Abstract
Breakdown of triple-helical interstitial collagens is essential in embryonic development, organ morphogenesis and tissue remodelling and repair. Aberrant collagenolysis may result in diseases such as arthritis, cancer, atherosclerosis, aneurysm and fibrosis. In vertebrates, it is initiated by collagenases belonging to the matrix metalloproteinase (MMP) family. The three-dimensional structure of a prototypic collagenase, MMP-1, indicates that the substrate-binding site of the enzyme is too narrow to accommodate triple-helical collagen. Here we report that collagenases bind and locally unwind the triple-helical structure before hydrolyzing the peptide bonds. Mutation of the catalytically essential residue Glu200 of MMP-1 to Ala resulted in a catalytically inactive enzyme, but in its presence noncollagenolytic proteinases digested collagen into typical 3/4 and 1/4 fragments, indicating that the MMP-1(E200A) mutant unwinds the triple-helical collagen. The study also shows that MMP-1 preferentially interacts with the α2(I) chain of type I collagen and cleaves the three α chains in succession. Our results throw light on the basic mechanisms that control a wide range of biological and pathological processes associated with tissue remodelling.
Keywords: collagenase, enzyme mechanism, matrix metalloproteinase, protein unfolding, triple helix
Introduction
Collagens are the major structural proteins of connective tissues such as skin, tendon, bone, cartilage, blood vessels and basement membranes. Interstitial collagens I, II and III are the most abundant and they provide the scaffolding of the tissue and guide cells to migrate, proliferate and differentiate. The degradation of these macromolecules is therefore an integral part of many biological processes such as embryogenesis, organ morphogenesis, tissue remodelling, angiogenesis and wound healing (Cawston, 1996; Woessner, 1998; Sternlicht and Werb, 2001). Recent studies have shown that collagenase-cleaved products of collagen I alter cellular activity by expressing cryptic biological functions: for example, activation and recruitment of osteoclasts during bone remodelling (Zhao et al, 1999), epithelial cell migration during wound healing (Pilcher et al, 1997) and apoptosis of amniotic fibroblasts at the term pregnancy (Lei et al, 1996). Thus, collagenase does not simply function to degrade and remove collagen fibrils but also controls cellular behavior during tissue remodelling. Aberrant collagenolysis, on the other hand, is associated with progression of diseases such as arthritis, cancer, atherosclerosis, aneurysm and fibrosis (Woessner, 1998; Brinckerhoff and Matrisian, 2002).
Interstitial collagens consist of three α chains of approximately 1000 residues with repeating Gly–X–Y triplets, where X and Y are often proline and hydroxyproline, respectively. Because of the high imino-acid content and the tripeptide unit repeats, the α chain adopts a left-handed poly-Pro II-like helix, and three left-handed α chains intertwine with each other to form a right-handed superhelix (Ramachandran and Kartha, 1955; Rich and Crick, 1961; Fraser et al, 1979; Kramer et al, 2001). This triple-helical conformation makes interstitial collagens resistant to most proteinases. In vertebrates, enzymes that can cleave the triple-helical structure are collagenases (Visse and Nagase, 2003) and cathepsin K produced by osteoclasts (Garnero et al, 1998). While cathepsin K cleaves collagen I in an acidic environment specialized primarily in bone resorption, all other collagenolytic enzymes act at neutral pH and are members of the matrix metalloproteinase (MMP) family, and are produced by many cell types including stromal cells, epithelial cells, macrophages and leukocytes (Sternlicht and Werb, 2001). The latter group includes collagenases (MMP-1, MMP-8, MMP-13 and MMP-18), MMP-2 (gelatinase A) (Aimes and Quigley, 1995; Patterson et al, 2001) and membrane-type 1-MMP (MMP-14) (Ohuchi et al, 1997). These MMPs cleave the three α chains of native triple-helical type I, II and III collagens after Gly in a particular sequence (Gln/Leu)–Gly#(Ile/Leu)–(Ala/Leu) (# indicates the bond cleaved) located approximately three quarters away from the N-terminus of the collagen molecule. The action of these enzymes is critical for the initiation of collagen breakdown, as once collagens are cleaved into 3/4 and 1/4 fragments they denature at body temperature and are degraded by gelatinases and other nonspecific tissue proteinases.
