Abstract
The mechanism by which conventional kinesin walks along microtubules is poorly understood, but may involve alternate binding to the microtubule and hydrolysis of ATP by the two heads. Here we report a single amino-acid change that affects stepping by the motor. Under low force or low ATP concentration, the motor moves by successive 8-nm steps in single-motor laser-trap assays, indicating that the mutation does not alter the basic mechanism of kinesin walking. Remarkably, under high force, the mutant motor takes successive 16-nm displacements that can be resolved into rapid double 8-nm steps with a short dwell between steps, followed by a longer dwell. The alternating short and long dwells under high force demonstrate that the motor stepping mechanism is inherently asymmetric, revealing an asymmetric phase in the kinesin walking cycle. Our findings support an asymmetric two-headed walking model for kinesin, with cooperative interactions between the two heads. The sensitivity of the 16-nm displacements to nucleotide and load raises the possibility that ADP release is a force-producing event of the kinesin cycle.
Keywords: double steps, kinesin, motor mutant, stepping mechanism
Introduction
The kinesin motor proteins bind to ATP and microtubules, and use the energy of nucleotide hydrolysis to move along the microtubule. The first discovered or conventional kinesin is a highly processive motor that takes more than a hundred steps each time it binds to a microtubule (Howard et al, 1989; Svoboda et al, 1993; Hackney, 1995). Steps by conventional kinesin along a microtubule are tightly coupled to ATP hydrolysis—the motor takes a single 8-nm step for each ATP it hydrolyzes (Schnitzer and Block, 1997; Coy et al, 1999).
The mechanism by which kinesin walks along a microtubule is not well understood, although several models have been postulated. One model involves a hand-over-hand stepping mechanism in which the two heads of the motor alternate in binding to the microtubule and hydrolyzing ATP, each head in turn taking a step to advance the motor towards the microtubule plus end (Howard, 2001; Schief and Howard, 2001). This model is supported by the finding that processivity of kinesin requires two heads (Hancock and Howard, 1998). It accounts for the observed cooperativity of binding to nucleotide and microtubules by the two heads of kinesin—the finding that only one head of the dimeric motor binds to a microtubule and releases ADP in the absence of ATP, and that ATP hydrolysis by the bound head is required for the other head to bind to the microtubule (Hackney, 1994)—by postulating that both heads participate in ATP hydrolysis in an alternating manner. If both heads hydrolyze ATP, the tight coupling between ATP hydrolysis and steps by the motor requires that both heads be involved in producing force and taking steps along the microtubule.
A hand-over-hand model could involve either a symmetric mechanism in which the rear head always steps to the same side of the forward head to take the next step along the microtubule (Howard, 1996), or an asymmetric mechanism in which the rear head steps to either side of the forward head (Hirose et al, 2000; Hoenger et al, 2000; Schliwa, 2003). The rationale underlying the proposal of a symmetric hand-over-hand mechanism is that the two heads of the dimeric motor are functionally equivalent and should undergo the same movements and conformational changes during the nucleotide hydrolysis cycle (Howard, 1996). The only available crystal structure of dimeric kinesin shows a rotational symmetry of the two heads around the axis of the coiled-coil stalk that causes the microtubule-binding regions of the heads to be on opposite sides of the motor (Kozielski et al, 1997). Thus, a symmetric hand-over-hand model would require that the motor rotate ∼180° each time an unbound head binds to the microtubule to take a step. This should produce rotations during processive movement that are detectable experimentally.
Failure to observe the rotational movement predicted by a hand-over-hand mechanism has led to the proposal of an ‘inchworm' model in which only one head binds to the microtubule and hydrolyzes ATP, dragging the second head along (Hua et al, 2002). This model is consistent with the failure to observe 180° rotations of microtubules bound to single kinesin motors, but does not account for the cooperativity of nucleotide and microtubule binding by the two heads of the motor (Hackney, 1994), which requires that both heads of the motor hydrolyze ATP and produce force. Further, the experimental results do not compel an inchworm model, as they could also be accounted for by an asymmetric hand-over-hand model in which conformational changes of the neck linker or another structural element, together with stepping by the rear head to either side of the forward head in successive steps, produce net rotations too small to be detected in the previous experiments (Hua et al, 2002).
