Abstract
RNA-binding proteins HuR and AUF1 bind to many common AU-rich target mRNAs and exert opposing influence on target mRNA stability, but the functional interactions between HuR and AUF1 have not been systematically studied. Here, using common target RNAs encoding p21 and cyclin D1, we provide evidence that HuR and AUF1 can bind target transcripts on both distinct, nonoverlapping sites, and on common sites in a competitive fashion. In the nucleus, both proteins were found together within stable ribonucleoprotein complexes; in the cytoplasm, HuR and AUF1 were found to bind to target mRNAs individually, HuR colocalizing with the translational apparatus and AUF1 with the exosome. Our results indicate that the composition and fate (stability, translation) of HuR- and/or AUF1-containing ribonucleoprotein complexes depend on the target mRNA of interest, RNA-binding protein abundance, stress condition, and subcellular compartment.
Keywords: exosome, mRNA stability, polysome, RNA-binding protein, RNA motif
Introduction
Post-transcriptional processes such as RNA splicing, and mRNA export, stability, and translation are emerging as critical mechanisms of gene regulation in mammalian cells. These regulatory programs are primarily governed by RNA-binding proteins (RBPs) that associate with pre-mRNAs and mRNAs and ensure their proper processing (splicing, 5′ end and 3′ end modifications, export) as well as their subcytoplasmic transit, half-life, and translation rate. Specialized RBPs that bind specific mRNA subsets have received increasing attention in recent years, given their influence on the expression of genes that are essential to the stress response, cell division, immune function, and tumorigenesis. Many such RBPs have been described that selectively recognize and bind to mRNAs bearing U- or AU-rich sequences (collectively termed AU-rich elements (AREs)), and modulate their translation and/or stability (Ross, 1995). ARE-binding proteins include BRF1, AUF1 (hnRNP D), tristetraprolin (TTP), TIAR, TIA-1, KSRP, and the Hu proteins HuB, HuC, HuD, and HuR (Zhang et al, 1993; Antic and Keene, 1997; Min et al, 1997; Carballo et al, 1998; Gueydan et al, 1999; Stoecklin et al, 2002). The influence of RBPs on mRNA turnover has been studied most extensively for two ubiquitous proteins: HuR, which promotes mRNA stabilization, and AUF1, which enhances mRNA decay (Loflin et al, 1999; Brennan and Steitz, 2001).
HuR binds with high affinity and specificity to target mRNAs bearing AREs and modifies their expression by either enhancing their stability, altering their translation, or performing both functions (Brennan and Steitz, 2001; Kullmann et al, 2002; Mazan-Mamczarz et al, 2003). HuR is predominantly nuclear (>90%), but a variety of stimuli can trigger its translocation to the cytoplasm (Keene, 1999; Wang et al, 2000a; Gallouzi and Steitz, 2001). While the hypothesis that HuR exports target mRNAs to the cytoplasm awaits experimental demonstration, HuR's influence on target mRNA stabilization and translation is robustly linked to its cytoplasmic presence. Through its post-transcriptional effects on target mRNAs (such as those encoding cyclin A, cyclin B1, c-fos, VEGF, TNF-α, β-catenin, c-myc, cyclooxygenase-2, myogenin, MyoD, GM-CSF, interleukins, p21, p27, p53, and hsp70), cytoplasmic HuR is emerging as a major regulator of various cellular responses, including cell division, carcinogenesis, muscle cell differentiation, replicative senescence, immune cell activation, and the stress response (Atasoy et al, 1998; Wang et al, 2000b, 2001; Kullmann et al, 2002; Figueroa et al, 2003).
An ARE-binding protein causing transcript destabilization, AUF1, is expressed in four isoforms (p37, p40, p42, and p45) arising through alternative splicing of a common pre-mRNA (Wilson and Brewer, 1999). Some differences in the activity of the various AUF1 isoforms have been documented, but all isoforms appear to enhance target mRNA decay, a process that is closely linked to the ubiquitination and targeting of AUF1 to the proteasome (Laroia et al, 1999; Loflin et al, 1999). Like HuR, AUF1 target mRNAs also encode mitogenic, immune response, cancer-associated, stress response, and cell cycle regulatory proteins (such as c-fos, c-jun, c-myc, egr-1, interleukins, p21, hsp70, MnSOD, catalase, cyclin D1, and cdc25) (Ross, 1995; Bhattacharya et al, 1999; Lin et al, 2000).
Given that HuR and AUF1 overlap in their tissue distribution, preferentially localize in the nucleus, and influence the expression of many common target mRNAs, functional links between the two RBP families have been postulated (Gao et al, 1994; Bhattacharya et al, 1999; Chen et al, 2002; Lu and Schneider, 2004). According to a widely established view, HuR/ELAV proteins can compete with AUF1 (Park et al, 2000; Cok et al, 2004) or with other decay-promoting RBPs (Ming et al, 2001; Stoecklin et al, 2002) for binding to the same ARE on specific target mRNAs, but other regulatory schemes have not been formally tested. Moreover, no studies have directly assessed the endogenous association of HuR and AUF1 with target mRNAs. In the present work, we provide evidence that endogenous HuR and AUF1 are capable of associating with the same target RNAs on nonoverlapping sites, both in intact cells and in vitro. We propose that whether HuR and AUF1 bind concurrently or competitively to a given mRNA is critically influenced by several factors, including the target RNA sequence, the abundance of each RBP, the stress condition of the cell, and the subcellular compartment in which the ribonucleoprotein (RNP) complex is investigated.
Results
Unique and shared mRNA targets for HuR and AUF1
In order to investigate functional links between RBPs HuR and AUF1, we set out to identify collections of endogenous mRNAs to which each protein associated in human cervical carcinoma HeLa cells. Immunoprecipitation (IP) reactions were performed to isolate mRNA subsets bound to HuR and AUF1 using specific antibodies. The nonspecific association of mRNAs with IP reagents was determined by parallel incubations with IgG1. The identification of mRNAs in each IP material was achieved by reverse transcription (RT) followed by cDNA array hybridization (Tenenbaum et al, 2002; López de Silanes et al, 2004). Representative array fields illustrate the relative abundance and specificity of signals from HuR IPs, AUF1 IPs, as well as control IgG1 IPs (Figure 1A). As shown (Figure 1B), 201 HuR-specific target transcripts, 194 AUF1-specific target transcripts, and 267 transcripts shared by HuR and AUF1 were identified on this array. The association of transcripts with either HuR or AUF1 was deemed specific when Z ratios were ⩾1.00 in comparisons of signals in HuR IPs versus IgG1 IPs, or signals in AUF1 IPs versus IgG1 IPs, respectively. See http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1361 for complete array results. In each mRNA subset, AREs of either Class I or III were found in ∼80% of the transcripts tested.
Figure 1.

