Abstract
Bacteriophage T4 AsiA is a versatile transcription factor capable of inhibiting host gene expression as an ‘anti-σ′ factor while simultaneously promoting gene-specific expression of T4 middle genes in conjunction with T4 MotA. To accomplish this task, AsiA engages conserved region 4 of Eschericia coli σ70, blocking recognition of most host promoters by sequestering the DNA-binding surface at the AsiA/σ70 interface. The three-dimensional structure of an AsiA/region 4 complex reveals that the C-terminal α helix of region 4 is unstructured, while four other helices adopt a completely different conformation relative to the canonical structure of unbound region 4. That AsiA induces, rather than merely stabilizes, this rearrangement can be realized by comparison to the homologous structures of region 4 solved in a variety of contexts, including the structure of Thermotoga maritima σA region 4 described herein. AsiA simultaneously occupies the surface of region 4 that ordinarily contacts core RNA polymerase (RNAP), suggesting that an AsiA-bound σ70 may also undergo conformational changes in the context of the RNAP holoenzyme.
Keywords: AsiA, NMR, RNA polymerase, Sigma70, structure
Introduction
T4 bacteriophage development in Escherichia coli is characterized by an intricate program of activation and repression of specific genes (Karam, 1994). Temporal expression of T4 genes takes advantage of the intrinsic adaptability of RNA polymerase (RNAP) to recognize diverse promoters, since T4 phage does not code for its own enzyme. In the first few minutes following infection, host RNAP transcribes a set of early phage genes that irreversibly diverts normal transcription programs of the host to preferential expression of phage genes (Mosig and Hall, 1994). Modification of the host RNAP is required for phage gene expression, which includes ribosylation of the RNAP α subunits and the binding of phage-encoded activators to the σ70 subunit (Brody et al, 1995; Kolesky et al, 1999). One of these factors, Anti-σ factor A (AsiA), was one of the earliest proteins to be identified that could directly interact with σ70 (Orsini et al, 1993; Ouhammouch et al, 1994).
AsiA binds tightly to the C-terminal conserved region 4 (SR4) of σ70 and blocks expression of genes whose promoters are dependent on recognition of the −35 consensus DNA sequence (Stevens, 1977; Orsini et al, 1993, Adelman et al, 1997; Severinova et al, 1998). Early observation of AsiA's potent ability to block host gene expression defined AsiA as the archetype of an anti-σ factor (Hughes and Mathee, 1998). Further work revealed that AsiA's primary role is more of a co-activator for phage gene expression, with inhibition of host expression occurring as a consequence of redirecting host RNAP to transcribe a subset of phage genes (Ouhammouch et al, 1995; Hinton et al, 1996; Adelman et al, 1998; Pal et al, 2003). Stimulation of gene expression by AsiA occurs in conjunction with the T4 early gene product MotA (Hinton, 1991; Schmidt and Kreuzer, 1992). MotA, AsiA and a modified RNAP holoenzyme are sufficient to transcribe phage middle genes. Supported by evidence of MotA/AsiA/σ70 complexes (Hinton et al, 1996; Sharma et al, 1999; Pande et al, 2002), it would seem that AsiA serves as an essential part of a transcription complex that controls the middle gene transcriptional program in addition to its ability to block host gene expression. Thus, AsiA supports opposing transcriptional outcomes for host and phage gene expression (Lambert et al, 2001; Minakhin et al, 2003). Recent structural (Lambert et al, 2001; Urbauer et al, 2001; 2002) and biochemical studies (Severinova et al, 1996; Adelman et al, 1997; Pahari and Chatterji, 1997; Colland et al, 1998; Severinova et al, 1998; Sharma et al, 1999; Lambert et al, 2001; Minakhin et al, 2001; 2003) established that AsiA has a novel fold capable of forming a tight association with σ70 and remains bound to the holoenzyme during expression of phage genes.
The three-dimensional structure of SR4 from a variety of σ factors and bound to a diverse group of binding partners suggests a common fold for this conserved domain (Campbell et al, 2002; 2003; Vassylyev et al, 2002; Sorenson et al, 2004) (Figure 1). These structures, in conjunction with that of σA SR4 from Thermotoga maritima reported herein, all suggest that SR4 adopts a single conformation that is not typically influenced by its binding partners (Campbell et al, 2002; 2003; Sorenson et al, 2004) or other conserved regions of the σ factor itself (Camarero et al, 2002; Campbell et al, 2003; Sorenson et al, 2004). Thus, it is remarkable that the structure of EcSR4 observed in the complex with AsiA is dramatically different from those previously determined. The C-terminal helix of region 4.2 is unstructured in the EcSR4/AsiA complex and the remaining four helices of regions 4.1 and 4.2 are completely reorganized. AsiA's interaction with EcSR4 occurs simultaneously with regions 4.1 and 4.2, forming a single species in solution. The interface between AsiA and EcSR4 encompasses residues of the σ domain responsible for both DNA and core RNAP binding. Thus, the interaction of AsiA with SR4 should displace SR4 from its location in the holoenzyme, resulting in a change in the relative orientation of σ70 domains within RNAP. By contrast, AsiA itself does not appear to adopt an altered conformation in the complex with EcSR4, and the controversy over the two disparate structures of AsiA (Lambert et al, 2001; Urbauer et al, 2002) is now resolved (see Correction in this issue). While σ70 has long been thought to be a protein composed of structured domains separated by malleable linkers, the structure of AsiA/EcSR4 suggests that the plasticity of σ factors can extend into the structured domains themselves.
Figure 1.
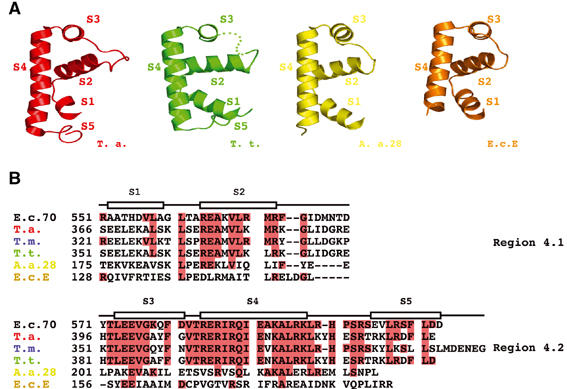
Comparison of the three-dimensional structures of SR4 determined in different contexts. (A) Ribbon diagrams tracing the backbone topology of the structure of SR4 determined in four different contexts. Conserved helices corresponding to helices S1–S5 are labeled. The structure of SR4 from Thermus aquaticus (T. a.) bound to DNA (residues 370–438) is colored red (PDB ID 1KU7; Campbell et al, 2002); SR4 from the RNAP holoenzyme of T. thermophilus (T. t.) (residues 351–423) is colored green (PDB ID 1IW7; Vassylyev et al, 2002); SR4 from the Aquifex aeolicus (A. a.) (residues 175–236) bound to the anti-σ factor FlgM is colored yellow (PDB ID 1RP3; Sorenson et al, 2004); and SR4 (residues 128–187) from E. coli σE (E. c. E.) bound to the anti-σ factor Rse-A is colored orange (PDB ID 1OR7; Campbell et al, 2003). (B) Amino-acid sequence alignment of SR4. Red shading indicates sequence identity between SR4 domains from T. aquaticus, T. maritima (T. m.), T. thermophilus, A. aeolicus, E. coli σE and E. coli σ70 (E. c. 70). The helical elements observed in the solution structure of T. maritima σA SR4 are indicated as boxed segments. The sequences are divided according to the conserved subdomains of E. coli σ70 SR4, region 4.1 and region 4.2.