Here we report a single amino-acid change of conventional kinesin that affects stepping by the motor. The mutant motor moves processively along the microtubule by taking successive 8-nm steps, like wild-type kinesin, but under high force the motor moves by 16-nm displacements instead of 8-nm steps. Analysis of the 16-nm displacements reveals that the motor stepping mechanism is asymmetric. The asymmetry is not stochastic, but recurs in a regular pattern that compels an asymmetric mechanism for kinesin walking. The load sensitivity of the 16-nm displacements identifies a potential force-producing event of the kinesin cycle.
Results
Mutant design
The kinesin-T94S mutant was designed to make the highly conserved nucleotide-binding motif or P-loop of Drosophila kinesin heavy chain, GQTSSGKT, resemble more closely the P-loop of the myosins, GESGAGKT, in amino-acid sequence. The change of T94 to S causes only a small predicted structural change in the motor (Figure 1); however, T94 interacts with the β-phosphate of ADP in the kinesin motors and may help stabilize ADP binding. The point mutation was thus expected to open the nucleotide-binding cleft and permit more rapid nucleotide binding or release by the motor. The mutant motor was expressed in bacteria as a fusion to a biotin-binding protein and tested for ADP release in biochemical assays and velocity of movement along microtubules in in vitro motility assays.
Figure 1.
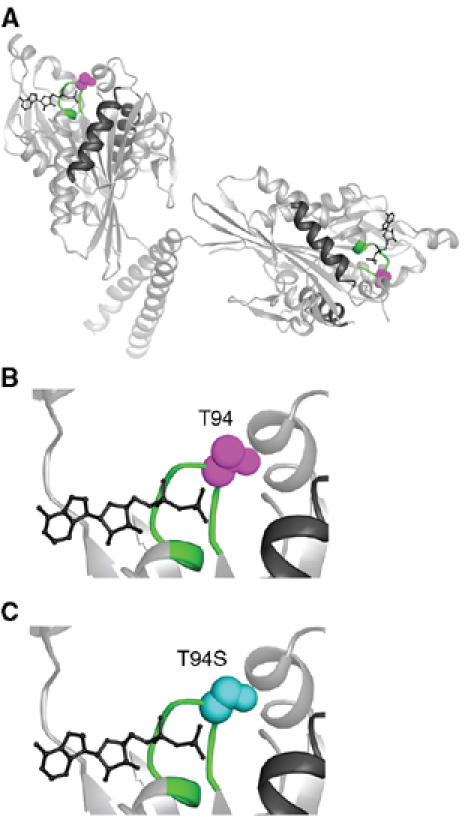
Kinesin-T94S mutant. (A) Structure of wild-type kinesin. The atomic structure of the dimeric motor is shown as a ribbon diagram (rat kinesin heavy chain, PDB 3KIN) (Kozielski et al, 1997). The conserved T94 in the nucleotide-binding P-loop is space-filled (purple) and the P-loop (GQTSSGKT) is green. (B) Close-up of the active site with the residue corresponding to Drosophila kinesin T94 in purple. (C) The active site with S94 (cyan) modeled into the wild-type structure in place of T94. ADP, wire diagram; helices α4 and α5, black.
ADP release assays
Single-turnover mant-ADP release experiments using FPLC-purified mutant or wild-type motor protein without microtubules showed that, upon addition of 500 μM Mg·ATP, the kinesin-T94S mutant releases ADP ∼3.6-fold faster (0.0128±0.0013 s−1, n=11) than wild-type kinesin (0.00353±0.00017 s−1, n=12) (Table I). When no nucleotide was added, the mutant motor released ADP at the same rate (0.00319±0.00079 s−1, n=4) as the wild-type motor with added Mg·ATP, indicating that release of ADP by the kinesin-T94S motor is not dependent on binding of ATP or microtubules. The wild-type motor without added nucleotide released ADP more slowly (0.00154±0.00080 s−1, n=3), with poorer curve fits (linear correlation coefficient, R>0.75) than the kinesin-T94S mutant (R>0.95).
Table 1.