Identification of HuR and AUF1 target mRNAs. (A) RNAs bound to either HuR or AUF1 in HeLa whole-cell lysates were isolated by IP assay using the corresponding antibodies, and the reverse-transcribed radiolabeled products were used for cDNA array hybridization (Materials and methods). Control IP reactions were performed using IgG1. Representative fields of each array are shown (black arrows, specific signals enriched in samples obtained by either HuR IP or AUF1 IP; white arrows, signals enriched in both HuR and AUF1 IP materials). (B) Partial list of genes encoding transcripts that were significantly enriched in the HuR IP only (left), in the AUF1 IP only (center), and in both the HuR and AUF1 IPs (right, left value corresponds to Z ratio for HuR, right value for AUF1). Parentheses, Z ratios. Complete array data are available (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1361). (C) For validation of HuR and AUF1 target mRNAs, HeLa whole-cell lysates were prepared and endogenous target transcripts were detected by RT–PCR assay of the IP material. PCR products were visualized on 1% agarose gels. Amplification of housekeeping transcripts encoding GAPDH and SDHA, bound at low levels with the IP material, showed equal loading of IP samples.
The validity of the target mRNA identification scheme was tested experimentally by monitoring the presence of several randomly chosen target transcripts in each category (unique HuR targets, unique AUF1 targets, and shared HuR and AUF1 targets) by IP assay followed by RT–PCR amplification (IP+RT–PCR) using sequence-specific primers. As shown, HuR targets were effectively amplified only from HuR IPs, AUF1 targets only from AUF1 IPs, while targets of both proteins were amplified from both IP groups (Figure 1C). Typically, IgG1 IPs showed undetectable or low-level amplification signals in these PCR reactions, performed at ∼25 cycles; control mRNAs encoding GAPDH and SDHA, which were not targets of either RBP, served to monitor background binding of mRNAs to the IP material. According to the Z ratio cutoff of ⩾1.00 chosen for this analysis, transcript enrichments were calculated for several sample mRNAs by quantitative real-time RT–PCR: enrichment was ∼6-fold for a Z ratio of 1.00 (e.g., TP53 mRNA in AUF1 IPs), ∼60-fold for a Z ratio of 1.90 (e.g., CCND1 mRNA in HuR IPs), and ∼1000-fold for a Z ratio of 4.13 (e.g., PTMA mRNA in HuR IPs) (data not shown). This analysis revealed that 42% of AUF1-bound mRNAs were uniquely associated with AUF1 and not with HuR, while 43% of HuR-bound mRNAs were uniquely associated with HuR and not with AUF1. Conversely, 58% of AUF1 target mRNAs were also targets of HuR; a comparable number of HuR targets (57%) were found to be targets of AUF1 also, revealing an extensive and previously unrecognized collection of mRNAs that were common putative targets of both proteins.
RNA-dependent association of HuR and AUF1
The discovery that a large proportion of HuR target mRNAs were also targets of AUF1 (and vice versa) led us to formally examine the possibility that both HuR and AUF1 could simultaneously bind to a given RNA molecule. To begin to address this hypothesis, we performed IP reactions to ascertain the joint presence of both proteins in the same RNP complex. As shown in Figure 2A, IP reactions using an anti-AUF1 antibody yielded abundant quantities of HuR (Ctrl., top), while a reciprocal IP assay using an anti-HuR antibody was capable of yielding AUF1 (Ctrl., middle), albeit less efficiently, possibly due to a decrease in AUF1 binding caused by the HuR antibody itself, or to other as yet unidentified reasons. A control antibody (IgG1) failed to show either protein in the IP material. In additional control groups, IP reactions were supplemented with heparin, a polyanion used extensively as a competitor of RNA to dissociate complexes formed by weak electrostatic interaction, and hence possibly in a nonspecific fashion (Piñol-Roma et al, 1988). The presence of heparin (+Heparin) failed to disrupt the association between HuR and AUF1, supporting the notion that the AUF1–HuR presence in common RNP complexes was specific. Moreover, a brief incubation with RNase A (+RNase) caused HuR to dissociate from AUF1, indicating that HuR and AUF1 likely associated through their joint binding to common target RNAs or RNP complexes, and did not form direct protein–protein complexes. Evidence that these RNP interactions were also likely to occur in intact cells was obtained by using either formaldehyde or irradiation with short-wavelength ultraviolet light (UVC), each capable of crosslinking RNP complexes in living cells. The analysis of lysates prepared after exposure of cells to crosslinking agents (Figure 2A, bottom) further supported an association between HuR and AUF1 in intact cells. Additional control IP assays (Figure 2B) revealed that these RNP complexes also encompassed other general RBPs involved in RNA processing, such as hnRNP A1 and hnRNP C1/C2 (Dreyfuss et al, 1993). Together, these data supported the view that HuR and AUF1 do not associate through protein–protein interactions, but can simultaneously associate in common RNA-containing complexes.
Figure 2.
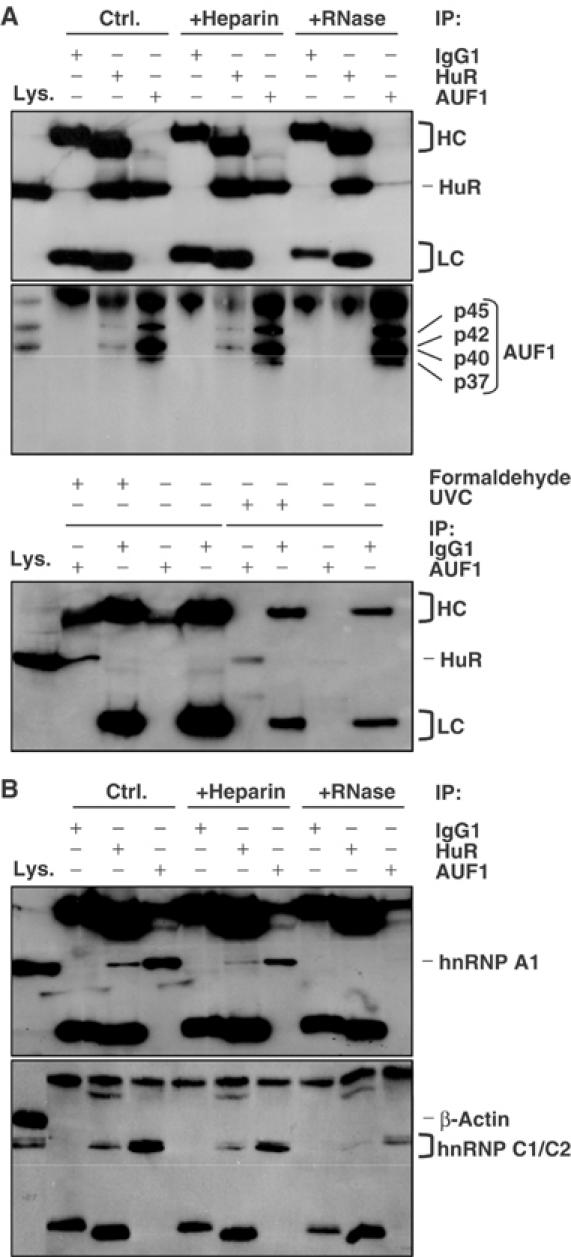
Joint presence of HuR and AUF1 within the same RNP complexes. IP assays were carried out using whole-cell lysate from HeLa cells and either anti-HuR antibody, anti-AUF1 antibody, or IgG1. IP reactions were performed without further treatment (Ctrl.), or in the presence of either heparin (+Heparin) or RNases (+RNase). (A) The presence of HuR (top) and AUF1 (four isoforms, middle) in the IP materials was monitored by Western blotting. Lys., 10 μg of whole-cell lysate, included as Western blotting control; HC, heavy chain; LC, light chain. Bottom: RNPs were crosslinked in intact cells by using either UVC irradiation or formaldehyde (Supplementary material), then subjected to IP and Western blotting. (B) The levels of hnRNP A1 and hnRNP C1/C2 in the IP materials were assessed by Western blotting.