Results
Three-dimensional structures of SR4 suggest a common overall fold
SR4 is one of four highly conserved domains, which are found in the primary or Group 1 σ factors exemplified by E. coli (Ec) σ70 (Figure 1) (Lonetto et al, 1992). σ70 and fragments derived from σ70 are generally capable of reconstituting biochemical functions, but have resisted most attempts at structure determination. Many fragments of σ70 appear to be poorly folded on their own and are overexpressed in inclusion bodies in bacteria (Severinova et al, 1996). To circumvent this problem, the three-dimensional structures of SR4s from a number of closely related primary (Campbell et al, 2002; Vassylyev et al, 2002), alternative (Sorenson et al, 2004) and extracytoplasmic (Campbell et al, 2003) σ's have been solved by X-ray methods in a variety of contexts. Superposition of these SR4 structures reveals a conserved, four α helix fold, to include a canonical helix–turn–helix (HTH) element responsible for DNA recognition (Figure 1A). The high sequence conservation of these closely related proteins is consistent with their similar three-dimensional structures (Figure 1B). However, even the most distantly related σ factor by sequence, E. coli σE, has a similar overall fold. Considering the diverse binding partners to which these SR4s were bound, there is a strong implication that the core, four-helix fold of SR4 is conserved and not significantly influenced by DNA, anti-σ's, core RNAP, nor even other domains of the σ factor itself. The most structurally diverse element of the SR4 fold is the C-terminus, which appears to form a short, fifth α helix that plays a significant role in docking SR4 to the β subunit of core RNAP (Vassylyev et al, 2002), and is implicated in interacting with MotA in an AsiA-modified holoenzyme (Pande et al, 2002). This fifth helix is only partially seen (Campbell et al, 2002) or not seen at all (Campbell et al, 2003; Sorenson et al, 2004) in several of the known structures, a consequence of poor electron density and/or inter-protein crystal contacts. Prompted by the need to clarify the structure and/or orientation of this helix, the structure of SR4 from T. maritima (TmSR4) was determined by multi-nuclear NMR spectroscopy in solution (Figure 2, Table I). Residues 313–393 of TmSR4 are equivalent to residues 533–613 of E. coli σ70 and share 65% sequence identity with the E. coli domain (Figure 1B). The structure of TmSR4 is, as expected, very similar to the structures of SR4 from other σ factors (Figure 2D). The pairwise root-mean-square deviation (r.m.s.d.) between Cα atoms of TmSR4 and all of the X-ray derived structures ⩽3 Å over helices 1–4 of the domain. The C-terminal fifth helix is clearly visible for TmSR4 in solution and packs against the N-terminus and helix S2, enclosing the small hydrophobic core of the domain (Figure 2C). The fifth helix is attached to a disordered loop (residues 379–383), poorly defined by the NMR data. Inherent flexibility may account for the poor electron density of this helix in many of the crystalline states. The structure of TmSR4, along with the structure of SR4 bound to a variety of binding partners, permits assessment of the structural reorganization of SR4 observed when AsiA engages the domain.
Figure 2.
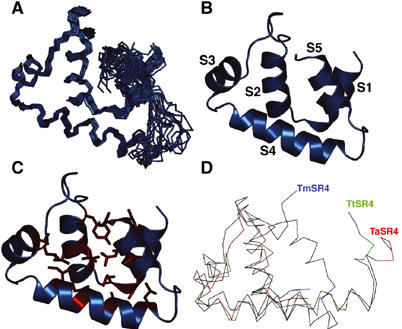
Three-dimensional structure of T. maritima σA SR4. (A) Superposition of the NMR structure family. Chemical shifts for residues 379–383 (the loop between S4 and S5) were not identified and this segment is relatively disordered in the family. (B) Helix arrangement in TmSR4. Helix S1 (residues 322–329), helix S2 (residues 334–342), helix S3 (residues 353–361), helix S4 (residues 364–377) and helix S5 (residues 385–393) were identified from secondary 13Cα/β shifts, dNN (i, i+1) NOEs and 3JNHα coupling constants. (C) Hydrophobic core of TmSR4 formed from nonpolar residues found in all the five helices. (D) Superposition of TmSR4 (blue), the T. aquaticus σA SR4 in its DNA-bound conformation (red) (Campbell et al, 2002) and the T. thermophilus σA SR4 in its holoenzyme conformation (green) (Vassylyev et al, 2002).
Table 1.
Structural statistics for T. maritima σA SR4
| R.m.s. deviations from experimental restraintsa | |
| NOEs (510) | 0.069±0.009 Å |
| φ, ψ, χ1 (160) | 0.372±0.017° |
| 3JNHα (53) | 0.850±0.041 Hz |
| Coordinate precisionb | |
| Backbone (residues 328–378+385–395) | 0.70±0.10 Å |
| All non-hydrogen (residues 328–378+385–395) | 1.2±0.11 Å |
| Quality factorsc | |
| % Residues in the most favorable region of Ramachandran plot | 84.6% |
| Bad contacts/structure |
2±1 |
| aNumbers in parentheses indicate the total number of restraints. No NOEs were included for atoms separated by three chemical bonds, with the exception of those involving backbone atoms. | |
| bResidues 328–378+385–398. | |
| cCalculated using the programs PROCHECK and PROCHECK_NMR (Laskowski et al, 1996) for the residues over which the coordinate precision was calculated. | |
AsiA can exist in two states in solution
AsiA is functionally monomeric, but, when overexpressed in E. coli, was found to strongly self-associate and was isolated as a homodimer (Kd=600 nM) (Lambert et al, 2001; 2003; Urbauer et al, 2001). The homodimeric interface is remarkably hydrophobic and involves nearly one-half of the residues in each monomer. In order to bind EcSR4, the homodimer must dissociate (Lambert et al, 2001), as nearly every residue at the homodimer interface has been implicated in contacting residues in EcSR4 (Lambert et al, 2001; Minakhin et al, 2003). Two different three-dimensional structures of the AsiA dimer have been reported (Lambert et al, 2001; Urbauer et al, 2002) and the distinction between these structures has now been resolved, revealing that the structure of Urbauer et al (2002) is the correct structure for AsiA (see Correction in this issue). Given that AsiA dimer must dissociate to bind EcSR4, the conformation of AsiA in its predominantly monomeric form (hereafter referred to as the AsiA monomer) was pursued to determine if any change in the AsiA structure occurs upon binding EcSR4. Unexpectedly, the addition of a cleavable, N-terminal hexahistidine (His6) tag was sufficient to express AsiA in E. coli predominantly as a monomer, which was readily identified by NMR spectroscopy (see Supplementary Figure 1) and confirmed by analytical centrifugation, with an apparent equilibrium dissociation constant of 300 μM (see Supplementary Figure 2). NMR spectroscopy of the AsiA monomer suggests that the conformation is closely related to that of AsiA bound to EcSR4 (see Supplementary Figure 1).
The structure of the free AsiA monomer was solved using 594 distance (NOEs), 39 angular (3JNHα) and 129 global orientational restraints (residual dipolar couplings (RDCs)) (Figure 3, Table I). The paucity of observed NOEs for free AsiA monomer, only 1/3 of the total number of NOEs observed for AsiA dimer, reflects the relatively poor behavior of AsiA in this state in solution. Overall, the positions of the six α helices in the AsiA monomer are similar to those observed in one monomer of AsiA dimer (Figure 3), and to the conformation of AsiA bound to EcSR4 (Figures 4 and 5). However, the structure of free AsiA monomer is not nearly as precisely defined as in the other two states. In particular, the C-terminal helix–loop–helix element encompassing helices 5 and 6 was very poorly defined by the experimental monomer data and several residues could not be observed (72–75 and 85), probably due to exchange broadening of this segment of AsiA in the free monomer. This segment of AsiA is also dispensable to its interaction with EcSR4 (Sharma et al, 2002; Pande et al, 2002), suggesting that it is loosely tethered to the balance of the molecule. Despite these limitations, the structures of the monomer and dimer forms of AsiA suggest that AsiA does not undergo functionally significant conformational changes upon binding EcSR4.
Figure 3.
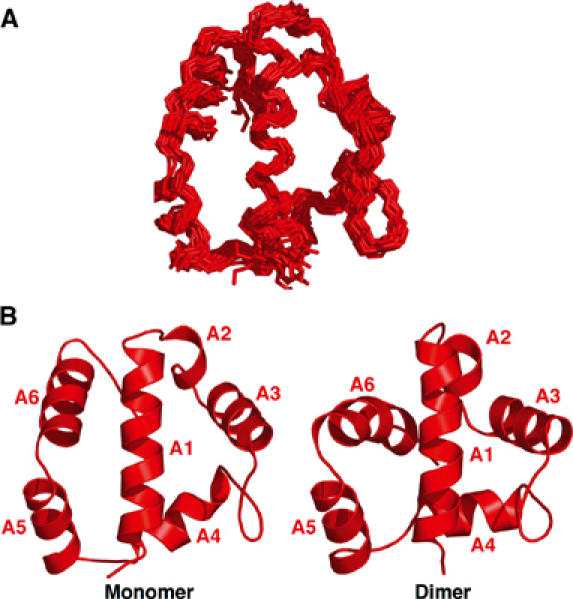
Three-dimensional structure of AsiA monomer. The three-dimensional structure of AsiA monomer is shown in (A) and compared to one molecule of the AsiA dimer (Urbauer et al, 2002) in (B). Structural statistics are given in Table II. The structures are quite similar, with the exception of an altered position of helix A6 between monomer and dimer. In the dimer, numerous NOEs between A6 and the balance of the monomer are seen, while NMR spectra of the monomer fail to show these contacts. Overall, the NOE density is considerably lower in the monomer (Table II), but the relative orientations of helical segments in the two states are quite similar.