Kinetic properties of the kinesin-T94S and wild-type kinesin motors
| Kinesin-T94S | Wild-type kinesin | |
|---|---|---|
| kd, ADP release−ATP | 0.00319±0.00079 s−1, n=4, R>0.95 | 0.00154±0.00080 s−1, n=3, R>0.75 |
| kd, ADP release+ATP | 0.0128±0.0013 s−1, n=11, R⩾0.90 | 0.00353±0.00017 s−1, n=12, R>0.93 |
| kd, ADP release+MTs | 0.0287±0.0187 s−1, n=9, R⩾0.73 | 0.0162±0.0062 s−1, n=8, R⩾0.65 |
| Velocity, MT gliding | 255±13 nm/s, n=20 | 853±25 nm/s, n=19 |
| Values are the mean±s.e.m.; R, linear correlation coefficient for the curve fit (values closer to 1.00 indicate a better fit); MT, microtubules. | ||
Single-turnover assays with microtubules were performed by mixing 0.2 μM kinesin-T94S or wild-type kinesin with 1 μM microtubules and monitoring the release of mant-ADP in a fluorometer. Results of these assays showed an acceleration of ADP release by microtubules (9- to 10-fold), which was comparable for the mutant and wild type. The dissociation rate for the kinesin-T94S motor (0.0287±0.0187 s−1, n=9) was higher than that of wild-type kinesin (0.0162±0.0062 s−1, n=8) (Table I), but the overall values did not differ significantly due to the variability from assay to assay. Attempts to measure the kcat of the mutant and wild-type motors in ATPase assays with microtubules gave variable values that were lower than normal for wild type and somewhat higher for the mutant. The high variability in mant-ADP release rates in the presence of microtubules and microtubule-stimulated kcat values may be due to the effects of the BIO fusion protein on motor–microtubule interactions when the motor is not bound to a glass surface or bead, as motor velocities in gliding and laser-trap assays were in the normal range for wild-type kinesin (see below). The kcat values with microtubules are not reported here to avoid misinterpretation of the mutant effects. Detailed studies of nonfusion mutant and wild-type motors in the presence of microtubules will be required to determine the effects of the T94S mutation on motor binding to nucleotide in the presence of microtubules. The altered rate of ADP release by the kinesin-T94S mutant in the absence of microtubules and the initial data presented here for assays with microtubules raise the possibility that ADP release in the presence of microtubules is also altered.
Gliding assays
Microtubule gliding assays (Song et al, 1997) were performed with lysates of the kinesin-T94S or wild-type motor. The assays showed good binding to microtubules by the motors attached to the coverslip, but a slower gliding velocity by ∼3.3-fold for the mutant (255±13 nm/s, n=20) compared to wild type (853±25 nm/s, n=19) (Table I).
Laser-trap assays
The increased ADP release rate in the absence of microtubules, but decreased velocity in gliding assays, suggested that stepping of the kinesin-T94S mutant along microtubules might be altered. Traces of single kinesin-T94S motors in laser-trap assays showed slow movement compared to wild type (Figure 2A and B). The velocity of the mutant at low load (∼1 pN) was 250 nm/s, which was about one-third the velocity of wild type, 760 nm/s, consistent with the gliding assays. The stall force of ∼8 pN for the mutant was similar to wild type. The mutant was processive with a step size of 8 nm at low load (<∼4 pN) (Figure 2C). Remarkably, at high load, the mutant showed frequent 16-nm displacements, which could be resolved into two rapid 8-nm steps with a short dwell between steps followed by a longer dwell, with the short and long dwells alternating between successive 8-nm steps (Figure 2C and E). The wild-type motor showed 8-nm steps, as reported by others (Svoboda et al, 1993; Nishiyama et al, 2002), with variable dwells between steps (Figure 2D and F).
Figure 2.