A typical collagenase is synthesized as a pre-proenzyme and secreted as an inactive proenzyme consisting of a propeptide, a catalytic domain, a short linker region rich in proline and a C-terminal hemopexin (Hpx) domain. Clark and Cawston (1989) first reported that the cleavage of triple-helical collagen by MMP-1 (collagenase 1) requires the C-terminal Hpx domain. The catalytic domain alone retains proteolytic activities on noncollageneous proteins and peptides, but it fails to cleave collagen (Clark and Cawston, 1989; Murphy et al, 1992). The importance of the Hpx domain was also shown for other collagenolytic MMPs such as MMP-2 (Patterson et al, 2001), MMP-8 (Knäuper et al, 1993), MMP-13 (Knäuper et al, 1997) and MMP-14 (Ohuchi et al, 1997). Nevertheless, structurally similar MMP-3 (stromelysin 1) and MMP-10 (stromelysin 2) are unable to cleave collagen I or II (Woessner and Nagase, 2000). Replacement of the Hpx domain in MMP-3 with the Hpx of MMP-1 did not make MMP-3 collagenolytic, indicating that the Hpx is not the sole component responsible for the expression of collagenolytic activity (Murphy et al, 1992). The crystal structure of porcine MMP-1 indicated that the catalytic domain and the Hpx domain are tandemly connected through the linker peptide (Li et al, 1995), but understanding how the Hpx domain assists in the cleavage of collagen is elusive. Therefore, the structural basis for collagen-degrading specificity among certain members of MMPs is not clearly understood. An additional enigma is the mechanism by which collagenases cleave triple-helical collagens when the dimensions of the collagenase active site and the structure of interstitial collagens are considered (Bode, 1995). The substrate-binding site of MMP-1 forms a deep cleft with the catalytic zinc located at the bottom, and the entrance of this groove is only 5 Å wide, sufficient to accommodate only a single polypeptide chain. Type I collagen, on the other hand, consisting of two α1(I) chains and one α2(II) chain, is 3000 Å in length and 15 Å in diameter. Thus, the triple-helical collagen does not fit into the active site cleft of the enzyme. Our molecular docking attempts to place the triple-helical model of Kramer et al (2001) to the crystal structure of porcine MMP-1, indicated that the closest susceptible peptide bond is at least 7 Å away from the catalytic zinc atom (Figure 1). In addition, because each α chain forms a poly-Pro II-like helix, the spatial orientation of side chains in a single α chain is unique and dissimilar to that of a single-peptide chain substrate that fits in the substrate-binding cleft by forming β strand-like hydrogen bonds. This means that either the active site of MMP-1 undergoes large conformational changes or that the triple-helical collagen needs to be unwound so that a single α chain can fit into the active site of the enzyme.
Figure 1.
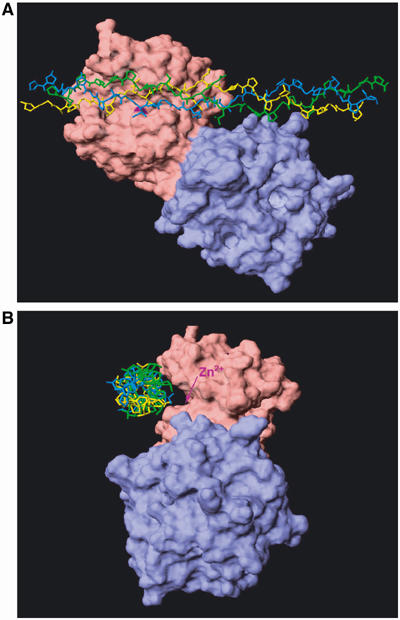
Alignment of the triple-helical peptide with the active site of MMP-1. (A) Collagen triple-helical peptides described by Kramer et al (2001) were manually aligned into the active site of the catalytic domain of porcine MMP-1 determined by Li et al (1995) using Insight II/Discover and the image was produced with Swiss PDB view (Guex and Peitsch, 1997). (B) The alignment model of MMP-1 and the triple-helical peptides in (A) were rotated 90° to the left. The location of the catalytic Zn2+ is indicated by an arrow. The active site shown as a cleft is unoccupied by the triple-helical peptide substrate. Pink, catalytic domain; blue, Hpx; purple, zinc ion.
In this report, using collagenase 1 (MMP-1) as a typical collagenase, we present evidence that collagenase locally unwinds triple-helical collagen before it hydrolyzes the peptide bonds. To our knowledge, this is the first demonstration that a single polypeptide proteinase induces significant conformational changes of the protein substrate before it cleaves specific peptide bonds.
