role as a chaperone to localise and stimulate separase (Kumada et al, 1998; Jallepalli et al, 2001; Jensen et al, 2001; Hornig et al, 2002). The finding that securin is dispensable for cell growth suggests that other levels of regulation control cohesin cleavage. In budding yeast, cohesin's Scc1 subunit is phosphorylated in mitosis by Polo-like kinase (Polo), which is important for efficient Scc1 cleavage (Alexandru et al, 2001; Stemmann et al, 2001).
An aspect of cohesin cleavage that has so far received little attention is the apparent ability of separase to discriminate between cohesin that is bound to chromosomes in metaphase and cohesin in the soluble cellular fraction. In vertebrates a large fraction of cohesin is removed from chromosomes already in prophase by a mechanism that does not involve Scc1 cleavage (Losada et al, 2000; Waizenegger et al, 2000). This ‘prophase pathway' of cohesin removal is thought to facilitate individualisation and condensation of chromosome arms as cells enter mitosis. Only a relatively small fraction of cohesin is still bound to chromosomes in metaphase, providing crucial cohesion at centromeres and between condensed sister arms. At anaphase onset, only a similarly small fraction of the total cellular cohesin is cleaved by separase (Waizenegger et al, 2000), and this fraction therefore most likely corresponds to chromatin-bound cohesin. During fission yeast anaphase, an equally small fraction of the cellular cohesin appears to be cleaved (Tomonaga et al, 2000). Targeted cleavage of only chromatin-bound cohesin would make the best use of separase activity to promptly separate sister chromatids at anaphase onset.
In budding yeast metaphase, approximately two-thirds of the total cohesin is bound to chromatin, while one-third is found in a soluble cellular fraction (Tóth et al, 1999; Weitzer et al, 2003). In contrast to higher eukaryotes, budding yeast chromosomes condense only slightly in mitosis (Guacci et al, 1994), which may allow a relatively large fraction of cohesin to remain chromatin bound. Consistently, in anaphase, the majority of, but not all, Scc1 is cleaved by separase (Uhlmann et al, 1999). It is not known whether budding yeast separase differentiates chromosomal and soluble cohesin. Here, we establish an assay to compare the rates of separase cleavage of chromatin-bound versus soluble cohesin. We find that chromatin-bound Scc1 is cleaved 2–3 times faster by separase compared to soluble cohesin. We analyse the reason behind this difference and provide evidence that Polo preferentially phosphorylates chromatin-bound cohesin to facilitate its cleavage. This demonstrates for the first time differential post-translational modification of chromosome-bound versus soluble cohesin, providing a paradigm that might be relevant to cohesin regulation in organisms other than budding yeast.
Results
Chromatin-bound Scc1 is cleaved faster than soluble Scc1
To address whether chromatin-bound budding yeast cohesin might be preferentially cleaved by separase over cohesin in the soluble cellular fraction, we established an in vitro assay to compare the cleavage rates of Scc1 in these two pools of cohesin. We prepared chromatin and soluble fractions from two metaphase-arrested cultures of budding yeast cells in which endogenous Scc1 was fused to an HA or myc epitope tag, respectively. Chromatin and soluble fractions containing reciprocal Scc1 epitope tags were mixed. This resulted in two reactions, one containing chromatin-bound Scc1-HA and soluble Scc1-myc, and one with chromatin-bound Scc1-myc and soluble Scc1-HA (see scheme of the experiment in Figure 1). Scc1 cleavage in the mixtures was induced by adding cell extracts containing overexpressed separase and maintaining the reactions at 25°C. Samples of the cleavage reaction were taken every minute to follow Scc1 cleavage. This allowed cleavage of chromatin-bound and soluble cohesin to be compared within one reaction, distinguished by their differential epitope tags.
Figure 1.
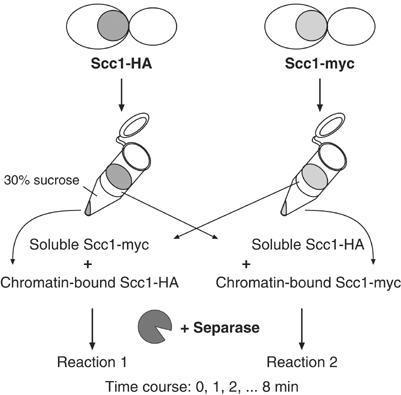
Experimental outline to compare the rates of Scc1 cleavage in chromatin-bound versus soluble cohesin. Whole-cell extracts of nocodazole-arrested cells were fractionated by centrifugation through a 30% sucrose cushion into chromatin-bound and soluble proteins. Chromatin and soluble fractions containing differentially tagged Scc1 were mixed and cell extract containing overexpressed separase was added. The two reciprocal reactions were incubated at 25°C and aliquots retrieved at 1 min intervals for analysis by quantitative Western blotting.
The amount of remaining full-length Scc1 with either epitope tag at each time point was analysed by quantitative Western blotting (Figure 2A). Scc1 cleavage over time adhered closely to the characteristics of a first order reaction, consistent with a bimolecular reaction in which Scc1 is turned over by a constant concentration of separase (Figure 2B). Comparison of the derived first order rate constants for Scc1 cleavage in each reaction showed that chromatin-bound Scc1 was cleaved significantly faster than soluble Scc1. Chromatin-bound Scc1-HA was cleaved six times faster than soluble Scc1-myc, and chromatin-bound Scc1-myc was cleaved 1.5 times faster than soluble Scc1-HA. Therefore, in both reactions, chromatin-bound cohesin was preferentially cleaved by separase. The discrepancy as to the extent of the difference was most likely caused by the differential epitope tags. When compared side by side, we consistently noticed that cleavage of Scc1-myc occurred at a reduced rate compared to corresponding fractions containing Scc1-HA (Supplementary Figure S1). The reason for this is unclear, but could be due to steric hindrance by the larger myc epitope tag (18 tandem repeats of the 10-amino-acid myc epitope, compared to six repeats of the nine-amino-acid HA epitope). Together, this suggests that the chromatin context enables Scc1 to be cleaved faster as compared to Scc1 in soluble cohesin.
Figure 2.