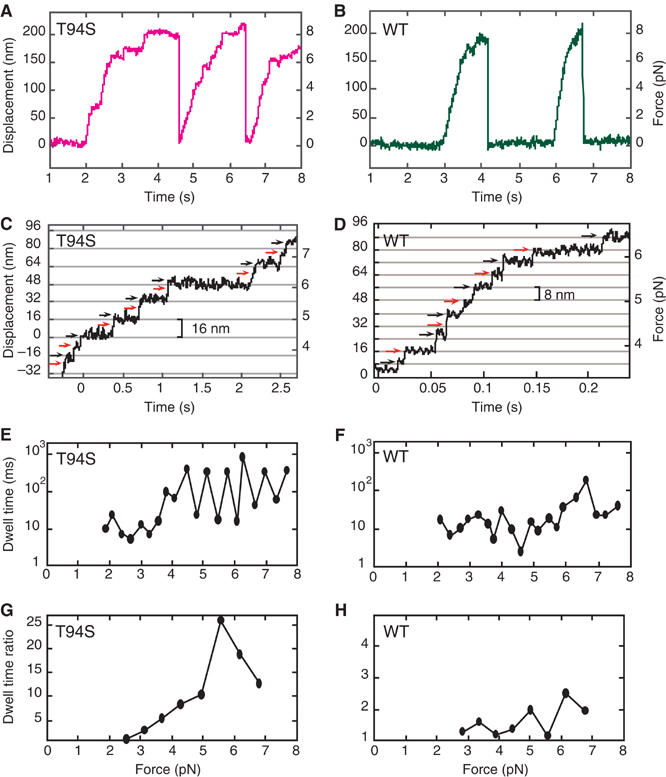
Single-molecule laser-trap assays of kinesin-T94S and wild-type kinesin. (A, B) The traces show processive movement by single kinesin-T94S (magenta) and wild-type kinesin (green) motors. (C, D) Single kinesin-T94S motors showed successive 8-nm steps at forces <∼4 pN, but exhibited successive 16-nm displacements, consisting of rapid double 8-nm steps, at forces >∼4 pN. Single wild-type kinesin motors under the same conditions showed sequential 8-nm steps. (E, F) A plot of dwell time versus force shows alternating short and long dwells following sequential 8-nm steps by the kinesin-T94S mutant. The wild type shows variable dwells following sequential 8-nm steps. (G, H) The ratios of the long dwell times divided by the short dwell times at different forces, calculated by averaging overlapping sets of three successive odd and even steps (see Materials and methods) corresponding to the traces in (C, D). The dwell time ratios or limp factors L of the mutant are significantly greater than those of wild type at forces >∼4 pN. Note the difference in Y-axis scales in (G, H).
The dwell times between steps by the mutant and wild-type motors were analyzed statistically. Steps in traces were assigned as odd or even, and the dwell times for three sequential odd steps and three sequential even steps were averaged (total=6 steps). The longer mean dwell time was divided by the shorter one to obtain the ratio. Two steps were then slipped from the first step, the next dwell time ratio was calculated, and this process was reiterated for the length of the run. Analysis of the traces for the mutant and wild type in Figure 2C and D is shown in Figure 2G and H.
In all, 31 traces of the mutant and 20 traces of wild type were analyzed in this way without selecting for 8- or 16-nm displacements (Figure 3). The dwell time ratio, referred to as the limp factor L (Asbury et al, 2003), increased sharply for the mutant from 2.99 to 6.35 with 2.4–6.9 pN force (Figure 3A). By contrast, the limp factor of wild-type kinesin increased slowly from 1.88 to 2.57 with 2.4–6.7 pN force, remaining at a low ratio of <3 (Figure 3A).
Figure 3.
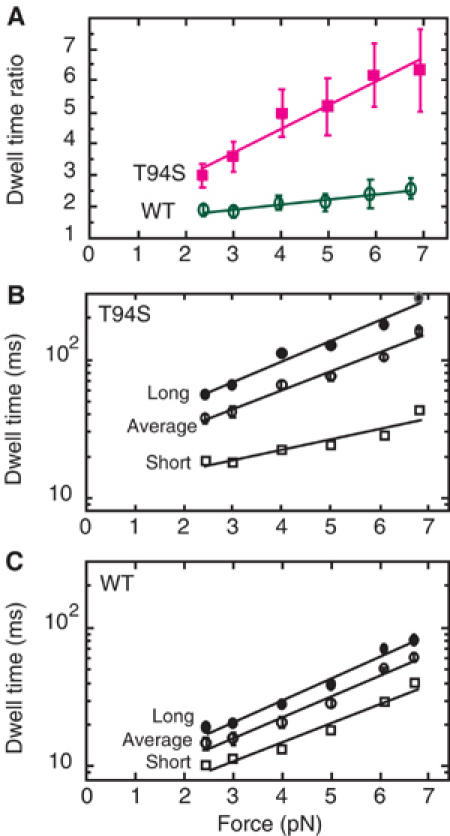
Dwell times versus force for steps by kinesin-T94S and wild-type kinesin. (A) The dwell time ratio or limp factor for 8-nm steps by the kinesin-T94S mutant (magenta) or wild-type kinesin (green) increases linearly with force, but the increase is significantly greater for the mutant than wild type. (B) The increase in long dwell times of the mutant with increasing force is paralleled by the increase in the average dwell times, while the short dwell times increase more slowly. (C) The long and short dwell times of wild-type increase in parallel with one another and with the average dwell times with increasing force. The error bars show the standard error of the mean (s.e.m.) for the dwell time ratios and average dwell times.