Distinct abundance of HuR and AUF1 in different cellular compartments
The association of HuR and AUF1 was further investigated in different subcellular fractions prepared from HeLa cells. As shown in Figure 3A, HuR and AUF1 associated extensively on common RNP complexes in the nucleus (note Western blotting signals in HuR and AUF1 IP materials, Nuc. lanes). By contrast, cytoplasmic HuR (typically 5–10% of cellular HuR) associated with AUF1 to a much lesser extent (note Western blotting signals on HuR and AUF1 IP materials in the Cyto. lanes). Accordingly, HuR and AUF1 appeared much less likely to bind to the same RNAs in the cytoplasm than in the nucleus, prompting further examination of the subcytoplasmic distribution of the two proteins. In keeping with earlier reports, HuR was found to colocalize primarily with low- and high-molecular-weight (LMW and HMW, respectively) polysome fractions, prepared by centrifugation of the cytoplasmic material through sucrose gradients (Figure 3B) (Antic and Keene, 1997; Gallouzi et al, 2000). By contrast, AUF1 was found to be largely restricted to the fractions devoid of all ribosomal components (unbound (Unb.), lanes 1 and 2), where cytosolic RBPs TTP, TIAR, and TIA-1, as well as β-tubulin, were also detected. Together, these data support the view that nuclear AUF1 and HuR associate through their joint binding to common target RNAs and/or RNPs, but this association is disrupted in the cytoplasm, where AUF1 is found in the cytosolic fraction and HuR with the polysome-bound fraction.
Figure 3.
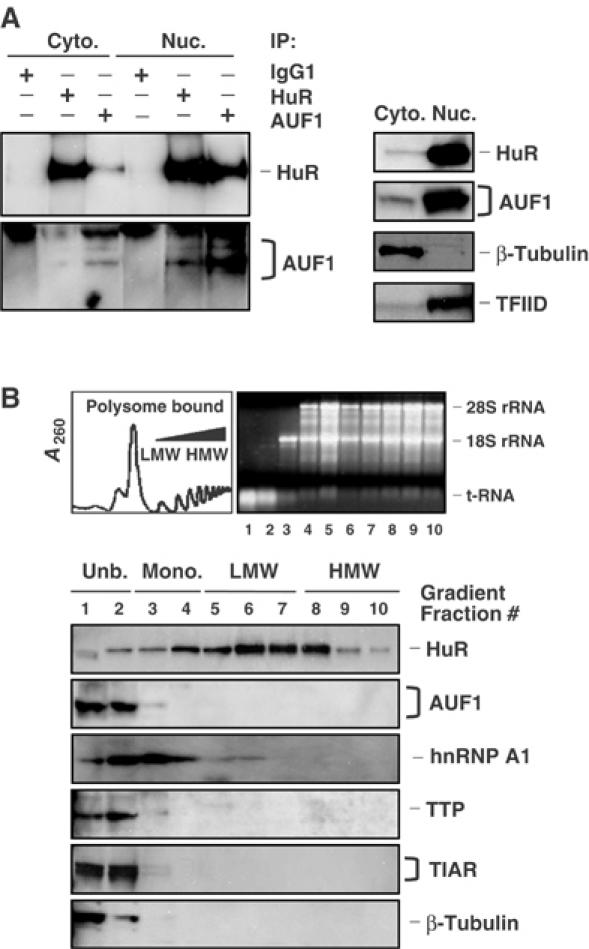
Subcellular distribution of HuR and AUF1. (A) Left: the relative abundance of HuR and AUF1 in RNP complexes obtained through IP (described in the legend of Figure 2) was assessed using either cytoplasmic (50 μg, Cyto.) or nuclear (10 μg, Nuc.) lysates. The relative abundance of HuR, AUF1, β-tubulin (a cytoplasm-specific protein), and TFIID (a nucleus-specific protein) was assessed by Western blotting using 10 μg of either cytoplasmic or nuclear lysates. (B) Representative polysome distribution profile (top left) and pattern of ethidium bromide-stained RNA (top right) from sucrose gradients (Materials and methods). From left to right: fractions lacked ribosomes or ribosome subunits (Unb.), contained ribosome subunits or single ribosomes (monosomes, Mono.), and spanned low-molecular-weight (LMW) and high-molecular-weight (HMW) polysomes. The levels of HuR, AUF1, hnRNP A1, TTP, TIAR, and β-tubulin were monitored by Western blotting using equal volumes of lysates from each fraction.
Binding of HuR and AUF1 to endogenous and synthetic mRNAs encoding p21 and cyclin D1
To obtain additional support for the joint RNA-binding model emerging from these findings, we focused on the analysis of specific mRNAs that were shared targets of both HuR and AUF1: the cyclin D1 mRNA (CCND1; Figure 1B; Lin et al, 2000) and the p21 mRNA (Joseph et al, 1998). The endogenous association of HuR and AUF1 with these two mRNAs was evidenced by IP assays using anti-HuR and anti-AUF1 antibodies followed by measurement of p21 and cyclin D1 mRNAs by RT–PCR. As indicated, both cyclin D1 and p21 mRNAs were found to be targets of HuR and AUF1; very little or no amplification was seen in the IgG1 IP lanes, and control GAPDH mRNA was only found as low-level contaminating material present in all of the IP reactions (Figure 4A). PCR amplification was performed after RT reactions using oligo-d(T), indicating that HuR and AUF1 bound polyadenylated cyclin D1 and p21 mRNAs (Figure 4A, mRNA). Importantly, however, the pre-mRNA forms (and possibly also excised introns) of these transcripts were also specifically bound by HuR and AUF1, as evidenced by the amplification of intron sequences of both transcripts in HuR and AUF1 IPs (in these RT reactions, random hexamers were used instead of oligo-d(T)) (Figure 4A, pre-mRNA). In all cases, samples were digested with DNase before RT and ‘no RT' control reactions were included to ensure that no genomic sequences were amplified (Figure 4A).
Figure 4.

Binding of HuR and AUF1 to distinct sites of the p21 and cyclin D1 mRNAs. (A) Following IP reactions using either whole-cell (1.5 mg, left), cytoplasmic (400 μg, Cyto., center), or nuclear (500 μg, Nuc., right) lysates and anti-HuR, anti-AUF1, or IgG1 antibodies, the binding of endogenous HuR and AUF1 to endogenous target mRNAs (mRNA) was detected by RT–PCR as explained above (Figure 1C); pre-mRNAs (pre-mRNA) were detected by IP of nuclear lysate followed by RT using random hexamers and PCR amplification using primers specific to intron sequences of the p21 and cyclin D1 genes; no amplification was seen in ‘No RT' control samples. (B) Pull-down assays to assess the ability of endogenous HuR and AUF1 to bind to biotinylated transcripts spanning the p21 and cyclin D1 mRNAs. The indicated biotinylated transcripts (1 μg each) were incubated with 40 μg of HeLa whole-cell lysate, whereupon their association with HuR or AUF1 was detected by Western blotting. Lys., 5 μg of whole-cell lysate; CR, coding region. Blackened box, CR; shaded, AREs. Bottom: schematic of proposed binding regions for HuR and AUF1.
