Abstract
Correction to: The EMBO Journal (2001) 20, 7149–7159. doi:10.1093/emboj/20.24.7149
Two different structures of the anti-σ factor T4 AsiA in its dimeric state have been reported, one by us and a second by Urbauer et al (2002). The principal distinction between these structures was in the monomer fold, which displayed an approximate mirror image relationship to one another. This difference prompted a re-examination of our AsiA structure in solution, leading us to conclude that our structure of the T4 AsiA dimer was incorrectly determined in the original report. The resolution of this discrepancy was driven by the solution of the AsiA structure in two new states: a free monomer and a monomer bound to conserved region 4 from Escherichia coli σ70 (EcSR4) (see report by Lambert et al in this issue (The EMBO Journal (2004) 23, 2952–2962). In each of these states, the chemical shifts for all atoms were similar to each other and very different from those observed in AsiA dimer, enabling a completely independent effort at the solution of the AsiA structure. The fold of AsiA determined in each of these new states was found to be very similar to that reported by Urbauer et al. Most convincing in this analysis were the 220 intermolecular NOEs observed between AsiA and EcSR4, which were only consistent with the fold reported by Urbauer et al. Both monomer states were further refined by residual dipolar coupling (RDC) analysis. We subsequently reanalyzed our original AsiA dimer NMR spectra and collected RDCs on the original dimer. No errors in chemical shift assignment were found and less than 4% of all NOEs needed to be reassigned for the fold of AsiA dimer to conform to the fold reported by Urbauer et al. This represented less than 3% of those NOEs between nonsequential residues in the polypeptide chain (17 NOEs in all); these NOEs were not previously appreciated to be ambiguous with respect to a mirror image fold. Figure 1 displays the revised structure and compares it to that of Urbauer et al. The coordinates of our structure with PDB accession code 1KA3 have been replaced. The PDB ID for the corrected AsiA dimer is 1TKV.
Figure 1.
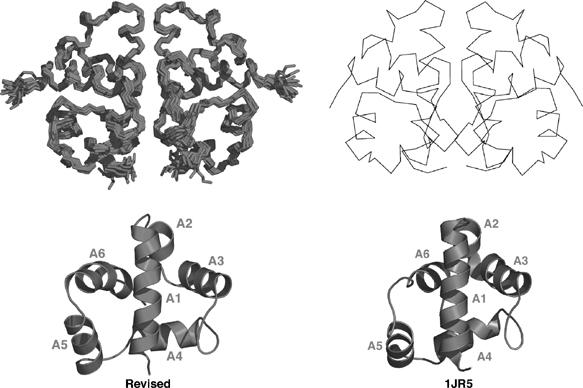
Comparison of recalculated structure of AsiA dimer to the structure of Urbauer et al (PDB accession code 1JR5). On the left is the recalculated structure family with a ribbon representation of the monomer shown at the bottom. On the right is the Cα trace of one model from 1JR5 and a ribbon representation at the bottom. The two structures are very similar to one another, with the exception of the position of helix A6. In 1JR5, the C-terminus is tucked against the back face of helices A1 and A4, while in our revised structures these restraints are not observed. Helix A6 in 1JR5 is also one turn shorter than that observed by us. Despite these differences, the two structures are in overall agreement.
References
- Urbauer JL, Simeonov MF, Urbauer RJB, Adelman K, Gilmore JM, Brody EN (2002) Solution structure and stability of the anti-σ factor AsiA: implications for novel functions. Proc Natl Acad Sci USA 99: 1831–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]