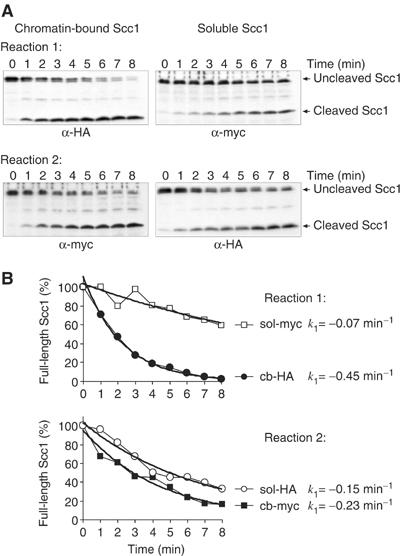
Scc1 in chromosomal cohesin is cleaved at a faster rate than Scc1 in soluble cohesin. (A) Western blot analysis of the two Scc1 cleavage reactions described in Figure 1. Reaction 1 compares cleavage of chromatin-bound Scc1-HA with soluble Scc1-myc, and reaction 2 compares cleavage of chromatin-bound Scc1-myc with soluble Scc1-HA. The strains were Y58 (MATa SCC1-myc18) and K8869 (MATa SCC1-HA6). Separase overexpression was in strain Y334 (MATa GAL-flag-ESP1-CBD). (B) Quantification of remaining full-length Scc1 in the chromatin-bound (cb) and soluble (sol) fraction over time. Bands on the Western blot in (A) were quantified using an IRDye800 coupled secondary antibody and an Odyssey fluorescence scanner (LI-COR). The graph was fitted with a first order reaction (bold line), and the first order rate constant k1 was derived.
Preferred cleavage of chromatin-bound Scc1 is maintained after its solubilisation
The faster cleavage of chromosomal Scc1 could be due to preferential separase targeting by the chromatin environment. Alternatively, chromatin-bound cohesin may acquire a property that facilitates its cleavage independently of the chromosomal context. To distinguish between these possibilities, we solubilised cohesin from chromatin by DNase I treatment. After DNase I digest, no DNA remained detectable and the released cohesin was found in a complex indistinguishable in size from soluble cohesin (Supplementary Figure S2; Weitzer et al, 2003). We then analysed the rate of cleavage of chromatin-released cohesin in the above assay. We compared cleavage of chromatin-released HA epitope-tagged Scc1 with either chromatin-bound or soluble myc epitope-tagged Scc1. Chromatin-released Scc1-HA was still four times faster cleaved than Scc1-myc from the soluble cellular fraction. It was also somewhat faster cleaved than Scc1-myc in chromatin-bound cohesin, but the latter difference was slight and probably due to the effect of the epitope tags (Figure 3A). This suggests that chromatin-released cohesin is cleaved at a similar rate to chromatin-bound cohesin. We also compared cleavage of chromatin-released myc epitope-tagged Scc1 with either chromatin-bound or soluble HA epitope-tagged Scc1. Chromatin-released Scc1-myc was cleaved at a similar rate to soluble Scc1-HA, and two-fold slower than chromatin-bound Scc1-HA (Figure 3B). Taking into account the effect of the myc epitope tag, this is consistent with the idea that cohesin released from chromatin is still cleaved at a faster rate than cohesin found in the soluble cellular fraction.
Figure 3.
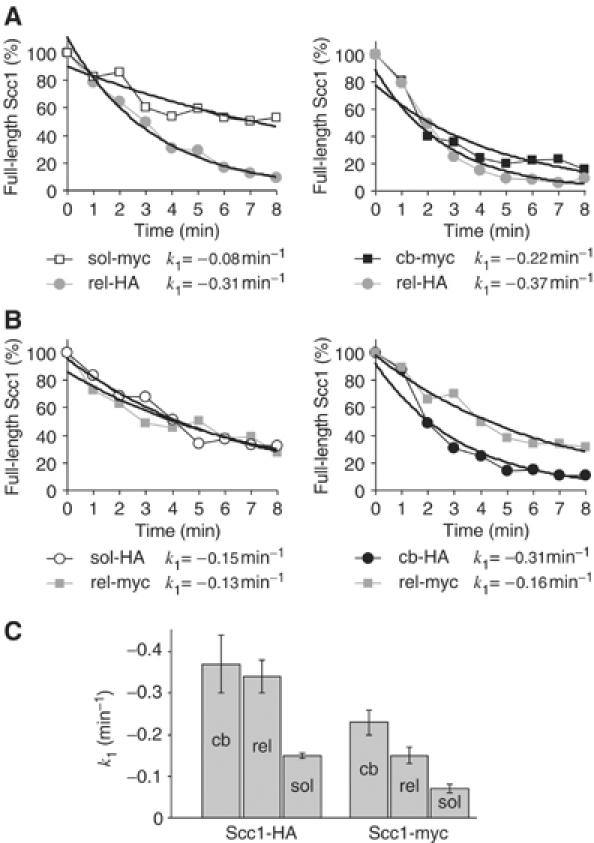
Scc1 in cohesin that was released from chromatin by DNase I treatment is cleaved at a similar rate to chromosomal Scc1. (A) Cleavage of chromatin-released Scc1-HA (rel) was compared in the same reaction with soluble (sol) or chromatin-bound (cb) Scc1-myc. (B) Cleavage of chromatin-released Scc1-myc was compared with soluble and chromatin-bound Scc1-HA. (C) Summary of rate constants obtained in parallel reactions for cleavage of Scc1 in chromosomal, chromatin-released, and soluble cohesin. All reactions contained the same volumes of soluble cell fractions and separase-enriched extract. Average rate constants are given together with error bars indicating the standard deviation (n=3 for chromatin-bound and soluble Scc1-HA and Scc1-myc, n=2 for chromatin-released Scc1).
The experiments so far compared cleavage of differentially epitope-tagged Scc1 within the same reaction mix. While providing a direct internal comparison, this method suffered from the differential effect of the two epitope tags. Throughout our experiments, we found that cleavage rates between individual reactions were highly reproducible, in particular when the sources of separase were aliquots of the same separase-enriched cell extract (compare e.g. Figures 2 and 3). We therefore compared the cleavage rates of chromatin-bound, chromatin-released, and soluble cohesin containing the same Scc1 epitope tag in parallel reactions. Figure 3C shows that both Scc1-HA and Scc1-myc in chromatin-released cohesin are cleaved 2.5 times faster than Scc1 with the same epitope tag in cohesin from the soluble cellular fraction. The cleavage of chromatin-released cohesin was comparable to chromatin-bound cohesin, but somewhat less efficient in the case of Scc1-myc. This suggests that the chromatin context may play some role in facilitating separase cleavage, but mainly that chromatin-bound cohesin has acquired an intrinsic property, maybe a post-translational modification, that allows its efficient cleavage even after chromatin release.