While the aforementioned results suggested that HuR and AUF1 were each able to form complexes with endogenous p21 and cyclin D1 mRNAs, it was important to assess whether the two proteins were capable of binding to each target on distinct, nonoverlapping sites of the same mRNAs, thereby permitting, at least theoretically, the joint binding of both proteins on each target (as we propose it occurs in the nucleus). Such in vitro analysis was performed by testing the ability of biotinylated transcripts encompassing various regions of the p21 and cyclin D1 mRNAs to associate with endogenous HuR or AUF1 (Figure 4B). Here, complex formation was visualized by ‘pull-down' of the RNP associations using streptavidin-coated beads and detection of RBPs on Western blots. As shown, various 3′ untranslated region (3′UTR) partial transcripts, but not coding region (CR) transcripts, were capable of pulling down both HuR and AUF1. Importantly, binding was found to occur at multiple discrete sites: for the p21 mRNA, HuR preferentially bound transcript 3′A, proximal to the CR, within an AU-rich stretch (shaded), while AUF1 displayed a preference for the distal region (3′C). Regarding the cyclin D1 3′UTR, HuR bound fragment 3′C, whereas AUF1 bound the proximal fragment 3′A but not the AU-rich stretch, as previously reported (Lin et al, 2000), and also bound the distal fragment 3′D (Figure 4B). The finding that AUF1 and HuR can bind to the same target RNAs on different, nonoverlapping regions supports the notion that the two proteins could, in principle, bind target transcripts simultaneously.
Exposure to UVC has opposing effects on the post-transcriptional regulation of p21 and cyclin D1
Given the ability of p21 and cyclin D1 mRNAs to associate with both HuR and AUF1, two RBPs exerting opposite effects on the stability of target transcripts, we sought to test if the steady-state levels of the two mRNAs were regulated in the same fashion in response to a given stimulus. UVC irradiation increased HuR–p21 mRNA associations in the cytoplasm, and consequently increased p21 mRNA stability and abundance in colon cancer cells (Wang et al, 2000a). In HeLa cells, UVC irradiation (15 J/m2) also induced p21 mRNA levels, and suppressed cyclin D1 mRNA levels (Figure 5A). Likewise, UVC elevated HuR cytoplasmic abundance, but the cytoplasmic association of HuR with AUF1 did not increase proportionately (Figure 5B). Interestingly, the binding of HuR and AUF1 to target mRNAs was influenced in opposite directions by UVC irradiation, as determined by IP+RT–(real-time)PCR assays: after UVC, HuR–p21 mRNA associations increased and HuR–cyclin D1 mRNA associations decreased, whereas AUF1–p21 mRNA associations were reduced and AUF1–cyclin D1 mRNA complexes were markedly elevated (Figure 5C).
Figure 5.
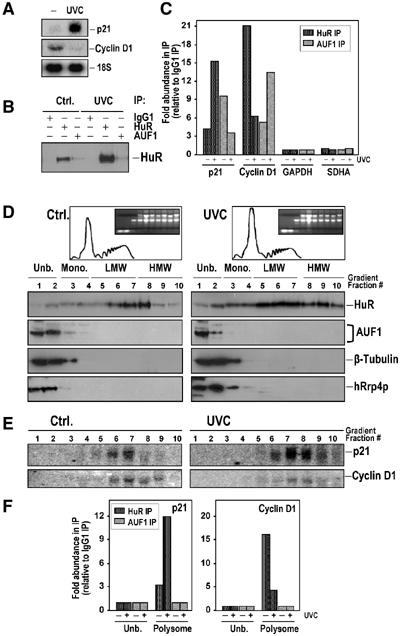
Effect of UVC irradiation on the expression of p21 and cyclin D1 mRNAs. At 5 h after exposure of HeLa cells to 15 J/m2 UVC, p21 and cyclin D1 mRNA abundance was assessed by Northern blotting (20 μg total RNA per lane) (A), the presence of HuR and AUF1 on common RNP complexes was monitored by IP and Western blot assays using cytoplasmic lysate, as described above (Figure 2) (B), and the presence of endogenous p21 and cyclin D1 mRNAs with each protein was assayed in the IP material from cytoplasmic lysates by real-time RT–PCR (C); fold differences in abundance were calculated after estimating the CT values (representing the number of PCR cycles required to reach a threshold set arbitrarily at 0.8) for each amplification curve (Materials and methods). Low-level amplification of GAPDH and SDHA served to monitor equal addition of RNA from IP materials. (D) Cytoplasmic lysates from either untreated (Ctrl.) or UVC-treated HeLa cultures were fractionated through sucrose gradients, whereupon aliquots from each fraction were used for Western blotting to detect HuR, AUF1, the exosome protein hRrp4p, and the control protein β-tubulin. (E) The relative presence of p21 and cyclin D1 mRNAs in each fraction from the sucrose gradient was determined by Northern blot analysis. (F) The relative abundance of p21 and cyclin D1 mRNAs bound to either HuR or AUF1 in pooled unbound fractions (1 and 2, Unb.) or pooled polysomal fractions (6–10, Polysome) from either untreated or UVC-treated cells was determined by IP+RT–(real-time)PCR.
UVC treatment reduced the polysomal content of the cell, but it actually caused HuR to increase its presence in the polysome-bound material (Figure 5D). By contrast, AUF1 remained almost exclusively localized in fractions 1 and 2, where other cytosolic components were identified, including the exosome protein hRrp4p. Northern blotting confirmed that p21 mRNA abundance increased in LMW and HMW fractions after UVC, while cyclin D1 mRNA levels were overall lower (Figure 5E). Experimental evidence for UVC-triggered changes in the composition of HuR–RNA complexes was obtained by IP+RT–(real-time)PCR analysis of pooled polysome fractions 6–10. Following UVC treatment, HuR–p21 mRNA complexes were found to be ∼4-fold more abundant in the polysomal fractions, while HuR–cyclin D1 mRNA complexes were ∼3.5-fold less abundant (Figure 5F). Testing of the unbound fractions (1 and 2) by IP+RT–(real-time)PCR using anti-HuR antibodies failed to amplify p21 or cyclin D1 products, likely due to the low abundance of HuR in these fractions (Figure 5F). While we cannot exclude the possibility that AUF1–RNA associations were disrupted by the fractionation process, no AUF1-bound RNAs were found in the polysome fractions (Figure 5F), where AUF1 was basically undetectable (Figure 5D), or in the unbound material (fractions 1 and 2, Figure 5F), where AUF1 reportedly associates with the exosome; such interaction has been linked to a rapid decay of AUF1-bound mRNAs (Laroia et al, 1999; Chen et al, 2001), and may explain our inability to amplify bound mRNAs.
siRNA-mediated reduction of HuR or AUF1 levels alters p21 and cyclin D1 expression
In order to determine the relative influence of HuR on the expression of p21 and cyclin D1, an RNA interference (RNAi)-based approach was devised. By 3 days after transfection of HeLa cells using an HuR-targeting siRNA (HuR siRNA), HuR abundance was reduced to ∼20% of the levels in the control siRNA-transfected group (C, Figure 6A). Cultures expressing reduced HuR showed markedly lower p21 and cyclin D1 expression levels than did control siRNA-transfected cells, as detected by both Western and Northern blotting (Figure 6A and B). As shown in Figure 6C, the decreased p21 and cyclin D1 mRNA steady-state levels were due, at least in part, to the lower stability of each mRNA in cells with reduced HuR expression, as determined using actinomycin D-based mRNA half-life measurements. When the control and HuR siRNA treatment groups were compared, p21 mRNA half-life declined from ∼4.6 to 2.7 h, cyclin D1 mRNA from ∼8 to 3.3 h, respectively (Figure 6C); by contrast, the stability of the long-lived GAPDH mRNA (inset) was >18 h in all transfection groups. These results underscore the stabilizing influence of HuR on target mRNAs, including those that encode p21 and cyclin D1. Interestingly, in lysates obtained from HuR siRNA-transfected cells, AUF1 prominently associated with the p21 3′A transcript and the cyclin D1 3′C transcript (Figure 6D), two sites that were preferentially bound by HuR in control lysates (Figure 4B), suggesting that HuR and AUF1 may compete for binding to these sites.