The average dwell time of the kinesin-T94S mutant was ∼2.5 times longer than that of wild type (mutant=37 ms and wild type=14.6 ms at 2.4 pN; mutant=159 ms and wild type=61 ms at 6.7–6.9 pN) (Figure 3B and C). For the mutant, the long dwell times increased sharply with force, whereas the short dwell times increased more gradually. The short dwell times of the mutant were longer than the average dwell times of wild type at low force, but they were shorter at high force.
The 16-nm displacements, characterized by short dwell times and high L values, appeared clearly at high force (⩾4 pN), where they were observed frequently for single kinesin-T94S motors (L>5, 32% of dwell time ratios, total=149, compared to 22% at <4 pN, total=74). The 16-nm displacements were observed infrequently for wild type (2% of dwell time ratios, total=148).
At low force, single kinesin-T94S motors showed 8-nm steps at higher frequency than at high force, rather than 16-nm displacements. The appearance of the 16-nm displacements at high force indicates the existence of a phase in the kinesin-T94S walking cycle that can be accelerated by force, resulting in the rapid double 8-nm steps. The 8-nm steps by the mutant were also observed at low ATP concentration (10 μM) (L=2.37 at 2.3–6.2 pN force). The low frequency of 16-nm displacements by the mutant at low ATP concentration (L>5, 5% of dwell times, total=99) is probably due to the time required to release ADP and bind ATP. The rate-limiting step of the kinesin cycle is thought to be the release of ADP (Hackney, 1988). But because the dwell time for the mutant at 10 μM ATP was ∼3 times that at 1 mM ATP, the rate-limiting step at low ATP concentration is likely to be ATP binding rather than ADP release, making the dwell time between steps by the mutant close to that of wild type.
Discussion
A kinesin stepping mutant
We report here a new mutant of Drosophila kinesin heavy chain that was rationally designed to make the nucleotide-binding P-loop resemble more closely that of the myosins. The mutant motor has a change of T94S in the P-loop, which was expected to open the nucleotide-binding cleft and increase the rate of nucleotide binding or release by the motor. This relatively minor structural change causes the motor to release ADP ∼3.6-fold faster than wild type in the absence of microtubules and to translocate microtubules in gliding assays with a velocity ∼3.3-fold slower than wild type. In single-molecule laser-trap assays, the mutant motor shows frequent 16-nm displacements under high force, which are resolvable into rapid double 8-nm steps, consisting of alternating slow and fast steps. At low force or low ATP concentration, successive 8-nm steps were observed instead of 16-nm displacements. The wild-type motor showed infrequent 16-nm displacements due to rapid double 8-nm steps. The frequency of rapid double 8-nm steps is thus enhanced in the mutant compared to wild type.
The 16-nm displacements arise by alternating fast and slow dwell times, causing alternate steps to be fast and slow. The structural basis of a slow step by the kinesin-T94S mutant that alternates with a fast step could be due to an inherent asymmetry of stepping to the right or left of the forward head caused by the handedness of the twist of the coiled coil. Stepping to the right might be constrained by the twist of the coiled coil, whereas stepping to the left might not be, and might be accelerated by interactions of the rear head with the forward head.
The successive 16-nm displacements in the kinesin-T94S traces with alternating short and long dwell times indicate that a step in the kinesin walking cycle is asymmetric, as predicted by asymmetric two-headed walking models, but not by symmetric ones such as hand-over-hand models in which the rear head always steps to the same side of the forward head (Howard, 1996), or inchworm models, in which only one head steps, dragging the other head along (Hua et al, 2002). Our results favor an asymmetric walking model in which the two heads of kinesin alternate in binding to the microtubule and hydrolyzing ATP.