Chromatin-bound Scc1 is hyperphosphorylated compared to soluble Scc1
Phosphorylation of Scc1, dependent on Polo-like kinase, has been shown to contribute to its efficient cleavage (Uhlmann et al, 2000; Alexandru et al, 2001). We therefore analysed whether chromatin-bound Scc1 may be preferentially phosphorylated. Most Scc1 from chromatin-bound cohesin in metaphase shows slow migration during electrophoresis, suggestive of phosphorylation (Alexandru et al, 2001; Figure 4A and B). In contrast, a significant fraction of Scc1 in soluble cohesin migrated faster, suggesting it was less phosphorylated (Figure 4B). It therefore appears that Scc1 in chromatin-bound cohesin is preferentially phosphorylated.
Figure 4.
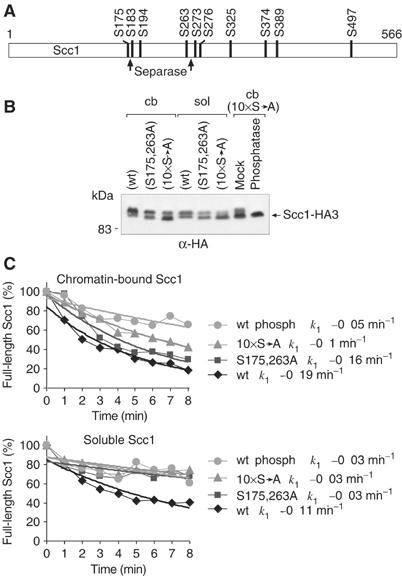
Scc1 in chromatin-bound cohesin is hyperphosphorylated compared to Scc1 in soluble cohesin. (A) Schematic representation of the 10 known Polo phosphorylation sites in Scc1. The two separase cleavage sites are indicated by arrows. (B) Migration of chromatin-bound (cb) and soluble (sol) Scc1, and the effect of mutations replacing serines S175 and S263 (S175,263A), or all 10 potential phosphoserine residues to alanine (10 × S → A). Chromatin-bound and soluble fractions of nocodazole-arrested strains Y1287 (MATa GAL-SCC1-myc18 SCC1promoter-SCC1-HA3), Y1296 (MATa GAL-SCC1-myc18 SCC1promoter-SCC1(S175,263A)-HA3), and Y1288 (MATa GAL-SCC1-myc18 SCC1promoter-SCC1(S175,183,194,263, 273,276,325,374,389,497A)-HA3) were analysed by Western blotting. Remaining slow migration of the chromatin-bound 10 × S → A mutant was resolved after treatment with λ-phosphatase. (C) Response of the Scc1 cleavage rate to phospho-site mutations and to dephosphorylation by λ-phosphatase. Scc1-HA cleavage was analysed in parallel reactions using chromatin-bound or soluble cohesin preparations.
Mass spectroscopy has identified 10 serine residues in Scc1 that can be directly phosphorylated by Polo (Alexandru et al, 2001; Figure 4A). In order to address the role of Scc1 phosphorylation in preferential cleavage of chromatin-bound cohesin, we mutated these 10 serines to alanine, individually and in combination. All mutant proteins were expressed under control of the endogenous SCC1 promoter and were able to complement for the deletion of the essential wild-type SCC1 gene. This indicates that the mutant proteins maintain the essential role of Scc1. Individual mutations showed only a minor effect (data not shown), but mutation of all 10 serines (10 × S → A) led to a significant shift in electrophoretic mobility of Scc1. A mutant changing two serines near the two separase cleavage sites was described previously (S175,263A) (Alexandru et al, 2001), and showed an intermediate mobility pattern (Figure 4B). Slower migrating bands, even though largely reduced, were still evident in the 10 × S → A mutant Scc1. These were due to residual phosphorylation as they could be removed by phosphatase treatment (Figure 4B). In all mutants, as with the wild-type protein, chromatin-bound Scc1 showed a pattern of slower mobility as compared to soluble Scc1. This indicates that Scc1 is preferentially phosphorylated when bound to chromatin, and that phosphorylation can occur at over 10 different sites within the protein.
Cleavage rate of chromatin-bound Scc1 is regulated by phosphorylation
To find out whether differential phosphorylation accounts for preferred cleavage of chromatin-bound cohesin, we compared the rates of cleavage of the phosphorylation site mutants. In two sets of experiments, we compared cleavage of wild-type, mutant, and phosphatase-treated wild-type cohesin, either chromatin bound or in the soluble fraction. Scc1 was tagged with HA epitopes in all cases, and the rates of cleavage were determined in parallel reactions. Myc epitope-tagged wild-type Scc1 was included in each reaction as an internal control. It was cleaved at the same rate in each reaction (data not shown). Figure 4C therefore directly compares cleavage of the HA epitope-tagged Scc1 variants.
The efficient cleavage of chromosomal Scc1-HA was reduced by 15% due to the S175,263A mutation, and was down by nearly half as a consequence of the 10 × S → A mutation. The reduction in cleavage was most likely due to the absence of phosphorylation, rather than other adverse effects of the mutations, because phosphatase treatment of wild-type chromosomal cohesin also caused a drastic reduction of cleavage. After phosphatase treatment, cleavage was a further two-fold slower than cleavage of the 10 × S → A mutant. This is consistent with the possibility that remaining phosphorylation even on the 10 × S → A mutant facilitates cleavage to some degree.
Scc1 in soluble wild-type cohesin was cleaved at a rate comparable to the chromatin-bound 10 × S → A mutant, and any phosphorylation site mutation reduced cleavage to rates similar to those of phosphatase-treated cohesin (Figure 4C). Together, these results suggest that the phosphorylation status of Scc1 is an important determinant of its rate of cleavage, and that the higher phosphorylation levels of Scc1 in chromatin-bound cohesin might be responsible for its faster cleavage.