Figure 6.

Effect of siRNA-mediated suppression of HuR levels on the expression and stability of p21 and cyclin D1 mRNAs. At 3 days after transfection with an siRNA that suppressed HuR protein levels by RNAi (HuR siRNA) or a control siRNA (C), the expression levels of HuR, cyclin D1, p21, and GAPDH (a control protein serving to monitor the equal loading and transfer of samples) were examined by Western blotting (A), and the abundance of p21 and cyclin D1 mRNAs (and control 18S rRNA) was assessed by Northern blotting (B). (C) A three days after transfection, the half-lives of p21 and cyclin D1 mRNAs in each siRNA group were assessed by using actinomycin D (2 μg/ml); mRNA half-lives (parentheses) were calculated from Northern blotting data (Materials and methods). Insets, representative Northerns, including signals of a stable mRNA encoding GAPDH; 18S signals revealed even loading of samples (not shown). Two independent Northern blotting experiments, yielding comparable results, were performed in order to calculate all mRNA half-lives. (D) The indicated biotinylated transcripts were incubated with 40 μg of lysates prepared from either control-transfected cells (C) or HuR siRNA-transfected cells expressing reduced HuR levels. Pull-down assays to assess the ability of endogenous AUF1 to bind biotinylated transcripts spanning the p21 and cyclin D1 mRNAs were performed as described in Figure 4B.
To study the effect of AUF1 on the expression of p21 and cyclin D1, we employed an RNAi approach based on the use of a plasmid expressing siRNA that targeted a region common to all four AUF1 isoforms (Supplementary material). At 3 days after transfection, AUF1 expression was reduced to ∼25% of the levels seen in control cells (C, Figure 7A). Cells displaying reduced AUF1 abundance exhibited markedly elevated levels of p21 and cyclin D1 proteins (Figure 7A), and mRNAs (Figure 7B). Again, the stability of the two transcripts was also strongly influenced by AUF1 abundance: when comparing the C group with the AUF1 siRNA group, p21 mRNA half-life increased from ∼2.0 to 4.4 h, while cyclin D1 mRNA half-life increased from ∼4 h to >8 h, respectively (Figure 7C); discrepancies in the half-lives of C populations (Figures 6C and 7C) were likely due to the different transfection reagents used. The stability of the long-lived GAPDH mRNA (Figure 7C, inset) was also >18 h in all transfection groups. These observations support the widely held view that AUF1 accelerates target mRNA decay. Furthermore, in lysates obtained from AUF1 siRNA-transfected cells, HuR can prominently bind the p21 3′C transcript and the cyclin D1 3′A transcript (Figure 7D), two sites that were preferentially bound by AUF1 in control lysates (Figure 4B), lending further support to the notion that HuR and AUF1 may also compete for binding to target sequences.
Figure 7.
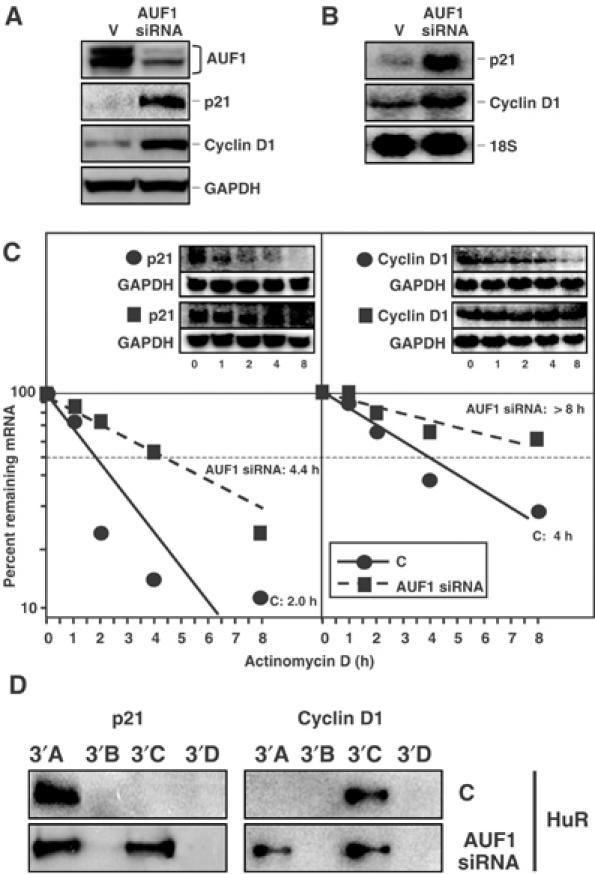
Effect of siRNA-mediated suppression of AUF1 levels on the expression and stability of p21 and cyclin D1 mRNAs. At 3 days after transfection with plasmid pSILENCER-AUF15, expressing a transcript capable of suppressing AUF1 abundance by RNAi (AUF1 siRNA), or the control plasmid (V), the levels of AUF1, cyclin D1, p21, and GAPDH were examined by Western blotting (A), and the abundance of p21 and cyclin D1 mRNAs (and control 18S rRNA) was assessed by Northern blotting (B). (C) At 3 days after transfection, the half-lives of p21 and cyclin D1 mRNAs in each siRNA group were assessed by using actinomycin D (2 μg/ml); mRNA half-lives (parentheses) were calculated from Northern blotting data that were processed as described in the legend of Figure 6. (D) The indicated biotinylated transcripts were incubated with 40 μg of lysates prepared from either control-transfected HeLa cells (C) or AUF1 siRNA-transfected cells expressing reduced AUF1 levels. Pull-down assays to assess the ability of endogenous HuR to bind biotinylated transcripts spanning the p21 and cyclin D1 mRNAs were performed as described in Figure 4B.
Based on the results obtained in this investigation, we propose a model whereby HuR and AUF1 can bind jointly as well as individually to common target mRNAs and influence their post-transcriptional fate (Figure 8 and Discussion).
Figure 8.
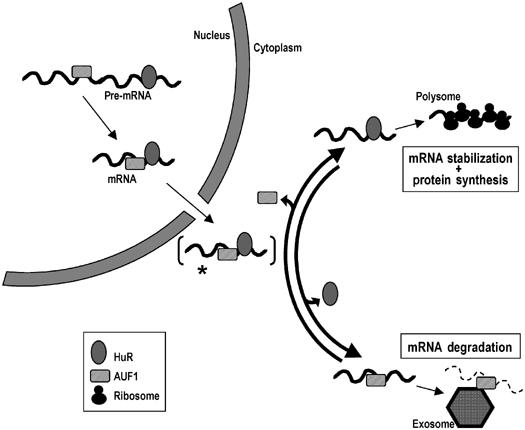
Schematic of the proposed model of HuR and AUF1 physical and functional interaction. See text for details.