Results
Unwound collagen chains are substrates of collagenase 1 (MMP-1)
It is generally considered that the triple-helical structure of collagen is critical for collagenases to cleave interstitial collagens (McCroskery et al, 1973). Based on the crystal structure of MMPs and our molecular modelling with a triple-helical peptide, we hypothesized that unwound collagen α chains, but not those in a triple-helical conformation, are the substrates of collagenases. We examined this possibility first by comparing the ability of MMP-1 to cleave native collagen I and heat-denatured collagen I (gelatin I) at different temperatures. At 37°C, collagen I was readily cleaved into the typical three-quarter and one-quarter fragments (Figure 2A). Gelatin I was also cleaved in a similar manner, but was less susceptible. In contrast, when the reaction temperature was reduced, gelatin I was more susceptible to cleavage than collagen I: At 10 and 4°C little collagen I hydrolysis was observed, but the activity for gelatin I was retained (Figure 2C and D). NH2-terminal sequencing of the 1/4 (TCB) fragments generated from gelatin I by MMP-1 indicated that enzyme cleaved the Gly775–Ile776 bond of the α1(I) chain and the Gly775–Leu776 bond of the α2(I) chain, sites identical to those cleaved in native collagen I. These results indicate that the conformational state of the substrate significantly influences the activity of MMP-1 and that unwound collagen is a better substrate at temperatures lower than 25°C, conditions that decrease the backbone mobility of the triple-helical structure. Based on these observations, we considered that unwinding of the triple helices may be a prerequisite for collagenase to cleave interstitial collagens.
Figure 2.

Digestion of collagen I and gelatin I by MMP-1. Collagen I (30 μg) and gelatin I (30 μg) were incubated with 6 nM (A, B) or 40 nM (C, D) active MMP-1 at various temperatures for up to 2 h. The reaction was terminated by addition of 20 mM EDTA and subjected to SDS–PAGE under reducing conditions. TCA and TCB are 3/4 fragments and 1/4 fragments of α1(1) and α2(1) chains, respectively.
Demonstration of collagen unwinding by MMP-1
If MMP-1 has an ability to unwind collagen, such activity might be separated from the activity that hydrolyzes peptide bonds. To investigate this possibility, we mutated Glu200, the residue essential for peptide hydrolysis, to Ala. We postulated that such a mutant would locally unwind collagen upon interaction with collagen, but would not cleave peptide bonds, and that the unwound collagen would then be susceptible to cleavage by a noncollagenolytic enzyme.
As shown in Figure 3A, the MMP-1(E200A) mutant was essentially inactive and unable to cleave the α1(I) and α2(I) chains of collagen I. As demonstrated previously (Clark and Cawston, 1989; Murphy et al, 1992), the catalytic domain of MMP-1 lacking the C-terminal Hpx domain (MMP-1(ΔC)) also could not cleave collagen I, even at high concentrations of the enzyme, whereas it readily cleaved gelatin I (Figure 3B and C). However, when collagen I was incubated with MMP-1(E200A) and MMP-1(ΔC) at 25°C, it was cleaved into the typical 3/4 (TCA) and 1/4 (TCB) fragments in a manner dependent on the concentration of MMP-1(ΔC) (Figure 3D) and MMP-1(E200A) (data not shown). MMP-3(ΔC) lacking the Hpx domain, full-length MMP-3 (data not shown) and human leukocyte elastase (HLE), all of which are unable to cleave the triple-helical region of collagen I, also generated 3/4 and 1/4 fragments in the presence of MMP-1(E200A) (Figure 4). In the case of HLE, most MMP-1(E200A) was split into catalytic and Hpx domains during incubation, but it retained unwinding activity. NH2-terminal sequencing of the TCB fragments indicated that MMP-1(ΔC) and MMP-3 cleaved the Gly775–Ile776 bond of the α1(I) chain and the Gly775–Leu776 bond of the α2(I) chain in the presence of MMP-1(E200A) (Figure 5). HLE cleaved the Val783–Gly784 bond of the α1(I) chain in the same locus. The HLE cleavage site of α2(I) chain was not identified, but potential sites are found in the same region. Incubation of collagen I with inactive full-length MMP-3(E202A) and MMP-1(ΔC) did not result in collagen cleavage, indicating that MMP-3 lacks the ability to unwind collagen. From these results, we have concluded that MMP-1(E200A) unwinds the triple-helical collagen I upon binding.
Figure 3.