Hyperphosphorylation of soluble Scc1 accelerates its cleavage
If the difference in the rate of cleavage was indeed due to preferential phosphorylation of chromatin-bound Scc1 over soluble Scc1, then enhancing Polo phosphorylation of soluble Scc1 should make it a likewise susceptible cleavage target. To test this, we used cells in which, in addition to endogenous Polo, levels of the kinase could be increased under control of the galactose-inducible GAL promoter. At 1 h after Polo induction in metaphase-arrested cells, the migration of soluble Scc1 had changed to a pattern similar to chromatin-bound Scc1, indicating increased phosphorylation (Figure 5A). We then compared the cleavage rates of soluble and chromatin-bound Scc1 before and after Polo induction. Scc1 was again tagged with HA epitopes, and cleavage rates were obtained in parallel reactions. Similar cleavage rates of Scc1-myc, contained in the reactions as an internal standard, were confirmed. Cleavage of soluble cohesin was 2.5 times accelerated after Polo induction, reaching 80% of the rate of chromatin-bound cohesin (Figure 5B). The cleavage rate of chromatin-bound cohesin increased only marginally, in keeping with its pattern of phosphorylation that did not significantly change after Polo induction. This suggests that the relative underphosphorylation of soluble cohesin is limiting its rate of cleavage. It also suggests that Polo phosphorylation of chromosomal cohesin is close to its maximum possible level in metaphase, thus ensuring fast cleavage.
Figure 5.
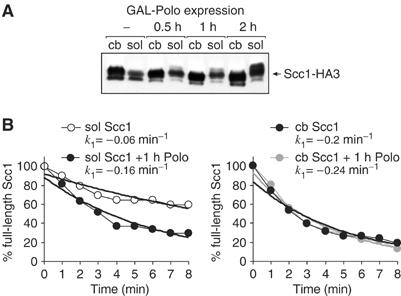
Phosphorylation of soluble Scc1 by overexpressed Polo makes its cleavage comparable to chromatin-bound Scc1. (A) Hyperphosphorylation of soluble Scc1 in strain Y1585 (MATa SCC1-HA3 CDC5 GAL-CDC5) after induction of Polo (encoded by CDC5) in metaphase-arrested cells. (B) Soluble and chromatin-bound cohesin was prepared 1 h after Polo induction, and its cleavage was compared to soluble and chromatin-bound cohesin from uninduced cells.
Scc1 phosphorylation enhances both recognition and cleavage by separase
We next addressed the mechanism responsible for preferential separase cleavage of phosphorylated Scc1. Separase might show enhanced affinity for phosphorylated Scc1, or phosphorylation might in another way facilitate the cleavage reaction. To investigate these possibilities, we first measured cleavage of recombinant, phosphorylated Scc1 by purified separase. Recombinant Scc1 can be isolated in a mitotically phosphorylated form after overexpression in insect cells that have been treated with the phosphatase inhibitor okadaic acid (Uhlmann et al, 2000). We compared cleavage of mitotically phosphorylated recombinant Scc1 to cleavage of the same protein after dephosphorylation with λ-phosphatase. Separase was purified from yeast via chitin affinity chromatography (Hornig et al, 2002). Saturating amounts of Scc1 were added to the reaction, and quantitative Western blotting revealed that after 10 min at 25°C separase had cleaved almost 10 times more mitotically phosphorylated Scc1 as compared to phosphatase-treated Scc1 (Figure 6A). The difference was due to the phosphorylation status of Scc1, rather than an effect of the phosphatase on separase activity, because preincubation of separase with λ-phosphatase did not change its activity (Figure 6A). This result is consistent with previous qualitative experiments that demonstrated a requirement of okadaic acid-induced or Polo-mediated phosphorylation for cleavage of recombinant Scc1 (Uhlmann et al, 2000; Alexandru et al, 2001).
Figure 6.
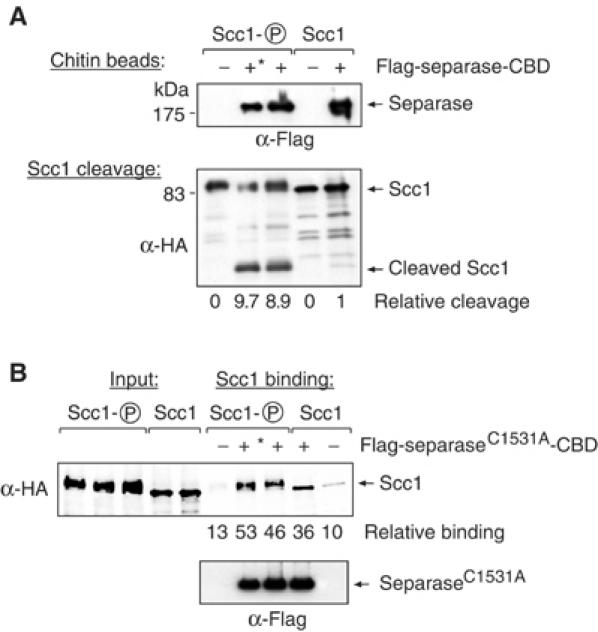
Fast cleavage of recombinant, phosphorylated Scc1 is partly due to higher affinity for separase. (A) Phosphorylated recombinant Scc1 (Scc1-P) is cleaved faster than phosphatase-treated Scc1 (Scc1). Recombinant Scc1 was incubated with separase and purified by chitin affinity chromatography from strain Y334. Cleavage was analysed by quantitative Western blotting, and relative amount of the cleavage product in each reaction is given. In one reaction, separase was phosphatase treated, denoted by *, before incubation with Scc1. (B) Separase binding to Scc1. Phosphorylated and phosphatase-treated recombinant Scc1 was bound to catalytically inactive separase (C1531A) that was affinity purified on chitin beads. Bound protein was analysed by quantitative Western blotting; binding is indicated relative to the respective input. A reaction in which separase was phosphatase treated is also included, denoted by *.