Discussion
Concurrent binding of HuR and AUF1 to common target mRNAs
The data presented here provide systematic evidence that HuR and AUF1, two RBPs exerting opposite influence on the post-transcriptional fate of target mRNAs, can concurrently bind to common target transcripts. A global search using cDNA arrays revealed that a prominent number of putative targets of each RBP (57 and 58%) were also targets of the other protein, and associations between the RBPs and 12 such targets were verified (Figure 1C). The physical association of HuR and AUF1 appeared to be RNA dependent and to occur predominantly in the nucleus, whereas in the cytoplasm, HuR and AUF1 appeared to favor binding to target mRNAs individually. Using cyclin D1 and p21 mRNAs as model target transcripts, our data support the notions that HuR-bound mRNAs undergo stabilization and engage with the protein translation machinery, whereas AUF1-bound transcripts likely follow a path of mRNA degradation.
The model emerging from these results (Figure 8) builds on previous characterization of the expression, function, and subcellular localization of these two RBPs by several groups, including our own. According to the present findings, we propose that HuR and AUF1 could simultaneously as well as competitively bind to a subset of common target mRNAs. This scheme complements a popular displacement model whereby decay-promoting and stability-promoting RBPs are thought to compete for the same binding site (Park et al, 2000; Ming et al, 2001; Stoecklin et al, 2002; Cok et al, 2004). The general association of RBPs to common target RNAs was seen predominantly in the nucleus, but was very reduced in the cytoplasm. One plausible hypothesis is that such target mRNAs are exported from the nucleus bound to both HuR and AUF1 (and likely additional RBPs), and the RNP complex contains HuR and AUF1 transiently upon reaching the cytoplasm (Figure 8, [*]) We propose that, shortly thereafter, either AUF1 is released and HuR remains bound, followed by the recruitment of the RNP complex to polysomes, or HuR is released and AUF1 remains bound, leading to the recruitment of the exosome to the RNP complex and the decay of the mRNA.
Cytoplasmic RNPs are influenced by the target transcript and the availability of HuR and AUF1
The factors that dictate whether a given target transcript will follow an HuR-governed path of stabilization and translation, or an AUF1-directed avenue of degradation are likely to be complex and multiple. First, we propose that the transcript itself and in particular the sequence and structure of the binding region are among the principal deciding variables. The relative affinities of HuR and AUF1 for target sites on p21 and cyclin D1 mRNAs, which are likely to differ significantly, remain to be tested. While HuR and AUF1 could bind each transcript on nonoverlapping sites (Figure 4B), a reduction in HuR levels caused AUF1 to associate more prominently with sites that were previously bound by HuR (Figure 6D), and the opposite effect was seen when AUF1 levels were reduced (Figure 7D). These results suggest that HuR and AUF1 may bind nonoverlapping sites on a given transcript and may also compete for binding to additional sites of the same mRNA. Given the limitations of biotin pull-down analyses, a systematic biochemical quantitation of these associations will likely yield important additional clues regarding the preferential binding of each RBP to a particular transcript. Further studies on the physiological relevance of 3′UTR sites governing the stability and translation of these mRNAs are warranted, particularly in the light of reports linking cyclin D1 3′UTR truncations to enhanced cyclin D1 abundance and cancer (Lebwohl et al, 1997; Hosokawa et al, 1998).
Second, we propose that the cytoplasmic complexes that eventually persist (HuR–mRNA or AUF1–mRNA) are influenced by the relative abundance of HuR and AUF1. Downregulation of HuR by an siRNA caused increased binding of AUF1 with p21 and cyclin D1 transcripts (Figure 6D), associated with the accelerated degradation of both transcripts and reduced expression of the corresponding proteins (Figure 6A–C). Conversely, siRNA-mediated decrease in AUF1 expression increased the binding of HuR to p21 and cyclin D1 transcripts (Figure 7D) and increased their stability and protein levels (Figure 7A–C). Moreover, the relative abundance of additional RBPs associating with these transcripts, such as hnRNP A1 (Figure 2B), RNA export proteins, poly(A)-binding proteins (PABPs), RNA helicases, or other RBPs, is likely to further influence their subcellular localization, stability, and translation. Translational inhibitory RBPs TIAR and TIA-1 were also found in the HuR IP material in experiments akin to those depicted in Figure 2 (not shown). A systematic analysis of all of the RBPs forming complexes with the p21 and cyclin D1 mRNAs is underway using large-scale pull-down approaches to identify the sets of proteins associating with biotinylated p21 and cyclin D1 transcripts.
Dynamic compartmentalization of RNP complexes
Third, the physiologic conditions of the cell are also likely to contribute decisively to the composition of the RNP complexes in each cell compartment. UVC and other stress agents, as well as treatment with proliferative and differentiation signals, all caused a transient enrichment in cytoplasmic HuR (Wang et al, 2000a, 2000b; Figueroa et al, 2003), while growth factor deprivation (Wang et al, 2000b) and endoplasmic reticulum stress (not shown) reduced cytoplasmic HuR levels. The subcellular localization of AUF1 is also influenced by stimuli such as heat shock, and its function is further modulated by signal-induced phosphorylation and ubiquitination (Laroia et al, 1999; Wilson et al, 2003). Additional layers of regulation are provided by nuclear or cytoplasmic proteins reported to interact with HuR (such as SETα/β, pp32, and APRIL; Gallouzi et al, 2001) or with AUF1 (including eIF4G, hsp70, and PABP; Laroia et al, 1999).
Finally, RNP complex composition, and whether it contains HuR, AUF1, or both proteins, is influenced by the subcellular compartment in which the complex is being investigated. In the nucleus, we propose that the two proteins can bind concomitantly to shared target transcripts. Nuclear HuR- and AUF1-containing RNP complexes likely assemble on pre-mRNAs, possibly recruited cotranscriptionally, as well as to mature mRNAs (Figure 4A). Moreover, HuR and AUF1 formed complexes with hnRNP C1/C2, as detected in nuclear lysates that had been prepared by sonication, and therefore contained large nuclear RNP complexes where pre-mRNAs reside (Figure 2B); hnRNP C1/C2 associate to nascent mRNAs and dissociate from them when mature mRNAs are released into the nucleoplasm for nuclear export (Mili et al, 2001).
In the cytoplasm, AUF1-lacking, HuR-containing RNP complexes are preferentially found to fractionate with the translation machinery. The analysis of sucrose gradients revealed the association of HuR with ribosomes and polysomes of increasing size, but failed to show the colocalization of AUF1 with such components; instead, AUF1 was largely restricted to the fractions lacking ribosomes or ribosome subunits (Figures 3 and 5). HuR was readily found to bind to p21 and cyclin D1 mRNAs in the polysome fractions, while AUF1-bound mRNAs were undetectable in the same assays (Figure 5F). Efforts to investigate directly HuR's role in the translation of these proteins by pulse 35S-labeling strategies such as those reported previously (Galbán et al, 2003; Mazan-Mamczarz et al, 2003) were unsuccessful due to technical limitations (including weak p21 and cyclin D1 detection by available antibodies and low expression levels of these proteins), although in human colorectal carcinoma cells expressing ∼10-fold higher levels of p21, UVC elevated the rate of p21 protein translation in an HuR-dependent manner (not shown).