Figure 4.
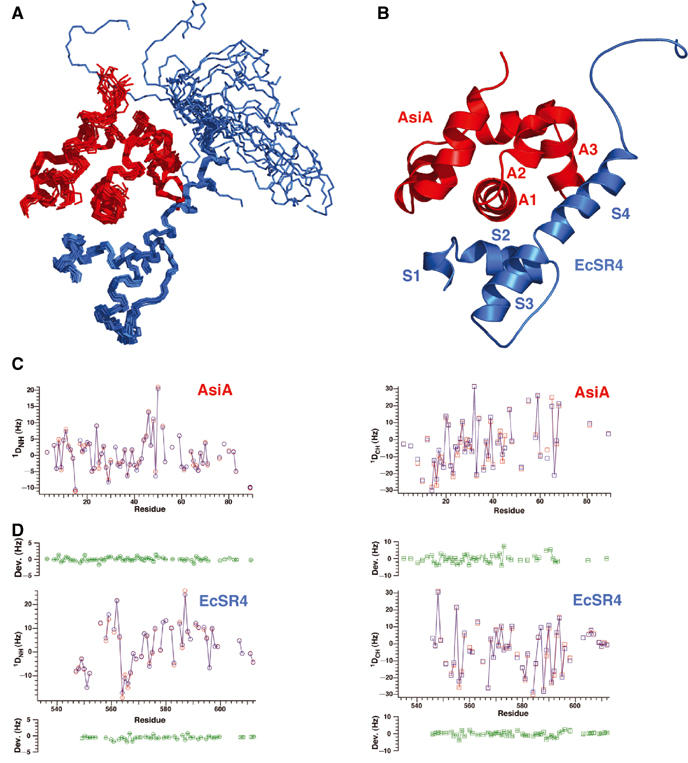
Three-dimensional structure of the AsiA/EcSR4 complex. (A) Superposition of the 20 conformers of the complex, with EcSR4 shown in blue (residues 545–613) and AsiA shown in red (residues 2–90). Residues 72–75 and residue 85 of AsiA were exchange broadened in the complex and not identified. Residues 533–544 of EcSR4 were disordered in the complex and are not shown. (B) Summary of the secondary structure elements of the AsiA/EcSR4 complex. (C) Comparison of observed (red) and calculated (blue) 1D couplings for NH (left panel) and CαH (right panel) as a function of residue number in AsiA. The mean deviation between calculated and observed couplings over the structure family is plotted in green±observed error from the mean for each measured coupling. (D) Comparison of observed (red) and calculated (blue) 1D couplings for NH (left panel) and CαH (right panel) as a function of residue number in EcSR4 displayed as in (C). The calculated couplings for NH and CαH were determined by SVD analysis with the program DC, as described in Materials and methods.
Figure 5.
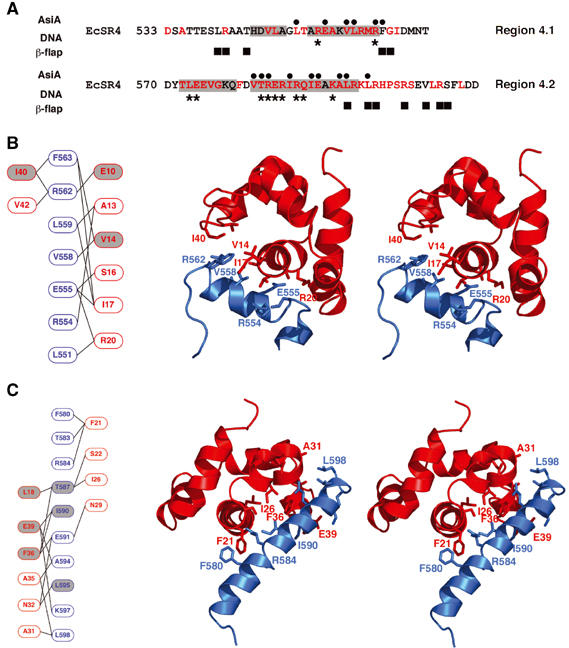
Detailed interaction between AsiA and EcSR4. (A) Comparison of AsiA-binding positions in the EcSR4 complex (black dots) versus the contact regions between SR4 and DNA (asterisks; Campbell et al, 2002) or the β-flap tip helix (black squares; Vassylyev et al, 2002). The interaction with AsiA overlaps both DNA and core RNAP-binding sites of SR4. The helices present in EcSR4 are indicated by gray shading and conserved amino acids are shown in red. (B) Subdomain I of the AsiA/EcSR4 interaction. Subdomain I is composed of helix A1 of AsiA and helix S2 of EcSR4. Schematic overview of contacting residues in the subdomain is shown in red and blue for AsiA and EcSR4, respectively, in the left panel. Gray shading indicates positions of AsiA, which are defective for EcSR4 binding in a pulldown assay (Lambert et al, 2001) or cellular toxicity experiment (Sharma et al, 2002; Pal et al, 2003). The right panel displays a stereoview of the subdomain, viewed down the axis of helix A1 in the complex, with selected side chains displayed on a ribbon representation of the backbone. The structure of the complex shown is a representative choice of one model from the structure family. (C) Subdomain II of the AsiA/EcSR4 interaction. Subdomain II is composed of the C-termini of helices A1, A3 and all of helix A2 of AsiA contacting helix S4 of EcSR4. A schematic overview of contacting residues in the subdomain is shown in red and blue for AsiA and EcSR4, respectively, in the left panel. Gray shading indicates positions of AsiA or EcSR4, which are defective for binding in a pulldown assay (Lambert et al, 2001) or cellular toxicity experiment (Sharma et al, 2002; Pal et al, 2003). The right panel displays a stereoview of the subdomain, viewed down the axis of helix A1 in the complex, with the side chains of the indicated residues in the schematic displayed on a ribbon representation of the backbone. The structure of the complex shown is an arbitrary choice of one model from the structure family. Note that the orientations of the stereo pairs in (B) and (C) differ by a 180° rotation about the horizontal axis.
Three-dimensional structure of the AsiA/EcSR4 complex
The structure of AsiA bound to EcSR4 was determined from 1486 experimental interproton distance restraints, to include 213 intermolecular NOEs between AsiA and EcSR4 (Figure 4A and B,; Table II). Since the structure of EcSR4 is significantly altered in the complex, RDCs were measured for both AsiA and EcSR4 and the model was refined against these global structural restraints (Table II; Figure 4C and D). This analysis confirmed the orientation and conformation of the individual molecules calculated from NOE, dihedral angle and 3JNHα coupling constants alone. Overall, the structure of AsiA bound to EcSR4 is very similar to that of AsiA monomer free in solution and to AsiA dimer. By contrast, EcSR4 adopts an unexpected conformation upon binding AsiA. Residues of EcSR4 that ordinarily interact with core RNAP, residues that form the hydrophobic core of EcSR4 and residues that one implicated in DNA binding are all involved in the interaction with AsiA, resulting in a dramatically altered conformation (Figures 5 and 6).
Table 2.
Structural statistics for the AsiA monomer
| R.m.s. deviations from experimental restraintsa | AsiA monomer |
| Intramolecular NOEs (594) | 0.075±0.008 Å |
| φ, ψ, χ1 (166) | 0.59±0.12° |
| 3JNHα b (39) | 1.04±0.05 Hz |
| 1DNH c (70) | 1.11±0.03 Hz |
| 1DCH c (59) | 2.45±0.09 Hz |
| Coordinate precision | |
| Backboneb | 0.88±0.12 Å |
| All non-hydrogenc | 1.46±0.1 Å |
| Quality factorsd | |
| % Residues in the most favorable region of Ramachandran plot | 84.9% |
| Bad contacts/structure |
0 |
| aNumbers in parentheses indicate the number of restraints for AsiA monomer in each category. No NOEs were included for atoms separated by three chemical bonds, with the exception of those involving backbone atoms. | |
| bCalculated using the program XPLOR-NIH 2.91 (Schwieters and Clore, 2001). | |
| cResidues 2–88. | |
| dCalculated using the programs PROCHECK and PROCHECK_NMR (Laskowski et al, 1996) for the residues over which the coordinate precision was determined. | |
Figure 6.