To analyse whether separase has a higher affinity for phosphorylated versus unphosphorylated Scc1, we repeated the experiment, but this time purified a proteolytically inactive separase protein containing a point mutation in the active site cysteine residue (C1531A). This prevented Scc1 cleavage in the reaction and thus enabled the examination of substrate binding. SeparaseC1531A was purified on chitin beads, similar amounts of mitotically phosphorylated or phosphatase-treated recombinant Scc1 were added, and bound Scc1 was quantified. Both phosphorylated and phosphatase-treated Scc1 associated with separase, but 1.5 times more phosphorylated Scc1 was recovered (Figure 6B). This small difference in association was reproducible, and in several repeats of this experiment 1.5–2 times more phosphorylated Scc1 bound to separase compared to phosphatase-treated Scc1. This indicates that separase possesses higher affinity for phosphorylated Scc1. However, discrimination against binding to unphosphorylated Scc1 is not large enough to explain the almost 10-fold difference in the efficiency of cleavage. Scc1 phosphorylation, in addition to increasing affinity to separase, might therefore facilitate cleavage more directly, maybe by providing substrate-induced catalysis.
Phosphorylation-dependent preferential cleavage of chromatin-bound Scc1 in vivo
So far we have shown that chromatin-bound cohesin is preferentially cleaved over soluble cohesin by separase in vitro, and that Scc1 phosphorylation is likely to play a role in determining this difference. We next addressed whether preferred cleavage of chromatin-bound cohesin takes place during budding yeast anaphase in vivo. We arrested cells in metaphase by depletion of the APC activator Cdc20 under control of the galactose-inducible GAL promoter, and released cells into synchronous anaphase by reinduction of Cdc20 (Uhlmann et al, 1999). Anaphase at the normal growth temperature of 25°C occurs very rapidly, therefore release was performed at 16°C to slow mitotic progression and facilitate analysis. Samples were taken every 5 min, soluble and chromatin-bound fractions separated, and levels of remaining full-length Scc1 over time were analysed by quantitative Western blotting. Full-length Scc1 started to disappear from the chromatin fraction 15 min after release into anaphase, but a reduction in the soluble fraction only became obvious after 25 min (Figure 7A). Therefore, chromatin-bound cohesin appears to be preferentially cleaved during budding yeast anaphase. This may aid the rapid resolution of sister chromatid cohesion at anaphase onset.
Figure 7.
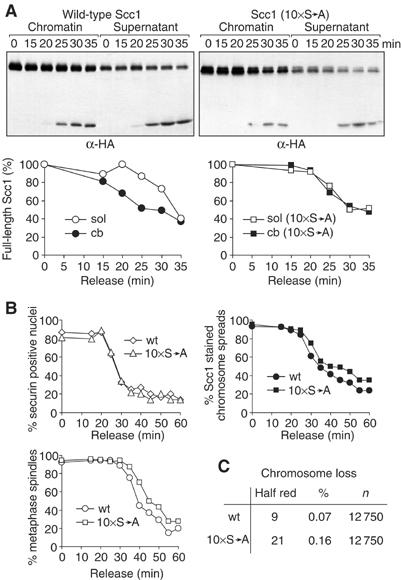
Phosphorylation accelerates cleavage of chromosomal Scc1 in vivo. (A) Faster cleavage of chromatin-bound Scc1 in vivo depends on phosphoserine residues. Strains Y1447 (MATa GAL-CDC20 PDS1-myc18 SCC1promoter-SCC1-HA3) and Y1449 (MATa GAL-CDC20 PDS1-myc18 SCC1promoter-SCC1(S175,183,194,263, 273,276,325,374,389,497A)-HA3) were arrested in metaphase by depleting Cdc20 and released into synchronous anaphase at 16°C. Soluble and chromatin-bound Scc1 was separated in samples taken at the indicated time points, and full-length Scc1 was quantified. Note that cleaved cohesin is released from chromatin, thus the Scc1 cleavage product in the soluble fraction originates partly from chromatin. (B) 10 × S → A mutant Scc1 dissociates from chromatin later and delays elongation of the anaphase spindle. As (A), but degradation of securin and spindle elongation were analysed by indirect immunofluorescence, and Scc1 binding to chromatin was visualised on chromosome spreads. (C) 10 × S → A mutant Scc1 confers increased chromosome loss. Strains Y1305 (MATa CFIII (CEN3.L.YPH278) URASUP11 SCC1promoter-SCC1-HA3) and Y1306 (MATa CFIII (CEN3.L.YPH278) URASUP11, SCC1promoter-SCC1-S175,183,194,263, 273,276,325,374,389,497A-HA3) were grown on medium lacking uracil to maintain a marker chromosome, and plated on rich medium. Half red sectored colonies were counted, indicative of chromosome loss in the first division after plating. Chromosome loss in the strain containing 10 × S → A Scc1 is larger than in wild type with a confidence level greater than 93.75%.
To investigate whether preferred cleavage of chromosomal cohesin in vivo depended on its phosphorylation, we repeated the experiment with a strain containing the Scc1 10 × S → A mutant as the sole source of Scc1. Cleavage of chromatin-bound cohesin was now delayed until 25 min after release into anaphase, and both chromatin-bound and soluble full-length Scc1 disappeared with similar kinetics. This suggests that the preferential cleavage of chromatin-bound cohesin in vivo depends on its phosphorylation. We also analysed whether the subcellular distribution of separase could help the preferential cleavage of chromatin-bound versus soluble cohesin. Approximately two-thirds of separase was found in the soluble cellular fraction and one-third was chromatin associated (Supplementary Figure S3). Therefore, a pool of separase exists on chromatin, but in contrast to cohesin most separase is found in the soluble cellular fraction.
Delayed cleavage of chromatin-bound cohesin slows anaphase progression and causes chromosomal instability
We next asked whether delayed cleavage of chromatin-bound cohesin had consequences for the faithful progression of mitosis. Previously, the Scc1 S175,263A mutation has been shown to cause lethality and delay cohesin cleavage by about 30 min in cells lacking securin (Alexandru et al, 2001), a situation in which separase is mislocalised and shows reduced activity (Hornig et al, 2002). The Scc1 10 × S → A mutation introduced a 10 min delay to cleavage of chromatin-bound cohesin in wild-type cells progressing through anaphase at 16°C. When we analysed chromosome segregation in these cells, compared to cells containing wild-type Scc1, we found a corresponding delay in the dissociation of cohesin from chromosomes, and of anaphase spindle elongation (Figure 7B). The time difference was small, but reproducible, and we confirmed that separase activation occurred at the exact same time in both strains as judged from the disappearance of nuclear securin.