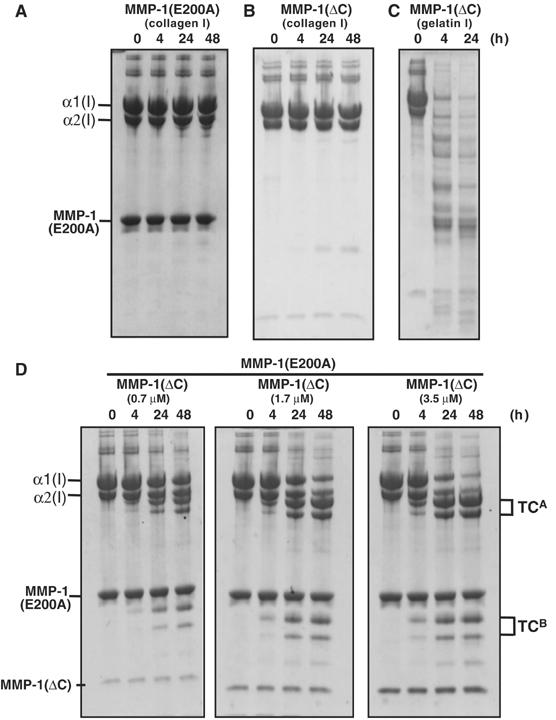
Digestion of collagen I in the presence of MMP-1(E200A) by MMP(ΔC). (A) Collagen I (30 μg) was incubated with 6 μM MMP-1(E200A) or (B) 0.7 μM MMP-1(ΔC) at 25°C for the indicated time. (C) Gelatin I (30 μg) was incubated with 0.7 μM MMP-1(ΔC). (D) Collagen I (30 μg) was made to react with an increasing amount of MMP-1(ΔC) in the presence of 6 μM MMP-1(E200A). The reaction products were analyzed as in Figure 2.
Figure 4.
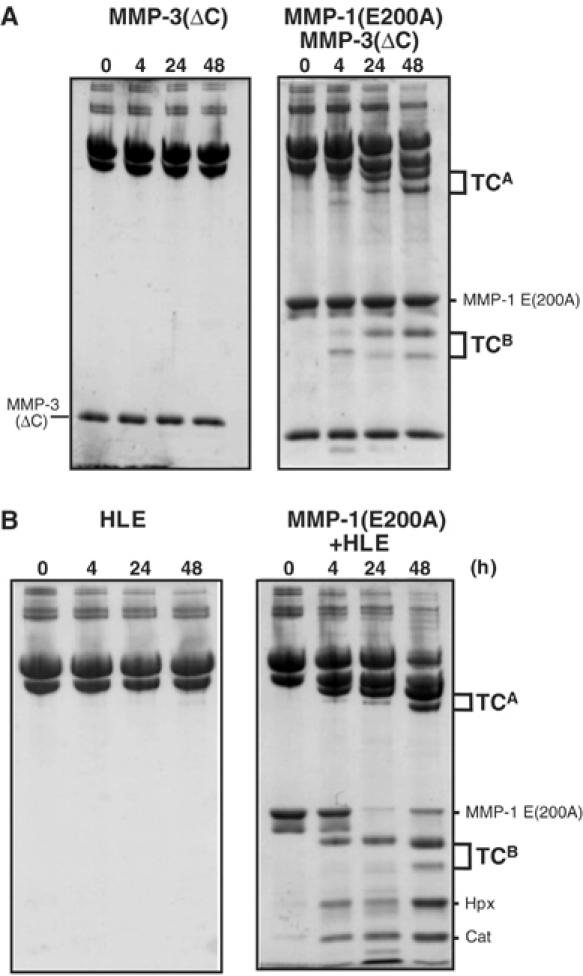
Digestion of collagen I by MMP-3(ΔC) or HLE in the presence of MMP-1(E200A). Collagen I (30 μg) was incubated with (A) 7.6 μM MMP-3(ΔC) or (B) 0.4 μM HLE in the presence of 6 μM MMP-1(E200A) at 25°C for up to 48 h. The reactions were terminated with 20 mM EDTA for MMP-3(ΔC) and 1 mM phenylmethylsulfonyl fluoride for HLE and the products were analyzed as in Figure 2.
Figure 5.