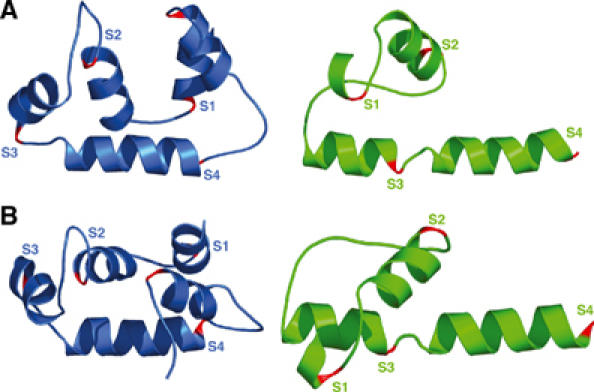
AsiA remodels the conformation of SR4. The structure of TmSR4 (blue) is compared to the structure of AsiA-bound EcSR4 (green) in two views. The C-termini of helices S1–S4 are colored red. Helix S5 seen in TmSR4 is unfolded in EcSR4 and the unfolded segment has been excluded from the views of EcSR4 for clarity. (A) Lateral view along helix S4 and (B) top view rotated 90° about the horizontal axis relative to (A). Apparent in both views, helix S2 in the complex is inverted N-to-C-terminus and helix S1 is repositioned by approximately 180° in the AsiA-bound conformation relative to the conformation in TmSR4. The DNA-binding HTH element (helices S3 and S4) is unfolded from its canonical orientation in TmSR4 and forms a pseudo-continuous helix in the AsiA-bound state.
The interaction between AsiA and EcSR4 can be divided into two subdomains. The first subdomain is formed with helix S2 in region 4.1 and the second subdomain is formed with helix S4 in region 4.2 of EcSR4. These contacts alone constrain an unambiguous docking orientation for AsiA on the protein domain. In subdomain I (Figure 5B), the N-terminal helix of AsiA (residues 5–21) forms an extensive set of contacts with a portion of region 4.1 (residues 551–563). The extensive set of AsiA contacts formed with this portion of region 4.1 buries 570 Å2. Although the majority of these contacts involve interactions between nonpolar side chains in each molecule, several salt bridges could be inferred, in particular between R562 of EcSR4 and E10 of AsiA. E10 of AsiA displays NOEs to V558, positioning the side chain of E10 near R562. Mutation of E10 to Lys (but not Ala) reduces AsiA toxicity when overexpressed in E. coli (Sharma et al, 2002). The functional deficiencies of deletion and point mutants introduced between residues 10 and 21 of AsiA are readily explained on the basis of the interactions observed in the complex (Lambert et al, 2001; Minakhin et al, 2001; 2003; Sharma et al, 2002; Pal et al, 2003).
Helix S4 in region 4.2 forms the second half of the contact surface with AsiA (Figure 5C). Here, the interaction surface is complex, composed of residues from the C-terminus of helix A1, helix A2 and the C-terminus of helix A3 of AsiA. This surface buries 752 Å2 of EcSR4 helix S4, principally by interactions formed between partially exposed hydrophobic residues of AsiA and elements of the hydrophobic core of EcSR4. It is these interactions between helix S4 of EcSR4 and AsiA that maintain the altered conformation of EcSR4 in the complex. Residues of the tight turn between SR4 helices S3 and S4 (Campbell et al, 2002; Vassylyev et al, 2002) form novel contacts in the complex (Figure 5C), connecting the C-terminus of helix S2 and the N-terminus of helix S4. These interactions are reinforced by the contacts between F563 of EcSR4 and AsiA residues in helix A1 (V14, I17) and helix A3 (I40, V42). Since these residues of AsiA and EcSR4 derive from both of the major subdomains of the interaction surface, the contact surfaces of AsiA and EcSR4 form an interlocking network to establish a stable AsiA/EcSR4 complex.
Despite the stability of the AsiA/EcSR4 complex, AsiA displays some behavior that could obscure contributions of the AsiA C-terminus to the interaction with EcSR4. Neither the backbone nor most of the side-chain chemical shifts of AsiA residues 72–75, nor residue 85, is observable in the complex; relatively few NOEs were observed between residues of this helix and other residues in AsiA to constrain its precise orientation. Similar observations were made in free AsiA monomer, suggestive of an intrinsic property of AsiA, which broadens the signals for this segment of the protein. Although deletion mutagenesis suggests that AsiA's C-terminus does not play a significant role in the interaction with EcSR4 (Sharma et al, 2002; Pal et al, 2003), the inability to examine fully the C-terminus of AsiA in the complex leaves open the question whether this region contributes to the interaction with EcSR4.
AsiA remodels EcSR4
The EcSR4 construct used in this study encompasses all of conserved regions 4.1 and 4.2 and extends into conserved region 3.2. As shown in Figure 1, SR4s from several different σ's adopt a conserved helical fold and do not appear to experience significant conformational changes in response to an interaction with DNA (Campbell et al, 2002), core RNAP (Vassylyev et al, 2002) or structurally diverse anti-σ's (Campbell et al, 2003; Sorenson et al, 2004). In addition, the fold of SR4 does not appear to be influenced by interaction with other structured domains in σ itself (Camarero et al, 2002; Vassylyev et al, 2002; Campbell et al, 2003; Sorenson et al, 2004). The disposition of the C-terminal fifth helix is less well defined in the known structures, but the existence of the helix in the presence of core RNAP (Vassylyev et al, 2002) and in solution (Figure 2) suggests that it, too, is a common structural feature of SR4. In stark contrast to these observations, EcSR4 does not adopt the canonical SR4 fold in the presence of AsiA. Although structural investigation of unbound EcSR4 is frustrated by unfavorable properties, the high sequence similarity of EcSR4 to sequences of SR4s whose structures are known (Figure 1) suggests that EcSR4 would adopt a fold similar to that reported for others. This is reinforced by the fact that the helical segments of EcSR4 bound to AsiA encompass the same segments seen in the known SR4 structures. Thus, it seems likely that AsiA induces a transition from the canonical fold of SR4 to the reorganized conformation observed in the complex.