The exosome is a pivotal component of the RNA degradation machinery, comprising primarily 3′ → 5′ exonuclease activity. The nuclear exosome has been mainly implicated in the processing of ribosomal RNA, small nuclear RNA, and small nucleolar RNA, but was recently postulated to participate also in nuclear processing of pre-mRNAs and the degradation of mRNAs, including those bearing AREs (Vasudevan and Peltz, 2003). In the cytoplasm, the principal function of the exosome is to carry out the degradation of mRNAs by 3′ → 5′ degradation (Mitchell et al, 1997). In addition to the exonucleases, a number of ancillary factors that recruit AU-rich mRNAs to the exosome, such as AUF1, KSRP, and TTP, have been documented (Chen et al, 2001). Despite intense efforts, we were unable to obtain direct evidence of AUF1-bound mRNAs associated with the exosome, likely because upon reaching the exosome, mRNA degradation proceeds rapidly. Supporting this notion was additional evidence that AUF1 and the exosome component hRrp4p overlapped in their distribution (Figure 5D), that no AUF1 target mRNAs were detected either by Northern or by AUF1 IP+RT–PCR in the soluble cytoplasmic fractions (fractions 1 and 2, Figure 5E), and that the cyclin D1 mRNA has been shown to be degraded by the exosome (Briata et al, 2003).
Conclusion
A large body of evidence has now established the antagonistic effects of mRNA stability-promoting RBPs like HuR and decay-promoting RBPs like AUF1. By contrast, our knowledge of their specific RNA recognition and binding sites is still nebulous, and whether one or several RBPs can bind to a given ARE-containing mRNA has not been systematically examined. Based on the data gathered from these studies, we propose a model (Figure 8) whereby HuR and AUF1 can jointly bind to common target mRNAs, likely in their pre-mRNA state, in the form of stable nuclear RNP complexes. In the cytoplasm, we hypothesize that a given mRNA will be preferentially found in association with HuR and/or AUF1 depending on a number of factors, including the target mRNA sequence, the relative abundance of HuR and AUF1, the influence of stress agents or possibly other stimuli, and the specific subcytoplasmic compartment in which the RNP is studied. Accordingly, a dynamic interplay among these elements will ultimately dictate the cytoplasmic fate of the mRNA: HuR-bound mRNAs will likely be subject to additional regulation by the translation machinery, whereas AUF1-bound mRNAs will likely undergo rapid exosome-mediated decay.
Materials and methods
Cell culture, cell fractionation, and RNA interference
Human cervical carcinoma HeLa cells were cultured in DMEM (Gibco-BRL) supplemented with 10% fetal bovine serum and antibiotics. Cytoplasmic and nuclear fractions were prepared as described (Feng et al, 1997), with minor modifications (Supplementary material). Whole-cell lysates were prepared essentially as described (Piñol-Roma et al, 1988). A total of 20 million cells were resuspended in 3 ml IP buffer (Supplementary material) and sonicated; 500-μl aliquots of the sonicated material were layered onto a 30% sucrose cushion (500 μl) in IP buffer and centrifuged (at 5000 g, 15 min) to remove insoluble cellular structures; the upper layer was designated as the whole-cell lysate.
For HuR RNAi analysis, siRNA duplexes (Supplementary material) were transfected with Oligofectamine (Invitrogen); for AUF1 RNAi analysis, pSILENCER-AUF15 plasmid and insert-less plasmid (Supplementary material) were transfected using Lipofectamine 2000 (Invitrogen). Cells were harvested 3 days after transfection and used for Northern or Western blotting.
Immunoprecipitation assays
For IP of endogenous RNA–protein complexes from whole-cell (1.5 mg), cytoplasmic (400 μg), or nuclear (500 μg) extracts, lysates were incubated (1 h, 4°C) with a 50% (v/v) suspension of Protein A–Sepharose beads (Sigma) that had been precoated with 30 μg of either IgG1 (BD Pharmingen), anti-HuR (Santa Cruz Biotech.), or anti-AUF1 (Upstate Biotech.) antibodies. Beads were washed using NT2 buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2, and 0.05% Nonidet P-40). Where indicated, 1 mg/ml heparin was added to the lysate, or lysate and beads were incubated with 1 μg RNase A and 10 U of RNase T1 per ml for 10 min at 30°C. RNP crosslinking by using UVC or formaldehyde is described in Supplementary material. Bound proteins were size-fractionated by SDS–PAGE and examined by Western blotting.
For the analysis of RNA in the IP material, beads were incubated with 100 μl NT2 buffer containing 20 U RNase-free DNase I for 15 min at 30°C, washed with NT2 buffer, and further incubated in 100 μl NT2 buffer containing 0.1% SDS and 0.5 mg/ml proteinase K (15 min, 55°C). RNA was extracted and precipitated in the presence of glycoblue (Ambion).
cDNA array analysis after IP
IP for genome-wide analysis of endogenous targets of HuR and AUF1 was performed using 3 mg of lysate and 30 μg of antibody (López de Silanes et al, 2004). RNA present in IP reactions using either anti-HuR, anti-AUF1, or IgG1 antibodies was reverse transcribed in the presence of [α-33P]dCTP and the radiolabeled product was used to hybridize cDNA arrays (http://www.grc.nia.nih.gov/branches/rrb/dna/index/dnapubs.htm#2, 9600 genes, MGC arrays). All of the data were analyzed using the Array Pro software (Media Cybernetics Inc.), normalized by Z score transformation (Cheadle et al, 2003) and used to calculate differences in signal intensities. Transcripts were considered to be enriched in HuR IPs or AUF1 IPs when Z ratios were ⩾1.00 in comparisons of signals in HuR IPs relative to IgG1 IPs, or in comparisons of signals in AUF1 IPs relative to IgG1 IPs, respectively. See http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1361 for complete array results.
Western blot, Northern blot, and mRNA stability assays
Proteins were resolved by 12% SDS–PAGE and transferred onto PVDF membranes. Primary and secondary antibodies are described (Supplementary material). Northern blotting was performed using RNA that was isolated either from whole cells (using STAT-60) or from each fraction of the sucrose gradients (below). [α-32P]dATP was used to end-label oligonucleotides used for detecting 18S rRNA and cyclin D1 and p21 mRNAs (Supplementary material) and to label GAPDH cDNA using random primers. For mRNA half-life assessments, actinomycin D (2 μg/ml) was added and total RNA prepared at the times shown; mRNA half-lives were calculated after measuring mRNA signals on Northern blots, normalizing to 18S rRNA signals, plotting values on logarithmic scales, and calculating the time period required for a given transcript to decrease to one-half of the initial abundance.
RT–PCR and real-time RT–PCR after IP of RNP complexes
For RT–PCR, 50% of the RNA isolated from the IPs was reverse transcribed using oligo-dT (for mRNA) or random hexamers (for pre-mRNA), and SSII RT (Invitrogen), and the resulting material was used for PCR amplification using gene-specific primer pairs (Supplementary material) and 94°C (30 s), 55°C (30 s), and 72°C (30 s) for 25–30 cycles, then 5 min at 72°C. As negative controls, 50% of the RNA was reverse transcribed without SSII RT, but otherwise processed similarly. For real-time PCR, amplification conditions were 50°C (2 min), 95°C (10 min), then 40 cycles of 95°C (15 s) and 60°C (1 min).