Stepping by heterodimeric kinesin
Recently, others have reported 16-nm displacements by a heterodimeric kinesin protein consisting of kinetically different mutant and wild-type motor subunits (Kaseda et al, 2003), providing evidence that the two heads of the heterodimeric motor alternate in stepping. The interactions of the two heads of a heterodimeric motor with one another could differ from that of the native motor, however, due to the mutation in one head or due to the different proteins to which they were fused. The results we report here differ from those of Kaseda et al (2003) in that we observe asymmetry of movement for a homodimeric kinesin motor. This substantiates these previous findings by showing that the asymmetric steps of the heterodimer must have been due to an inherent asymmetry of stepping by the motor and were not dependent on the differences in the motor subunits.
Limping by wild-type kinesin motors
Our findings differ from those reported recently by others (Asbury et al, 2003) for wild-type Drosophila and native squid kinesin. These workers found evidence for alternating slow and fast steps under high force (4 pN) for truncated Drosophila kinesin proteins, whereas we did not observe frequent 16-nm displacements for our truncated wild-type kinesin, which was expressed as a fusion to a 106-residue BIO protein with two linker residues. The basis of these conflicting results could be a difference in the kinesin preparations used in the assays, a dependence of limping on the total length of the proteins analyzed, such that shorter proteins show a greater tendency to limp because they interact asymmetrically with the surface of the bead rather than the microtubule, or the conditions of the laser-trap assays. In the present work, we used a kinesin motor fused to a BIO domain to prevent interactions of the motor with the bead.
The velocities reported by Asbury et al (2003) both for their truncated and native kinesin proteins were unusually slow, ∼65–165 nm/s (calculated from the mean fast and slow dwell times in their Figure 3C). These velocities are significantly slower than the velocities of 300 and ∼600 nm/s for squid kinesin at 4 pN reported previously by the same laboratory (Visscher et al, 1999; Block et al, 2003), and the velocity of 390 nm/s at 4.0 pN of our wild-type kinesin, DmK447-BIO. These unusually slow velocities could arise if the force-clamp assays were performed under higher actual force than the reported value of 4 pN, or the kinesin proteins used in the assays were heterogeneous, comprising some motors in which one head was slower than the other due to partial inactivation during purification. The slow velocities of the truncated and native kinesin motors reported by Asbury et al (2003) need to be accounted for and correlated with the limping behavior of the motors before attempting to reconcile our data with theirs.
Kinesin-T94S and stepping models
The results we report here are important in that they show that a kinesin mutant with a single amino-acid change in the nucleotide-binding P-loop, under high force, shows a high frequency of rapid double 8-nm steps, resulting in 16-nm displacements. Under low force or low ATP concentrations, the mutant takes 8-nm steps, like wild-type kinesin. This suggests that the mutation enhances a step in the wild-type kinesin stepping cycle that is sensitive to load and the nucleotide state of the motor. Load-sensitive conformational changes have not previously been identified for kinesin and are believed to correspond to the force-producing steps of the hydrolysis cycle. The finding that the mutant motor is altered in ADP release tentatively identifies ADP release as a force-generating event of the kinesin cycle, although it is still possible that a different step of the hydrolysis cycle is affected. Further biochemical studies will be required to determine the step in the cycle that is altered in the mutant and how it affects the motor stepping mechanism. The observation of rapid double 8-nm steps under high load with alternating long and short dwells implies that a phase of the stepping cycle is affected by the mutant that involves the alternative regulation or interactions of the two heads.
The disappearance of the 16-nm displacements at low force or low ATP concentration indicate that another rate-limiting step, probably ATP binding, suppresses the rapid double 8-nm steps by interposing a longer dwell between the steps, resolving them into two steps. The ability of the kinesin-T94S motor to take successive 8-nm steps demonstrates that the basic mechanism of walking by the kinesin motor is unaltered. The appearance of the rapid double 8-nm steps or 16-nm displacements at high force and their infrequent appearance in wild-type traces indicates that the mutant enhances a step in the walking cycle that is inherently asymmetric, revealing an asymmetric walking mechanism for kinesin.
Materials and methods
Plasmids
Plasmids to express the kinesin-T94S mutant or corresponding wild-type Drosophila kinesin heavy chain (KHC) motor fused to a biotin-binding protein were constructed by inserting DNA fragments synthesized by the polymerase chain reaction (PCR) into pMW172/KHC (Song and Endow, 1996). The plasmids encode M-G-S+KHC residues E4-Q447, linked by G-S to 106 residues of the Propionibacterium shermanii biotin-binding transcarboxylase (BIO) (Pinpoint Xa, Promega). PCR-synthesized regions were confirmed by DNA sequence analysis.