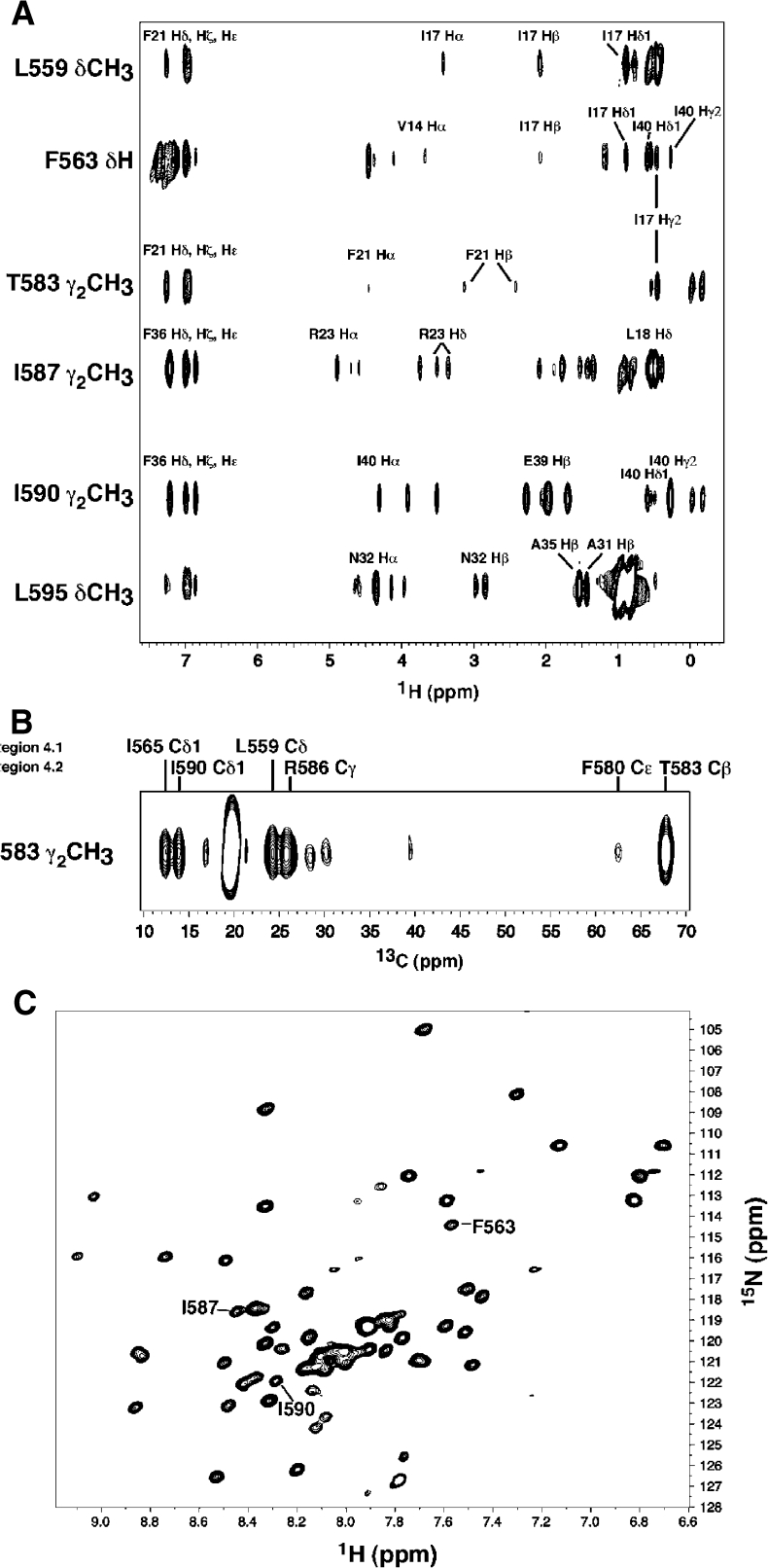
Four helices of AsiA-bound EcSR4 are organized into a drastically altered three-dimensional structure (Figure 6). The orientation of helix S2 relative to helices S3 and S4 is inverted, with the C-terminus helix S2 brought into close proximity to the N-terminus of helix S4. In the canonical fold of SR4, it is the N-terminus of helix S2 that is proximal to the N-terminus of helix S4 (illustrated by example with TmSR4 in Figure 6). NOEs to EcSR4 residues I565 (C-terminus of S2) and T583 (N-terminus of S4) illustrate some of the many contacts observed between helices S2 and S4 unique to the conformation of SR4 in the complex with AsiA (Figure 7). These amino acids simultaneously contact the same residue in AsiA (Figure 7A) and also reside near one another in the complex (Figure 7B). I17 of AsiA, for example, displays NOEs to F563 (region 4.1) and T583 (region 4.2) with comparable intensities (Figure 7A). This occurs in the complex even though F563 and T583 are more than 8 Å apart from one another in the canonical fold of SR4 (Figure 1). In the complex, F563 and T583 are brought into spatial proximity, which is clearly evident from NOEs between regions within SR4 itself (Figure 7B). Contacts to T583, a component of the DNA-binding surface of SR4 (Campbell et al, 2002), actually turn this residue inward in the complex. As a result, the chemical shift of the Cγ2 methyl protons of T583 shifts upfield to ≈−1 ppm due to the proximity of this residue to two phenylalanine residues, F580 in EcSR4 and F21 in AsiA. The NMR spectrum of the complex clearly indicates that regions 4.1 and 4.2 contact AsiA simultaneously; only a single species of EcSR4 is observed spectroscopically (Figure 7C).
Figure 7.
Spectroscopic analysis of AsiA/EcSR4. (A) Cross sections of a 13C-F1-edited/F2-filtered NOESY spectrum taken at the 13C/1H chemical shifts of the indicated residue positions in EcSR4. Selected assignments to the 1H chemical shifts in AsiA are shown. Residues in region 4.1 (L559 and F563) and region 4.2 (T583) contact the same position in AsiA (I17) with comparable intensities. (B) 13C-edited NOESY spectrum of the side-chain methyl group from T583 observed in 13C/15N-EcSR4 in the AsiA/EcSR4 complex. The 1H chemical shift of the T583 Hγ2 methyl group shifts upfield by ≈1.5 ppm in the complex relative to the chemical shift of this group in TmSR4. T583 forms unique contacts in the complex with the indicated residues in both regions 4.1 and 4.2 in EcSR4. The NOESY spectrum illustrates that this residue is turned inward in the complex, away from its position in the free TmSR4 domain. The portion of the spectrum shown displays the 13C chemical shifts in lieu of the 1H chemical shifts due to 13C frequency labeling of the protons during t1 in the experiment. (C) 1H–15N HSQC spectrum of EcSR4 bound to AsiA. The indicated residues in region 4.1 (F563) and region 4.2 (I587 and I590) specifically interact with AsiA (see Figure 5) and are clearly seen to be single peaks in the spectrum, indicative of a single species in solution.
Another striking transition evident from a comparison of the canonical SR4 fold to that of the AsiA-bound EcSR4 is the alteration of the DNA-binding surface itself. The HTH motif of EcSR4, formed by helices S3 and S4, is transformed into an extended, pseudo-continuous α-helix in the complex (Figure 6). Secondary 13Cα/β shifts for residues V582 and T583, residues which form the tight turn between helices S3 and S4, change from −2.92/+3.91 and −0.78/+2.16 ppm in TmSR4 to +3.65/−0.21 and +0.79/−1.26 ppm in the EcSR4 complex. The 3JNHα coupling constant for V582 decreases from 9.2 Hz in TmSR4 to 4.3 Hz in the EcSR4 complex. Both observations are consistent with the induction of helicity in the region of the protein between helices S3 and S4. The extended pseudo-continuous helix of the HTH element is in excellent agreement with RDC data (Figure 4, Table III).
Table 3.
Structural statistics for the AsiA/EcSR4 complex
| |
AsiA |
EcSR4 |
| R.m.s. deviations from experimental restraintsa | ||
| Intramolecular NOEs (825/448) | 0.045±0.004 Å | 0.05±0.004 Å |
| Intermolecular NOEs (213) | 0.046±0.009 Å | |
| φ, ψ (154/122) | 0.514±0.09° | 0.568±0.02° |
| 3JNHα b (−/47) | ND | 1.24±0.07 Hz |
| 1DNH c (53/66) | 0.84±0.04 Hz | 0.6±0.04 Hz |
| 1DCH c (61/49) | 1.7±0.09 Hz | 2.4±0.17 Hz |
| Coordinate precision | ||
| Backboned | 0.77±0.11 Å | 0.63±0.08 Å |
| All non-hydrogend | 1.3±0.16 Å | 1.2±0.08 Å |
| Quality factorse | ||
| % Residues in the most favorable region of Ramachandran plot | 90.0% | 88.4% |
| Bad contacts/structure |
0 |
0 |
| aNumbers in parentheses indicate the total number of restraints for AsiA/EcSR4. No NOEs were included for atoms separated by three chemical bonds, with the exception of those involving backbone atoms. | ||
| bND=not determined. | ||
| cCalculated using the program XPLOR-NIH 2.91 (Schwieters and Clore, 2001). | ||
| dResidues 2–71+76–84+86–89 for AsiA and residues 546–598 for EcSR4. | ||
| eCalculated using the programs PROCHECK and PROCHECK_NMR (Laskowski et al, 1996) for the residues over which the coordinate precision was determined. | ||
EcSR4 bound to AsiA lacks the C-terminal fifth helix (S5) observed in Tm and TtSR4. Comparison of secondary 13Cα/β chemical shifts, 3JNHα coupling constants and intramolecular NOEs of TmSR4 to EcSR4 suggests that S5 (residues 605–612) is unfolded in the EcSR4/AsiA complex, potentially by AsiA itself. Since deletion of the C-terminus of EcSR4 appears to have no effect on binding AsiA (Minakhin et al, 2001), the unfolding of S5 interpreted on the basis of these structural comparisons may serve a different purpose. One possibility is to provide access to this region of EcSR4 to MotA, which contacts EcSR4 in the vicinity of the C-terminus (Pande et al, 2002).