Cells containing Scc1 10 × S → A as the sole source of Scc1 were viable and, apart from a delay, anaphase progression appeared normal. To test more rigorously the effect of this mutation on chromosome segregation, we analysed the rate of chromosome loss using a sensitive colony sectoring assay (Spencer et al, 1990). This assay indicated that a marker chromosome was lost with twice the frequency of that observed in wild-type strains, suggesting that efficient cleavage of chromatin-bound cohesin at anaphase onset is important for faithful chromosome segregation (Figure 7C).
Polo kinase colocalises with Scc1 on chromatin
Our finding that Scc1 is more highly phosphorylated when bound to chromosomes offers a plausible explanation as to why chromatin-bound cohesin is preferentially cleaved by separase. But how is chromatin-bound Scc1 preferentially phosphorylated in a Polo-dependent manner? To address this, we analysed whether Polo may colocalise with cohesin on chromosomes. In a subcellular fractionation experiment, we found that a significant part of Polo, close to half of the total cellular pool, was associated with the chromatin fraction (Figure 8A). As a comparison, approximately 5% of the total cellular protein is found in this fraction (data not shown). Chromosome binding of Polo could be confirmed by immunostaining of mitotic chromosome spreads. Interestingly, the staining of Polo on mitotic chromosomes coincided with cohesin (Figure 8B). Both proteins were enriched around two foci that most likely correspond to the two clusters of centromeres in mitosis where cohesin is known to accumulate (Blat and Kleckner, 1999). Association of Polo-like kinase (Plx1) with chromatin has also been seen on chromosomes assembled in Xenopus egg extracts (Budde et al, 2001).
Figure 8.
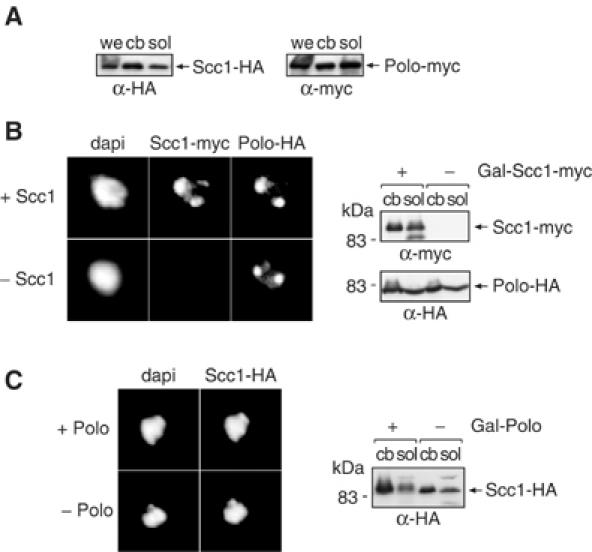
Chromatin provides a Polo-rich environment. (A) Polo in the chromatin-bound fraction. Whole extract (we), chromosomal (cb), and soluble (sol) cellular fractions were prepared from strain Y1570 (MATa SCC1-HA6 CDC5-myc18); Scc1 and Polo were detected by Western blotting. (B) Polo associates with chromatin independently of cohesin. Scc1 was depleted from strain Y1571 (MATa GAL-SCC1-myc18 CDC5-HA6) and cells were arrested in metaphase by nocodazole treatment. Chromatin binding of Scc1 and Polo was analysed by chromosome spreading, and levels of both proteins in soluble and chromatin-bound fractions were confirmed by Western blotting. (C) Cohesin binds chromatin independently of Polo. Cells were arrested by nocodazole treatment after depletion of Polo in strain Y8646 (MATa GAL-CDC5 SCC1-HA6). Polo depletion was confirmed by the fast mobility of hypophosphorylated Scc1. Cohesin binding to chromosomes was analysed by chromosome spreading and cellular fractionation.
Polo may associate with chromosomes as a consequence of binding cohesin. If this were the case, chromosomal cohesin would not have any advantage over soluble cellular cohesin with which the soluble pool of Polo could likewise associate. We therefore tested whether binding of Polo to chromosomes depended on cohesin. Scc1 was depleted from cells under control of the repressible GAL promoter, conditions under which none of cohesin's subunits binds to chromatin (Tóth et al, 1999; Weitzer et al, 2003), but the amount and pattern of Polo association with mitotic chromosomes were unaltered (Figure 8B). This suggests that Polo binds chromosomes independently of cohesin. We also tested whether cohesin binding to chromatin depended on Polo. After Polo depletion under control of the repressed GAL promoter, the levels of chromatin-bound Scc1 remained unchanged (Figure 8C). Therefore, Polo and cohesin bind to chromosomes independently of each other. The pool of chromosomal Polo may thus provide an environment for preferential phosphorylation of cohesin that binds within its vicinity.
Discussion
Anaphase onset is triggered when the Scc1 subunit of the chromosomal cohesin complex is cleaved by the protease separase. Only part of the total cellular cohesin is bound to chromosomes in metaphase, and it has been suspected that separase may specifically recognise and cleave only this fraction to separate sister chromatids, while leaving cohesin in the soluble cellular fraction intact. Direct evidence for preferential cleavage of chromosomal cohesin or suggestions as to a possible targeting mechanism have not been available. We now show that the rate of separase cleavage of chromosomal cohesin in budding yeast is indeed significantly greater when compared to cleavage of cohesin in the soluble cellular fraction, and that this is most likely due to preferential phosphorylation of chromatin-bound cohesin by Polo kinase.
In budding yeast, two-thirds of cohesin is chromatin bound in metaphase, and accordingly a relatively large fraction of cohesin is cleaved at anaphase onset. If cells are arrested in G1, or small G1 daughter cells are isolated, little or no uncleaved Scc1 remains detectable (Guacci et al, 1997; Michaelis et al, 1997). This suggests that in the course of mitotic progression most budding yeast Scc1, also in the soluble fraction, is cleaved. Nevertheless, preferential cleavage of chromosomal cohesin at anaphase onset will increase the promptness of sister chromatid separation. Some Scc1 may remain uncleaved and can be reused as large mother cells enter the subsequent S phase soon after mitosis. The origin of cohesin in the soluble cellular fraction in budding yeast metaphase is unclear. Not all cohesin may ever bind to chromosomes, and a soluble fraction of cohesin can be detected even during S and G2 phase (Weitzer et al, 2003). In higher eukaryotes, a large fraction of cohesin dissociates from chromatin during chromosome condensation in prophase, leading to a substantial pool of soluble cohesin in metaphase (Waizenegger et al, 2000). Whether a fraction of chromatin-bound cohesin becomes soluble during budding yeast chromosome condensation is not known.