Linear sucrose gradient fractionation
Linear sucrose gradient fractionations were performed as described (Feng et al, 1997) with minor modifications. A total of 50 million cells were incubated for 15 min with 100 mg/ml cycloheximide, and cytoplasmic extracts (500 μl), prepared as described above, were loaded onto the sucrose gradients (10–50% (w/v), 100 mM KCl, 20 mM Tris–HCl (pH 7.5), and 5 mM MgCl2). After centrifugation (Beckman SW41, 39 000 rpm, 90 min, 4°C), the material was fractionated into 1-ml aliquots using a gradient fractionator (Brandel) and monitored by optical density measurement (A254). For IP assays, pooled fractions 1 and 2 (unbound) or 6–10 (polysomal) were diluted with two volumes of NT2 buffer. For Northern blotting, each fraction was diluted with an equal volume of water and RNA was isolated using Trizol LS (Invitrogen); equal RNA volumes (50% of each fraction) were used.
In vitro transcription and biotin pull-down
In vitro transcription and biotin pull-down reactions were performed as described, except that whole-cell lysates (40 μg) were used (López de Silanes et al, 2004). Primers used for the preparation of templates for in vitro transcription and transcript sizes are provided (Supplementary material).
Supplementary Material
Supplementary Material
Acknowledgments
We thank G Dreyfuss for the anti-hnRNP A1 and anti-hnRNP C1/C2 antibodies, and CY Chen for the anti-hRrp4p antibody. We are grateful to KG Becker and D Teichberg for their assistance with the cDNA array data.
References
- Antic D, Keene JD (1997) Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am J Hum Genet 61: 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy U, Watson J, Patel D, Keene JD (1998) ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci 111: 3145–3156 [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Giordano T, Brewer G, Malter JS (1999) Identification of AUF-1 ligands reveals vast diversity of early response gene mRNAs. Nucleic Acids Res 27: 1464–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA (2001) HuR and mRNA stability. Cell Mol Life Sci 58: 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata P, Ilengo C, Corte G, Moroni C, Rosenfeld MG, Chen CY, Gherzi R (2003) The Wnt/beta-catenin → Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol Cell 12: 1201–1211 [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG (2003) Analysis of microarray data using Z score transformation. J Mol Diagn 5: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464 [DOI] [PubMed] [Google Scholar]
- Chen CY, Xu N, Shyu AB (2002) Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol Cell Biol 22: 7268–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cok SJ, Acton SJ, Sexton AE, Morrison AR (2004) Identification of RNA binding proteins in RAW 264.7 cells that recognize an LPS responsive element in the 3-UTR of the murine COX-2 mRNA. J Biol Chem 279: 8196–8205 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG (1993) hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 62: 289–321 [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST (1997) FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell 1: 109–118 [DOI] [PubMed] [Google Scholar]
- Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Gorospe M, Muñoz A (2003) Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol 23: 4991–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbán S, Martindale JL, Mazan-Mamczarz K, López de Silanes I, Fan J, Wang W, Decker J, Gorospe M (2003) Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol 23: 7083–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Steitz JA (2001) Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA 7: 1348–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA (2000) HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA 97: 3073–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Steitz JA (2001) Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294: 1895–1901 [DOI] [PubMed] [Google Scholar]
- Gao FB, Carson CC, Levine T, Keene JD (1994) Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc Natl Acad Sci USA 91: 11207–11211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V (1999) Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J Biol Chem 274: 2322–2326 [DOI] [PubMed] [Google Scholar]
- Hosokawa Y, Suzuki R, Joh T, Maeda Y, Nakamura S, Kodera Y, Arnold A, Seto M (1998) A small deletion in the 3′-untranslated region of the cyclin D1/PRAD1/bcl-1 oncogene in a patient with chronic lymphocytic leukemia. Int J Cancer 76: 791–796 [DOI] [PubMed] [Google Scholar]
- Joseph B, Orlian M, Furneaux H (1998) p21waf1 mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J Biol Chem 273: 20511–20516 [DOI] [PubMed] [Google Scholar]
- Keene JD (1999) Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA 96: 5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmann M, Gopfert U, Siewe B, Hengst L (2002) ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev 16: 3087–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G, Cuesta R, Brewer G, Schneider RJ (1999) Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284: 499–502 [DOI] [PubMed] [Google Scholar]
- Lebwohl DE, Muise-Helmericks R, Sepp-Lorenzino L, Serve S, Timaul M, Bol R, Borgen P, Rosen N (1997) A truncated cyclin D1 gene encodes a stable mRNA in a human breast cancer cell line. Oncogene 9: 1925–1929 [PubMed] [Google Scholar]
- Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M (2000) Downregulation of cyclin D1 expression by prostaglandin A2 is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell Biol 20: 7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loflin P, Chen CY, Shyu A-B (1999) Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev 13: 1884–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M (2004) Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA 101: 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Schneider RJ (2004) Tissue distribution of AU-rich mRNA binding proteins involved in regulation of mRNA decay. J Biol Chem 279: 12974–12979 [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Galbán S, López de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M (2003) RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA 100: 8354–8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Shu HJ, Zhao Y, Piñol-Roma S (2001) Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol Cell Biol 21: 7307–7319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Turck CW, Nikolic JM, Black DL (1997) A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev 11: 1023–1036 [DOI] [PubMed] [Google Scholar]
- Ming XF, Stoecklin G, Lu M, Looser R, Moroni C (2001) Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol Cell Biol 21: 5778–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Park S, Myszka DG, Yu M, Littler SJ, Laird-Offringa IA (2000) HuD RNA recognition motifs play distinct roles in the formation of a stable complex with AU-rich RNA. Mol Cell Biol 20: 4765–4772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, Choi YD, Matunis MJ, Dreyfuss G (1988) Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev 2: 215–227 [DOI] [PubMed] [Google Scholar]
- Ross J (1995) mRNA stability in mammalian cells. Microbiol Rev 59: 423–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C (2002) Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J 21: 4709–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum SA, Lager PJ, Carson CC, Keene JD (2002) Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods 26: 191–198 [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Peltz SW (2003) Nuclear mRNA surveillance. Curr Opin Cell Biol 15: 332–337 [DOI] [PubMed] [Google Scholar]
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook NJ, Gorospe M (2000a) HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 20: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lin S, Caldwell CM, Furneaux H, Gorospe M (2000b) HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J 19: 2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M (2001) Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol 21: 5889–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GM, Brewer G (1999) The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acids Res Mol Biol 62: 257–291 [DOI] [PubMed] [Google Scholar]
- Wilson GM, Lu J, Sutphen K, Suarez Y, Sinha S, Brewer B, Villanueva-Feliciano EC, Ysla RM, Charles S, Brewer G (2003) Phosphorylation of p40AUF1 regulates binding to A+U-rich mRNA destabilizing elements and protein-induced changes in ribonucleoprotein structure. J Biol Chem 278: 33039–33048 [DOI] [PubMed] [Google Scholar]
- Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G (1993) Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol 13: 7652–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