Discussion
The initiation of transcription is the point at which most bacterial genes are regulated. In bacteria, promoter specificity is partially determined by the σ subunit of RNAP. The primary σ factor of E. coli, σ70, is responsible for recognition of canonical ‘housekeeping' promoters whose architecture contains consensus DNA-binding sequences centered at −10 and −35 relative to the transcription start site. Promoter contacts by σ70 are made by the internal conserved region 2 and the C-terminal conserved region 4, which interact with the −10 and −35 promoter sequences, respectively (Lonetto et al, 1992). The DNA-contacting regions of σ factors are both separable and independent, which permits σ factors to bind to a wide range of promoter sequences (Helmann and Chamberlin, 1988). Biochemical and structural studies indicate that σ factors undergo a number of conformational changes (Dombroski et al, 1992; 1993; Murakami et al, 2002; Campbell et al, 2003; Sorenson et al, 2004). Thus, σ factors possess structurally conserved domains responsible for interaction with core RNAP and promoter DNA, with flexible linkers allowing access to regulatory proteins (Darst et al, 2002). The linker flexibility has been shown to be essential for the action of anti-σ factors like Rse-A and FlgM, which occlude the core RNAP-binding surfaces of their target σ factors without significant change to the structure of individual domains (Campbell et al, 2003; Sorenson et al, 2004). The structure of SR4 from a variety of σ factors, is not influenced by its binding partner in the holoenzyme (Vassylyev et al, 2002), by DNA (Campbell et al, 2002) or by other domains of σ itself (Vassylyev et al, 2002), even when other σ domains are brought into close proximity with SR4 when bound to some anti-σ factors (Camarero et al, 2002; Campbell et al, 2003; Sorenson et al, 2004). Thus, the intrinsic structure of SR4 is not easily altered by its interaction with its binding partners. It is therefore remarkable that SR4 adopts such a drastic reorganization upon binding to AsiA. Although the structure of free EcSR4 is not known, the wealth of structural data suggests that AsiA induces this transition rather than merely stabilizing a pre-existing altered form of SR4.
The interaction with AsiA converts the DNA-binding HTH element of EcSR4 into a pseudo-continuous α helix. This is most likely the principal cause for the loss of DNA binding by SR4 in an AsiA-bound holoenzyme (Orsini et al, 2001). Critical arginine residues for DNA binding in region 4.1 (R554 and R562) as well as region 4.2 (R584) are positioned away from the surface of EcSR4 where DNA would be located (Campbell et al, 2002; Murakami et al, 2002), forming new contacts with other SR4 regions. Other key residues for DNA interaction (e.g. T583) are partially buried at the AsiA/EcSR4 interface, leaving them inaccessible to the DNA. Complementing the blockage of core RNAP-binding sites, AsiA envelops the DNA-binding surface of SR4 almost in its entirety. The transformation of the DNA-binding surface of EcSR4 might seem at odds with the observation that an AsiA-modified polymerase can weakly support transcription at T4 and E. coli promoters under some conditions (Orsini et al, 2001; Pal et al, 2003). However, transcription in these cases is dependent on long-term incubation periods in the presence of DNA or incubation with agents that promote protein/DNA interactions. An AsiA-modified holoenzyme, for example, can weakly express RNA from the T4 middle promoter PuvsX in the absence of MotA if the polymerase is incubated for extended periods at 37°C in the absence of NTPs (Pal et al, 2003). Similar observations have been made for the E. coli lacUV5 promoter in the presence of potassium glutamate (Orsini et al, 2001). In both cases, stabilizing polymerase interactions with DNA are key to promoting transcription by the AsiA-modified holoenzyme. At PuvsX, high temperatures, long incubation times and the absence of NTPs provide an opportunity for competing binding modes to come into play, permitting a small quantity of the modified polymerase to form a closed complex and isomerize into an open complex. For lacUV5 in glutamate, this process is accelerated by the stabilization of protein/DNA interactions attributable to glutamate (Leirmo et al, 1987; Record et al, 1998). This does not imply, however, that −35 binding by SR4/AsiA is partially restored in glutamate or by long incubation times with DNA. The DNaseI footprint of the AsiA-modified holoenzyme on lacUV5 displays a loss of protection at −35 (Orsini et al, 2001), consistent with the structure of AsiA/EcSR4, which implies that no typical DNA binding is possible by the remodeled SR4 domain. The footprint of the AsiA-modified holoenzyme mirrors that observed for an AsiA-modified polymerase at galP1, a promoter which is not dependent on −35 DNA binding (Minakhin et al, 2003). Thus, bypass of the effects of AsiA is not inconsistent with obstruction of DNA binding by an AsiA/SR4 complex in the holoenzyme. Rather, footprinting suggests that the α-CTD (Orsini et al, 2001) may instead compensate for the loss of SR4/DNA interaction to aid in stabilizing the modified enzyme on DNA.
The dramatic structural rearrangements in EcSR4 seen in the complex may also explain why AsiA engages free σ70 rather than σ70 pre-associated with core polymerase (Hinton and Vuthoori, 2000). Since AsiA's interaction surface on EcSR4 is coincident with the core polymerase-binding surface (Vassylyev et al, 2002), it is logical that AsiA would prefer to bind free σ70. AsiA's binding site would be unobstructed in free σ70, but would require competing with core polymerase-binding sites in a pre-formed holoenzyme, something AsiA is inefficient at doing (Hinton and Vuthoori, 2000; Camarero et al, 2002). This implies that engagement of a preformed AsiA/σ70 complex with core polymerase should also result in an altered conformation for the RNAP holoenzyme, where the position of AsiA/SR4 on the surface of the enzyme is changed. This has recently been shown to be the case by measuring the distance between regions 2 and 4 in an AsiA-modified holoenzyme, which is closed by ≈10 Å (Simeonov et al, 2003).
The recent structural analyses of the Rse-A/σE and σ28/FlgM complexes suggested that one mechanism by which anti-σ factors inhibit RNAP is by physically blocking association of σ factor with core RNAP (Campbell et al, 2003; Sorenson et al, 2004). Rse-A ties up the σ factor into a condensed conformation, disabling it from participating in transcription until a stimulus dislodges anti-σ from the complex. The remodeling of SR4 by AsiA illustrates another mechanism by which an anti-σ factor could inhibit RNAP. AsiA remodels, rather than obstructs, the C-terminal DNA-binding domain of σ70. In this case, however, the purpose is not merely to inhibit RNAP, rather, AsiA also redirects the polymerase to phage genes; in a way, AsiA imposes a promoter choice on the enzyme.
Can the structural transformation observed for an isolated EcSR4 occur in the context of the holoenzyme? Limited evidence suggests that it is possible. Simeonov et al (2003) observed that the distance between conserved regions 2 and 4 is significantly reduced in an AsiA-modified RNAP. This is consistent with the notion that AsiA's interaction with EcSR4 blocks core polymerase-binding sites, forcing the AsiA/SR4 complex to be oriented differently on the surface of the enzyme. A change in the conformation of TmSR4 was also noted in a region 2–4 construct of σA when mixed with AsiA (Camarero et al, 2002), although TmSR4 does not form as stable a complex with TmSR4 as it does with EcSR4 (LJ Lambert and MH Werner, unpublished). The key question to be asked is what the implications are of the remodeled SR4 on activation of phage genes by MotA and whether there is a general role for remodeling in enforcing promoter choices upon RNAP.
Materials and methods
Proteins
EcSR4 and AsiA were expressed and purified as previously described (Lambert et al, 2001; 2003). cDNA for TmSR4 was inserted into the pQE-9 vector (Qiagen), overexpressed in HMS174 cells and purified by a combination of phosphocellulose and gel filtration chromatography in Tris–HCl/NaCl, pH 7.5. Complexes of EcSR4 with AsiA were prepared as previously described (Lambert et al, 2001; 2003). Monomeric AsiA was dialyzed into 10 mM sodium phosphate (pH 6.2), 50 mM NaCl, 1 mM benzamidine-HCl, and concentrated to 0.6–0.7 mM. For NMR analysis, the monomer was analyzed in the presence and/or absence of 2 mM CHAPS, which appeared to improve the spectral properties of the monomeric protein.