In vertebrates, it has been estimated that the amount of cohesin left on chromosomes after condensation is only about 5% of the total (Losada et al, 2000). The amount of cohesin cleaved at anaphase onset appears similarly small (Waizenegger et al, 2000), but it has not been formally shown that the cleaved cohesin in fact corresponds to chromosomal cohesin. This seems likely to be the case, because cohesin only promotes sister chromatid cohesion when bound to chromosomes, and cleavage of human cohesin is required for sister chromatid separation (Ciosk et al, 2000; Hauf et al, 2001). To achieve preferential cleavage, the discrimination of human separase for chromatin-bound cohesin is likely to be greater than the two- to three-fold faster cleavage of chromosomal cohesin observed in budding yeast. The same is probably true in fission yeast, where a relatively small fraction of the total cohesin is cleaved in anaphase (Tomonaga et al, 2000). Preferential cleavage of chromatin-bound cohesin in budding yeast depends on its phosphorylation, probably by a high local concentration of chromosome-associated Polo kinase. An alternative explanation for preferential phosphorylation might be that chromatin-bound cohesin is in a distinct conformational state that assists recognition by the kinase. This appears less likely, as overexpression of Polo led to hyperphosphorylation mainly of soluble rather than chromosomal cohesin. We therefore suggest that preferential modification of chromatin-bound cohesin is due to a high local concentration of a chromosome-associated enzyme. Such a mechanism might also contribute to distinguish chromosomal from soluble cohesin in other organisms.
Could Polo-like kinase be involved in determining preferential cleavage of chromosomal cohesin also in vertebrates? There, Polo promotes separase-independent cohesin removal from chromosomes during condensation in prophase (Losada et al, 2002; Sumara et al, 2002). It seems at first sight counterintuitive that Polo could promote cleavage-independent cohesin dissociation, and at the same time mark persisting chromosomal cohesin for separase cleavage. Another chromosomal kinase could therefore facilitate cohesin cleavage in vertebrates, or Polo could phosphorylate different cohesin subunits to promote removal in prophase and cleavage in anaphase. On the other hand, Polo may in fact provide a generic mark for all chromosomal cohesin. Polo may not be specifying cohesin for one of the two pathways of removal, but it may identify all chromosomal cohesin as ‘still chromosome bound'. Such a mark could help the prophase pathway as well as cohesin cleavage by separase. Once released from chromosomes, phosphorylation may no longer be maintained. This would ensure that all mechanisms of cohesin removal are specifically targeted at remaining chromosomal cohesin.
In vertebrate, as well as Drosophila and fission yeast cells, cohesin reassociates with chromosomes very soon after mitosis as chromosomes decondense in telophase (Losada et al, 2000; Waizenegger et al, 2000; Warren et al, 2000). Whether cohesin plays a specific role in chromosomes in telophase and the following G1 phase, before cohesion between sister chromatids must be again established in the next round of DNA replication, is not known. Nevertheless, maintenance of a pool of intact soluble cohesin will facilitate cohesin's fast reassociation with chromosomes when these cells exit from mitosis.
Materials and methods
Strains and plasmids
The SCC1 coding sequence, C-terminally fused to a triple HA epitope tag, was cloned into YIplac204 (Gietz and Sugino, 1988), under control of the SCC1 promoter (1100 nt of SCC1 5′ upstream sequence). Polymerase chain reaction (PCR) was used to generate fragments containing the 10 serine to alanine mutations to replace wild-type sequences in the above construct. Constructs were confirmed by nucleotide sequencing and integrated at the TRP1 locus of a host strain in which the promoter at the endogenous SCC1 locus was replaced for the GAL promoter. This allowed repression of endogenous wild-type Scc1 in glucose-containing medium after integration of the phosphorylation site mutants. Endogenous SCC1 and Polo (CDC5) loci were fused to epitope tags using PCR-based gene targeting (Knop et al, 1999). The SCC1(S175,263A) double mutant was previously described, as was the cdc5Δ GAL-CDC5 strain (Alexandru et al, 2001). Strains containing CDC20 under GAL control were described, as well as strains overexpressing separase or separaseC1531A fused to the chitin binding domain (Uhlmann et al, 1999, 2000).
Analysis of Scc1 cleavage kinetics
Exponentially growing cells were arrested in metaphase by addition of 5 μg/ml nocodazole. Soluble and chromatin fractions were prepared after spheroplast lysis as previously described (Uhlmann et al, 1999). The Scc1 in vitro cleavage assay was modified as follows: chromatin-bound and soluble fractions containing equivalent amounts of reciprocally tagged Scc1 were mixed, ensuring chromatin was thoroughly resuspended. Chromatin-released cohesin and phosphatase-treated chromosomal cohesin were also included, which were obtained as described (Uhlmann et al, 2000; Weitzer et al, 2003). An aliquot of this mix was taken at the time point 0. Then extract containing overexpressed separase was added and the mix was incubated at 25°C with light agitation. Aliquots were taken every minute and the reaction was stopped by injecting the samples directly into preheated SDS–PAGE loading buffer. Samples were subjected to quantitative Western blotting: after Western transfer, membranes were blocked with Odyssey buffer (LI-COR):PBS (1:1), and incubated with 12CA5 and 9E10 mouse monoclonal antibodies against the HA and myc epitope, respectively, and a goat anti-mouse IRDye800 coupled secondary antibody (LI-COR). The fluorescent signal was quantified using the Odyssey infrared imager (LI-COR) and Infrared Imaging System software. Scc1 at time point 0 was set to 100% and remaining full-length Scc1 was plotted over time. Scc1 cleavage by separase adhered closely to the characteristics of a first order reaction, and graphs were fitted with exponential regression (R2 typically >0.9) using Excel software.