Protein expression and purification
The kinesin-T94S and corresponding wild-type kinesin proteins fused to BIO were expressed in bacteria using the T7 RNA polymerase system (Studier et al, 1990). The fusion proteins were 123.4 kDa dimers of a 567-residue subunit. The kinesin region of the fusion protein corresponds to the conserved motor domain (E4-F326), the neck linker (G327-T344) and 103 residues of the coiled coil (A345-Q447). Proteins were purified for biochemical assays by chromatography on P11 phosphocellulose, followed by MonoQ and/or Superose 12 FPLC. The Superose 12 chromatography step was necessary to remove high-molecular-weight kinesin aggregates. The purified proteins were diluted extensively for use in laser-trap assays.
Motility assays
Microtubule gliding assays on motor-coated coverslips were performed as described (Song et al, 1997), using lysates of the kinesin-T94S or wild-type motors fused to BIO.
ADP release assays
Single-turnover mant-ADP release experiments were performed using FPLC-purified kinesin-T94S or wild-type protein fused to BIO by incubating 2 μM mant-ATP [2′(3′)-O-(N-methyl-anthraniloyl)-adenine 5′-triphosphate] with 0.5 μM motor for ⩾2 h on ice in the dark. Samples were warmed to room temperature for 5–10 min and fluorescence (excitation, 356 nm; emission, 446 nm) was recorded in a SPEX FluoroMax or SLM Aminco 8100 spectrofluorometer before and after addition of 500 μM ATP. Mant-ADP release assays in the presence of microtubules were performed by mixing 0.2 μM motor+mant-ATP after incubation on ice with 1 μM GTP-depleted microtubules in a cuvette and recording fluorescence before and after addition of the microtubules. Fluorescence was plotted versus time and data points were fit to a single exponential curve,
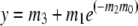 |
where y=fluorescence (cps), m0=time (s) and y=m3+m1 at t=0, using KaleidaGraph v 3.08d. Dissociation rate constants (kd=m2) were obtained from the curve fits. In some experiments, samples were spin-dialyzed to remove excess mant-ATP after incubation on ice and before recording fluorescence; rates were the same as assays in which mant-ATP was not depleted. Data for the experiments were therefore combined.
Laser-trap assays
Laser-trap assays were performed using highly diluted purified kinesin-T94S or wild-type motors expressed as fusions to BIO and attached to 0.2 μm avidin-coated beads in buffer+200 mM NaCl+1 mM ATP (Endow and Higuchi, 2000; Nishiyama et al, 2002). The data were derived from single motors, based on the statistics of binding of the beads to microtubules (Svoboda and Block, 1994; Endow and Higuchi, 2000). Assays performed at low ATP concentrations contained 10 μM ATP. Beads were trapped at a stiffness of 0.04 pN/nm and monitored for binding and movement on microtubules. Data were taken without a low-pass filter, then filtered through 50 Hz (Figure 2A and B) and 200 Hz (Figure 2C and D) low-pass filters for analysis.
Statistical analysis
Dwell times between steps of the mutant and wild type were analyzed statistically, as described by others (Asbury et al, 2003). Steps in a given trace were assigned as odd or even, and dwell times of three sequential odd steps and three sequential even steps were averaged. The longer mean dwell time was divided by the shorter one to obtain the dwell time ratio. Two steps were slipped along the trace from the first step, the next dwell time ratio was calculated from the next overlapping six steps, and this process was reiterated for the length of the run. The theoretical ratio for an exponential distribution of random dwell times generated by computer simulation was calculated to be 2.23 for analysis of overlapping sets of six sequential steps and 1.84 for 10 sequential steps. These values were close to the value of 1.79 obtained by analyzing dwell times from computer-simulated stepping records (Asbury et al, 2003).
Acknowledgments
We thank C Nicchitta and K Alexander for use of spectrofluorometers, and S Rosenfeld for the gift of mant-ATP. This work was supported by grants from the Japan Ministry of Education, Science, Sport & Culture to HH, Human Frontier Science Program to HH and SAE, St Jude Children's Research Hospital Cancer Center and American Lebanese Syrian Associated Charities to HWP, and NIH to SAE.