NMR spectroscopy
All NMR experiments were acquired at 36°C on either a Bruker DMX500, DMX600, or AVANCE 800 spectrometer equipped with a z-shielded gradient triple-resonance probe or a triple-resonance cryoprobe (at 600 and 800 MHz). Neither backbone nor side-chain resonances for AsiA residues 72–75 and residue 85 could be observed in AsiA bound to EcSR4. In TmSR4, the signals for residues 379–383 were not observed. 3JNH coupling constants were measured by quantitative J correlation spectroscopy (Vuister et al, 1999). NOEs involving protons of the protein were obtained from 3D 15N-separated or 3D 13C-separated NOE spectra (mixing times of 120 and 200 ms). Intermolecular restraints were identified from F1-edited, F2-filtered NOESY employing 13C–15N-SR4/12C–14N-AsiA, and verified from a NOESY spectrum in which the labeling pattern was reversed. RDCs for AsiA (monomer and complex) and EcSR4 (complex) were measured in the NMR sample buffer to which the filamentous phage Pf1 was added to a final concentration of ≈9 mg/ml. N–H, Cα–H and Cα–C′ couplings were measured as described (Bax et al, 2001). Initial values for the axial component of the dipolar coupling tensor and for the rhombicity were determined from a normalized histogram of the RDCs (Clore et al, 1998).
Structure calculations
The structures were calculated with the program X-PLOR-NIH 2.91 employing IVM dynamics (Schwieters and Clore, 2001), as adapted to incorporate pseudo-potentials for 3JNHα coupling constants (Garrett et al, 1994), secondary 13Cα and 13Cβ chemical shifts (Kuszewski et al, 1995), a conformational database potential for dihedral angles (Kuszewski and Clore, 2000) and a harmonic potential for the refinement against RDCs (Tjandra et al, 1997). Initial refinement began from an extended polypeptide chain for each protein and the fold established with an unambiguous set of NOE, φ, NH RDCs and/or CαH RDCs employing a torsion angle restraint protocol provided in the XPLOR-NIH release. The conformation of each molecule was independently established by this approach and did not rely on the interaction with a partner protein. Iterative refinement was performed by systematic analysis of NOE spectra with structural models and chemical shifts until >95% of the observable NOEs had been incorporated into the refinement. For the AsiA/EcSR4 complex, the proteins were manually docked on the basis of 50 intermolecular NOEs involving both subdomains of the complex. RDCs were included throughout with the force constant set to a final value of 0.5 or 1.0 as previously described (Tjandra et al, 1997). The agreement between expected and calculated RDC values was determined by SVD analysis with the program DC provided in the nmrPipe spectral analysis package. The final ensemble of 23 TmSR4 conformers, 20 AsiA monomer conformers and 20 EcSR4/AsiA conformers was selected on the basis of lowest energy and least number of restraint violations (TmSR4, AsiA monomer, AsiA/EcSR4) and the lowest r.m.s. difference between calculated and observed RDCs (AsiA monomer, AsiA/EcSR4); these structures had no distance restraint violations >0.5 Å and no dihedral angle violations >5°. AsiA monomer and AsiA/EcSR4 were further refined with explicit waters as described by Spronk et al (2002). Structure quality was assessed with PROCHECK and PROCHECK_NMR (Laskowski et al, 1996). Structures were displayed and analyzed using the program PYMOL (DeLano 2002).
PDB codes
The PDB accession codes are 1TLG for the structure of the T4 AsiA monomer, 1TLH for the T4 AsiA/sigma70 complex and 1TTY for the T. maritima region 4.
Supplementary Material
Supplementary Data
Acknowledgments
We are grateful to Alicia Dombroski, Richard Ebright and Debbie Hinton for many useful discussions during the course of this work. This work was supported by NIH grant GM63793 to MHW and NSF grant DBI-9974819 to BD. MHW is a Distinguished Young Scholar of the WM Keck Foundation.
References
- Adelman K, Brody EN, Buckle M (1998) Stimulation of bacteriophage T4 middle transcription by the T4 proteins MotA and AsiA occurs at two distinct steps in the transcription cycle. Proc Natl Acad Sci USA 95: 15247–15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Orsini G, Kolb A, Graziani L, Brody EN (1997) The interaction between the AsiA protein of bacteriophage T4 and the σ70 subunit of Escherichia coli RNA polymerase. J Biol Chem 272: 27433–27435 [DOI] [PubMed] [Google Scholar]
- Bax A, Kontaxis G, Tjandra N (2001) Dipolar couplings in macromolecular structure determination. Methods Enzymol 339: 127–174 [DOI] [PubMed] [Google Scholar]
- Brody EN, Kassavetis GA, Ouhammouch M, Sanders GM, Tinker RL, Geiduschek EP (1995) Old phage, new insights: two recently recognized mechanisms of transcriptional regulation in bacteriophage T4 development. FEMS Microbiol Lett 128: 1–8 [DOI] [PubMed] [Google Scholar]
- Camarero JA, Shekhtman A, Campbell EA, Chlenov M, Gruber TA, Bryant DA, Darst SA, Cowburn D, Muir TW (2002) Autoregulation of a bacterial σ factor explored by using segmental isotopic labeling and NMR. Proc Natl Acad Sci USA 99: 8536–8541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA (2002) Structure of the bacterial RNA polymerase promoter specificity subunit. Mol Cell 9: 527–539 [DOI] [PubMed] [Google Scholar]
- Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp SS, Gross CA, Darst SA (2003) Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-σ-RseA. Mol Cell 11: 1067–1078 [DOI] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM, Bax A (1998) A robust method for determining the magnitude of the fully asymmetric alignment tensor of oriented macromolecules in the absence of structural information. J Magn Reson 133: 216–221 [DOI] [PubMed] [Google Scholar]
- Colland F, Orsini G, Brody EN, Buc H, Kolb A (1998) The bacteriophage T4 AsiA protein: a molecular switch for σ70-dependent promoters. Mol Microbiol 27: 819–829 [DOI] [PubMed] [Google Scholar]
- Darst SA, Opalka N, Chacon P, Polyakov A, Richter C, Zhang G, Wriggers W (2002) Conformational flexibility of bacterial RNA polymerase. Proc Natl Acad Sci USA 99: 4296–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System http://www.pymol.org
- Dombroski AJ, Walter WA, Gross CA (1993) Amino-terminal amino acids modulate σ-factor DNA-binding activity. Genes Dev 7: 2446–2455 [DOI] [PubMed] [Google Scholar]
- Dombroski AJ, Walter WA, Record MT Jr, Siegele DA, Gross CA (1992) Polypeptides containing highly conserved regions of transcription initiation factor 70 exhibit specificity of binding to promoter DNA. Cell 70: 501–512 [DOI] [PubMed] [Google Scholar]
- Garrett DS, Kuszewski J, Hancock TJ, Lodi PJ, Vuister GW, Gronenborn AM, Clore GM (1994) The impact of direct refinement against three-bond HN-CαH coupling constants on protein structure determination by NMR. J Magn Reson B 104: 99–103 [DOI] [PubMed] [Google Scholar]
- Helmann JD, Chamberlin MJ (1988) Structure and function of bacterial σ factors. Annu Rev Biochem 57: 839–872 [DOI] [PubMed] [Google Scholar]
- Hinton DM (1991) Transcription from a bacteriophage T4 middle promoter using T4 motA protein and phage modified RNA polymerase. J Biol Chem 266: 18034–18044 [PubMed] [Google Scholar]
- Hinton DM, March-Amegadzie R, Gerber JS, Sharma M (1996) Characterization of pre-transcription complexes made at the bacteriophage T4 middle promoter: involvement of the T4 MotA activator and the T4 AsiA protein, a σ70 binding protein, in the formation of the open complex. J Mol Biol 256: 235–248 [DOI] [PubMed] [Google Scholar]
- Hinton DM, Vuthoori S (2000) Efficient inhibition of Escherichia coli RNA polymerase by the bacteriophage T4 AsiA protein requires that AsiA binds first to free σ70. J Mol Biol 304: 731–739 [DOI] [PubMed] [Google Scholar]