Cleavage sites in collagen I by MMP-1(ΔC), MMP-3(ΔC) and HLE detected in the presence of MMP-1(E200A). The N-terminal amino-acid sequence of the TCB fragments of α1(I) and α2(I) chain was determined as described in Materials and methods. In the case of HLE, only the α1(I) chain fragment was determined. The residues in the α2(I) chain indicated by brackets are the predicted cleavage sites of HLE based on enzyme specificity.
To determine the equilibrium- binding constant of MMP-1(E200A) with guinea-pig collagen I, various concentrations of MMP-1(E200A) (1–6 μM) were incubated with collagen I in the presence of a very high concentration of a ‘cutter' proteinase MMP-1(ΔC), under the conditions in which the rate of collagen cleavage was dependent on the concentration of MMP-1(E200A). This allowed us to determine that the KD of MMP-1(E200A) for collagen I was 1.6 μM. This value is similar to the Km reported by Welgus et al (1981) for human MMP-1 and guinea-pig collagen I. This indicates that the mode of MMP-1(E200A) interaction with collagen I is essentially the same as the native MMP-1.
Preferential interaction of MMP-1 with the α2(I) chain and the requirement of the active site for collagen unwinding
It was notable that the α1(I) chain was cleaved more rapidly by noncollagenolytic proteinases in the presence of MMP-1(E200A) compared with the active MMP-1 alone (compare Figures 3D and 4 with Figure 2A and B). This suggests that the ‘unwinder' MMP-1(E200A) preferentially interacts with the α2(I) chain, which renders the α1(I) chain more exposed and susceptible to a ‘cutter' proteinase.
When MMP-1(E200A) and collagen were first incubated with 10 μM GM6001X, an active site-directed synthetic MMP inhibitor, and then made to react with the serine protease HLE, no collagen digestion was detected (Figure 6). This indicates that the unwinding activity of MMP-1(E200A) requires an unoccupied active site which presumably accommodates the α2(I) chain.
Figure 6.
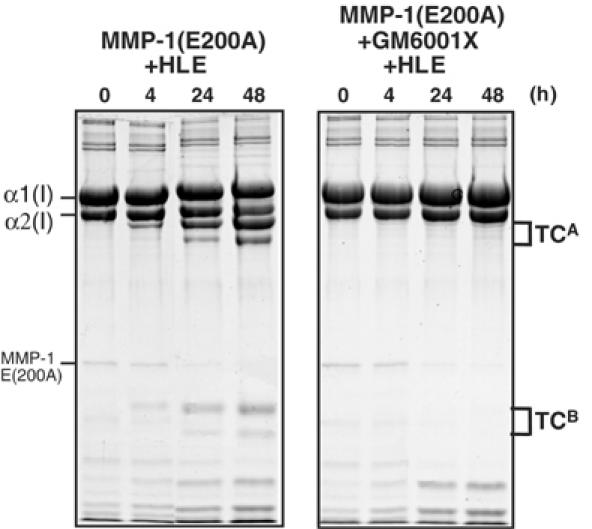
MMP-1(E200A) bound to GM6001X is unable to unwind collagen I. Collagen I (30 μg) was incubated with 2.5 μM MMP-1(E200A) and 0.4 μM HLE in the absence or presence of 10 μM GM6001X at 25°C for up to 48 h. The reaction of MMP-1(ΔC) and HLE was terminated by 1 mM phenylmethylsulfonyl fluoride and the products were analyzed as in Figure 4.
MMP-1 unwinds collagen I only locally
To examine the extent of collagen unwinding by MMP-1, we measured the melting temperature, Tm, of guinea-pig collagen I in the absence and presence of MMP-1(E200A) or MMP-3(E202A). MMP-3(E202A) served as control since it does not unwind collagen I. As shown in Figure 7, there was no significant change in Tm (∼40°C) even in the presence of MMP-1(E200A) or MMP-3(E202A). This suggests that the unwinding of collagen by MMP-1 takes place only locally, and it does not affect the overall triple-helical structure. To confirm this, we employed the property of chymotrypsin, which cleaves denatured collagens but not the native triple-helical collagens. As shown in Figure 8, chymotrypsin rapidly cleaved gelatin I into small fragments, but not collagen I. Incubation of collagen I with active MMP-1 at 25°C in the presence or absence of chymotrypsin generated the typical collagenase fragments, which are indistinguishable from those generated by collagenase alone. These results indicate that MMP-1 unwinds collagen only locally at the site where the collagenolytic cleavage takes place.
Figure 7.