Hyperphosphorylation by Polo of Scc1 in soluble cohesin
Cells containing an extra copy of the CDC5 gene, encoding Polo kinase, under control of the galactose-inducible GAL promoter (Sullivan and Uhlmann, 2003), were grown in raffinose and arrested in metaphase by addition of 5 μg/ml nocodazole for 2.5 h. During this period, Polo expression was induced by adding 2% galactose 2, 1, or 0.5 h before the final arrest point.
In vivo cleavage of chromatin-bound versus soluble Scc1
Arrest in metaphase and release into synchronous anaphase using CDC20 under GAL promoter control was as described (Uhlmann et al, 1999), but cultures were cooled to 16°C before release. Aliquots of 5 × 108 cells were harvested at 5 min intervals and resuspended in ice-cold wash buffer (50 mM HEPES/KOH pH 7.5, 100 mM KCl, and 2.5 mM MgCl2). After completion of the time course, cells were resuspended in 200 μl EB buffer (50 mM HEPES/KOH pH 7.5, 100 mM KCl, 2.5 mM MgCl2, 1 mM DTT, and protease inhibitors), added to 500 μl glass beads and broken on an IKA-Vibrax shaker for 10 min. The crude cell extract was retrieved and separated into chromatin-enriched and supernatant fractions by centrifugation at 12 000 g for 10 min. Small chromatin fragments in the soluble fraction were removed by a second round of centrifugation at 100 000 g for 10 min. The chromatin pellet was washed in EB buffer supplemented with 0.25% Triton X-100 to remove detergent soluble material, and resuspended in 200 μl of the same buffer for analysis by quantitative Western blotting as above.
Separase binding and cleavage of recombinant Scc1
Purification of phosphorylated Scc1 after overexpression in baculovirus-infected insect cells, treated with okadaic acid, was as described (Uhlmann et al, 2000). Dephosphorylation of purified Scc1 was performed using λ-phosphatase (New England Biolabs). Separase and separaseC1531A were purified after overexpression in yeast by chitin affinity chromatography as described (Hornig et al, 2002). Scc1 was added to separase-decorated chitin beads, and cleavage or binding was analysed after 10 min at 25°C. SDS–PAGE loading buffer was directly added to stop the cleavage reaction, while the beads were washed extensively with EB buffer containing 0.25% Triton X-100 before bound Scc1 was analysed in SDS–PAGE loading buffer.
Other techniques
In situ immunofluorescence and chromosome spreading were performed as previously described (Michaelis et al, 1997), as was the assay to determine the frequency of chromosome loss (Spencer et al, 1990).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Acknowledgments
We thank all the members of the laboratory for sharing reagents and ideas, and Chris Lehane and Sara Nakielny for critical comments on the mansucript. FU is supported by the EMBO Young Investigator Programme.
References
- Alexandru G, Uhlmann F, Poupart M-A, Mechtler K, Nasmyth K (2001) Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105: 459–472 [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N (1999) Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98: 249–259 [DOI] [PubMed] [Google Scholar]
- Budde PP, Kumagai A, Dunphy WG, Heald R (2001) Regulation of Op18 during spindle assembly in Xenopus egg extracts. J Cell Biol 153: 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 1–20 [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K (1998) An Esp1/Pds1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93: 1067–1076 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D (1994) Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol 125: 517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A (1997) A direct link between sister chromatid cohesion and chromosome condensation revealed through analysis of MCD1 in S. cerevisiae. Cell 91: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Nasmyth K (2003) Building and breaking bridges between sister chromatids. BioEssays 25: 1178–1191 [DOI] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters J-M (2001) Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293: 1320–1323 [DOI] [PubMed] [Google Scholar]
- Hornig NCD, Knowles PP, McDonald NQ, Uhlmann F (2002) The dual mechanism of separase regulation by securin. Curr Biol 12: 973–982 [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Lengauer C (2001) Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer 1: 109–117 [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters J-M, Kinzler KW, Vogelstein B, Lengauer C (2001) Securin is required for chromosomal stability in human cells. Cell 105: 445–457 [DOI] [PubMed] [Google Scholar]
- Jensen S, Segal M, Clarke DJ, Reed SI (2001) A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J Cell Biol 152: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Kumada K, Nakamura T, Nagao K, Funabiki H, Nakagawa T, Yanagida M (1998) Cut1 is loaded onto the spindle by binding to Cut2 and promotes anaphase spindle movement upon Cut2 proteolysis. Curr Biol 8: 633–641 [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T (2002) Cohesin release is required for sister chromatid resolution but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev 16: 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T (2000) Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol 150: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J, Huang X, Zhang P (2001) Securin is not required for cellular viability but is for normal growth of mouse embryonic fibroblasts. Curr Biol 11: 1197–2101 [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Nasmyth K (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35: 673–745 [DOI] [PubMed] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P (1990) Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW (2001) Dual inhibition of sister chromatid separation at metaphase. Cell 107: 715–726 [DOI] [PubMed] [Google Scholar]
- Sullivan M, Uhlmann F (2003) A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat Cell Biol 5: 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters J-M (2002) The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell 9: 515–525 [DOI] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, Nasmyth K, Yanagida M (2000) Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev 14: 2757–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K (1999) Yeast Cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 13: 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F (2001) Secured cutting: controlling separase at the metaphase to anaphase transition. EMBO Rep 2: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F (2003) Chromosome cohesion and separation: from men and molecules. Curr Biol 13: R104–R114 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400: 37–42 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart M-A, Koonin EV, Nasmyth K (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters J-M (2000) Two distinct pathways remove mammalian cohesin complexes from chromosome arms in prophase and from centromeres in anaphase. Cell 103: 399–410 [DOI] [PubMed] [Google Scholar]
- Warren WD, Steffensen S, Lin E, Coelho P, Loupart M-L, Cobbe N, Lee JY, McKay MJ, Orr-Weaver T, Heck MMS, Sunkel CE (2000) The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr Biol 10: 1463–1466 [DOI] [PubMed] [Google Scholar]
- Weitzer S, Lehane C, Uhlmann F (2003) A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol 13: 1930–1940 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D (1996) Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J Cell Biol 133: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M (2000) Cell cycle mechanism of sister chromatid separation; roles of Cut1/separin and Cut2/securin. Genes Cells 5: 1–8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3