- Hughes KT, Mathee K (1998) The anti-σ factors. Annu Rev Microbiol 52: 231–286 [DOI] [PubMed] [Google Scholar]
- Karam JD (ed) (1994) The Molecular Biology of Bacteriophage T4. Washington, DC: American Society for Microbiology Press [Google Scholar]
- Kolesky S, Ouhammouch M, Brody EN, Geiduschek EP (1999) ∑ Competition: the contest between bacteriophage T4 middle and late transcription. J Mol Biol 291: 267–281 [DOI] [PubMed] [Google Scholar]
- Kuszewski J, Clore GM (2000) Sources of and solutions to problems in the refinement of protein NMR structures against torsion angle potentials of mean force. J Magn Reson 146: 249–254 [DOI] [PubMed] [Google Scholar]
- Kuszewski J, Qin J, Gronenborn AM, Clore GM (1995) The impact of direct refinement against 13Cα and 13Cβ chemical shifts on protein structure determination by NMR. J Magn Reson B 106: 92–96 [DOI] [PubMed] [Google Scholar]
- Lambert LJ, Schirf V, Demeler B, Cadene M, Werner MH (2001) Flipping a genetic switch by subunit exchange. EMBO J 20: 7149–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert LJ, Schirf V, Demeler B, Werner MH (2003) Molecular analysis of activator engagement with RNA polymerase. Methods Enzymol 370: 505–521 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK_NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Leirmo S, Harrison C, Cayley DS, Burgess RR, Record MT Jr (1987) Replacement of potassium chloride by potassium glutamate dramatically enhances protein–DNA interactions in vitro. Biochemistry 26: 2095–2210 [DOI] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA (1992) The σ70 family: sequence conservation and evolutionary relationships. J Bacteriol 174: 3843–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakhin L, Camarero JA, Holford M, Parker C, Muir TW, Severinov K (2001) Mapping the molecular interface between the σ70 subunit of E. coli RNA polymerase and T4 AsiA. J Mol Biol 306: 631–642 [DOI] [PubMed] [Google Scholar]
- Minakhin L, Niedziela-Majka A, Kuznedelov K, Adelman K, Urbauer JL, Heyduk T, Severinov K (2003) Interaction of T4 AsiA with its target sites in the RNA polymerase σ70 subunit leads to distinct and opposite effects on transcription. J Mol Biol 326: 679–690 [DOI] [PubMed] [Google Scholar]
- Mosig G, Hall DH (1994) Gene expression: a paradigm of integrated circuits. In Molecular Biology of Bacteriophage T4, Karam JD (ed), pp 127–131. Washington, DC: American Society for Microbiology Press [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002) Structural basis of transcription initiation: an RNA polymerase holoenzyme–DNA complex. Science 296: 1285–1290 [DOI] [PubMed] [Google Scholar]
- Orsini G, Kolb A, Buc H (2001) The Escherichia coli RNA polymerase·anti-70 AsiA complex utilizes -carboxyl-terminal domain upstream promoter contacts to transcribe from a −10/−35 promoter. J Biol Chem 276: 19812–19819 [DOI] [PubMed] [Google Scholar]
- Orsini G, Ouhammouch M, Le Caer JP, Brody EN (1993) The asiA gene of bacteriophage T4 codes for the anti-σ70 protein. J Bacteriol 175: 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouhammouch M, Adelman K, Harvey SR, Orsini G, Brody EN (1995) Bacteriophage T4 MotA and AsiA proteins suffice to direct Escherichia coli RNA polymerase to initiate transcription at T4 middle promoters. Proc Natl Acad Sci USA 92: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouhammouch M, Orsini G, Brody EN (1994) The asiA gene product of bacteriophage T4 is required for middle mode RNA synthesis. J Bacteriol 176: 3956–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahari S, Chatterji D (1997) Interaction of bacteriophage T4 AsiA protein with Escherichia coli 70 and its variant. FEBS Lett 411: 60–62 [DOI] [PubMed] [Google Scholar]
- Pal D, Vuthoori M, Pande S, Wheeler D, Hinton DM (2003) Analysis of regions within the bacteriophage T4 AsiA protein involved in its binding to the 70 subunit of E. coli RNA polymerase and its role as a transcriptional inhibitor and co-activator. J Mol Biol 325: 827–841 [DOI] [PubMed] [Google Scholar]
- Pande S, Makela A, Dove SL, Nickels BE, Hochschild A, Hinton DM (2002) The bacteriophage T4 transcription activator MotA interacts with the far-C-terminal region of the 70 subunit of Escherichia coli RNA polymerase. J Bact 184: 3957–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record MT Jr, Courtenay ES, Cayley S, Guttman HJ (1998) Biophysical compensation mechanisms buffering E. coli protein-nucleic acid interactions against changing environments. Trends Biochem Sci 23: 190–194 [DOI] [PubMed] [Google Scholar]
- Schmidt RP, Kreuzer KN (1992) Purified motA protein binds the −30 region of a bacteriophage T4 middle-mode promoter and activates transcription in vitro. J Biol Chem 267: 11399–11407 [PubMed] [Google Scholar]
- Schwieters CD, Clore GM (2001) Internal coordinates for molecular dynamics and minimization in structure determination and refinement. J Magn Reson 152: 288–302 [DOI] [PubMed] [Google Scholar]
- Severinova E, Severinov K, Darst SA (1998) Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J Mol Biol 279: 9–18 [DOI] [PubMed] [Google Scholar]
- Severinova E, Severinov K, Fenyo D, Marr M, Brody EN, Roberts JW, Chait BT, Darst SA (1996) Domain organization of the Escherichia coli RNA polymerase 70 subunit. J Mol Biol 263: 637–647 [DOI] [PubMed] [Google Scholar]
- Sharma M, Marshall P, Hinton DM (1999) Binding of the bacteriophage T4 transcriptional activator, MotA, to T4 middle promoter DNA: evidence for both major and minor groove contacts. J Mol Biol 290: 905–915 [DOI] [PubMed] [Google Scholar]
- Sharma UK, Praveen PVK, Balganesh TS (2002) Mutational analysis of bacteriophage T4 AsiA: involvement of N- and C-terminal regions in binding to 70 of Escherichia coli in vivo. Gene 295: 125–134 [DOI] [PubMed] [Google Scholar]
- Simeonov MN, Urbauer RJB, Gilmore JM, Adelman KN, Brody EN, Niedziela-Majka A, Minakhin L, Heyduk T, Urbauer JL (2003) Characterization of the interactions between the bacteriophage T4 AsiA protein and RNA polymerase. Biochemistry 42: 7717–7726 [DOI] [PubMed] [Google Scholar]
- Sorenson MK, Ray SS, Darst SA (2004) Crystal structure of the flagellar σ/anti-σ complex σ28/FlgM reveals an intact σ factor in an inactive conformation. Mol Cell 14: 127–138 [DOI] [PubMed] [Google Scholar]
- Spronk CAEM, Linge JP, Hilbers CW, Vuister GW (2002) Improving the quality of protein structures derived by NMR spectroscopy. J Biomol NMR 22: 281–289 [DOI] [PubMed] [Google Scholar]
- Stevens A (1977) Inhibition of DNA-enzyme binding by an RNA polymerase inhibitor from T4 phage-infected Escherichia coli. Biochim Biophys Acta 475: 193–196 [DOI] [PubMed] [Google Scholar]
- Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A (1997) Use of dipolar 1H–15N and 1H–13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nat Struct Biol 4: 732–738 [DOI] [PubMed] [Google Scholar]
- Urbauer JL, Adelman K, Urbauer RJB, Simeonov MF, Gilmore JM, Zolkiewski M, Brody EN (2001) Conserved regions 4.1 and 4.2 of 70 constitute the recognition sites for the anti-factor AsiA, and AsiA is a dimer free in solution. J Biol Chem 276: 41128–41132 [DOI] [PubMed] [Google Scholar]
- Urbauer JL, Simeonov MF, Urbauer RJB, Adelman K, Gilmore JM, Brody EN (2002) Solution structure and stability of the anti-σ factor AsiA: implications for novel functions. Proc Natl Acad Sci USA 99: 1831–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S (2002) Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417: 712–719 [DOI] [PubMed] [Google Scholar]
- Vuister GW, Tessari M, Karini-Nejad Y, Whitehead B (1999) Pulse sequences for measuring coupling constants. In Biological Magnetic Resonance 16, Krishna NR, Berliner LJ (eds), pp 195–259. New York, NY, USA: Kluwer Press [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data