References
- Asbury CL, Fehr AN, Block SM (2003) Kinesin moves by an asymmetric hand-over-hand mechanism. Science 302: 2130–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM, Asbury CL, Shaevitz JW, Lang MJ (2003) Probing the kinesin reaction cycle with a 2D optical force clamp. Proc Natl Acad Sci USA 100: 2351–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy DL, Wagenbach M, Howard J (1999) Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J Biol Chem 274: 3667–3671 [DOI] [PubMed] [Google Scholar]
- Endow SA, Higuchi H (2000) A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature 406: 913–916 [DOI] [PubMed] [Google Scholar]
- Hackney DD (1988) Kinesin ATPase: rate-limiting ADP release. Proc Natl Acad Sci USA 85: 6314–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD (1994) Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci USA 91: 6865–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD (1995) Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature 377: 448–450 [DOI] [PubMed] [Google Scholar]
- Hancock WO, Howard J (1998) Processivity of the motor protein kinesin requires two heads. J Cell Biol 140: 1395–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K, Henningsen U, Schliwa M, Toyoshima C, Shimizu T, Alonso M, Cross RA, Amos LA (2000) Structural comparison of dimeric Eg5, Neurospora kinesin (Nkin) and Ncd head-Nkin neck chimera with conventional kinesin. EMBO J 19: 5308–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A, Thormählen M, Diaz-Avalos R, Doerhoefer M, Goldie KN, Muller J, Mandelkow E (2000) A new look at the microtubule binding patterns of dimeric kinesins. J Mol Biol 297: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Howard J (1996) The movement of kinesin along microtubules. Ann Rev Physiol 58: 703–729 [DOI] [PubMed] [Google Scholar]
- Howard J (2001) Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer Associates Inc [Google Scholar]
- Howard J, Hudspeth AJ, Vale RD (1989) Movement of microtubules by single kinesin molecules. Nature 342: 154–158 [DOI] [PubMed] [Google Scholar]
- Hua W, Chung J, Gelles J (2002) Distinguishing inchworm and hand-over-hand processive kinesin movement by neck rotation measurements. Science 295: 780–781 [DOI] [PubMed] [Google Scholar]
- Kaseda K, Higuchi H, Hirose K (2003) Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol 5: 1079–1082 [DOI] [PubMed] [Google Scholar]
- Kozielski F, Sack S, Marx A, Thormählen M, Schönbrunn E, Biou V, Thompson A, Mandelkow E-M, Mandelkow E (1997) The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell 91: 985–994 [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Higuchi H, Yanagida T (2002) Chemomechanical coupling of the forward and backward steps of single kinesin molecules. Nat Cell Biol 4: 790–797 [DOI] [PubMed] [Google Scholar]
- Schief WR, Howard J (2001) Conformational changes during kinesin motility. Curr Opin Cell Biol 13: 19–28 [DOI] [PubMed] [Google Scholar]
- Schliwa M (2003) Kinesin: walking or limping? Nat Cell Biol 5: 1043–1044 [DOI] [PubMed] [Google Scholar]
- Schnitzer MJ, Block SM (1997) Kinesin hydrolyses one ATP per 8-nm step. Nature 388: 386–390 [DOI] [PubMed] [Google Scholar]
- Song H, Endow SA (1996) Binding sites on microtubules of kinesin motors of the same or opposite polarity. Biochemistry 35: 11203–11209 [DOI] [PubMed] [Google Scholar]
- Song H, Golovkin M, Reddy ASN, Endow SA (1997) In vitro motility of AtKCBP, a calmodulin-binding kinesin protein of Arabidopsis. Proc Natl Acad Sci USA 94: 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185: 60–89 [DOI] [PubMed] [Google Scholar]
- Svoboda K, Block SM (1994) Force and velocity measured for single kinesin molecules. Cell 77: 773–784 [DOI] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM (1993) Direct observation of kinesin stepping by optical trapping interferometry. Nature 365: 721–727 [DOI] [PubMed] [Google Scholar]
- Visscher K, Schnitzer MJ, Block SM (1999) Single kinesin molecules studied with a molecular force clamp. Nature 400: 184–189 [DOI] [PubMed] [Google Scholar]