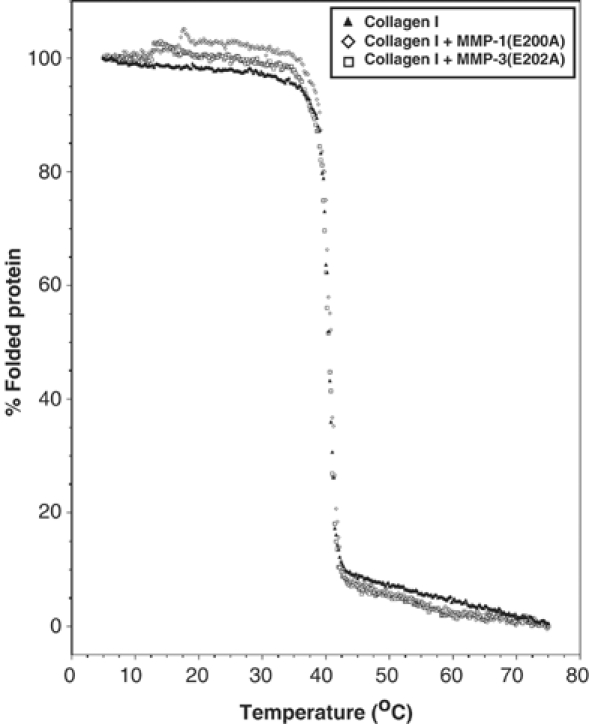
Thermal transition curve for guinea-pig type I collagen with or without MMP-1(E200A). Pepsin-treated guinea-pig type I collagen (3 μM) was incubated with or without an equimolar concentration of MMP-1(E200A) or MMP-3(E202A), and molar ellipticities were recorded at 222 nm while the temperature increased from 5 to 70°C at a rate of 35°C/h.
Figure 8.
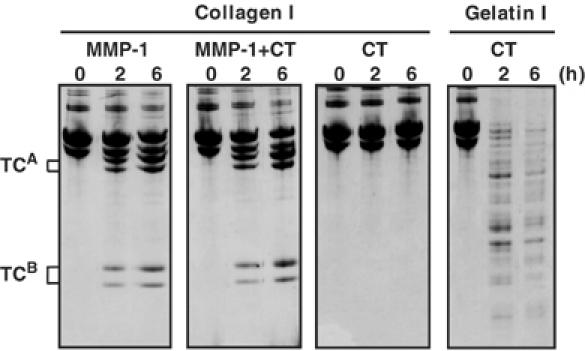
Unwinding of collagen by MMP-1 occurs only locally. Collagen I (30 μg) was incubated with 0.05 μM MMP-1 with or without chymotrypsin (CT) (20 μg/ml) at 25°C for up to 6 h. CT alone was incubated with collagen I and gelatin I as controls.
Reassociation of the catalytic domain and the Hpx domain of MMP-1 cleaves collagen I
The study with HLE showed that collagen-unwinding activity was retained even when MMP-1(E200A) was split into the catalytic domain and the Hpx domain (see Figure 4B), suggesting that the combination of MMP-1(ΔC) and the Hpx domain of MMP-1(HpxMMP-1) can unwind and cleave collagen. This was confirmed by adding MMP-1(ΔC) to the isolated HpxMMP-1 domain (data not shown). We then compared the collagen-unwinding ability of MMP-1(E200A) and HpxMMP-1 by incubating collagen I with various concentrations of an ‘unwinder' component (MMP-1(E200A) or HpxMMP-1) at various concentrations in the presence of a constant amount of a ‘cutter' MMP-1(ΔC). Figure 9 shows that 2 μM MMP-1(E200A) is approximately two-fold more effective than 4 μM HpxMMP-1. A similar level of collagenolysis was observed with only 0.01 μM of the full-length wild-type MMP-1 (data not shown). The combination of HpxMMP-1 and MMP-3(ΔC) did not cleave collagen I, suggesting that the correct pairing of the MMP-1 catalytic domain and the Hpx domain is essential for collagenolysis. The requirement of higher concentrations of the unwinder and the cutter to cleave collagen suggests that both components must simultaneously bind to the collagen substrate. In the case of HpxMMP-1 and MMP-1(ΔC), the ratio of the α1(I) to α2(I) chain cleavage products was similar to that of full-length MMP-1, suggesting that together they behave like a full-length collagenase most likely by associating with collagen in a similar manner. The high efficiency of the active full-length collagenase is due to the fact that the two essential elements are linked in a single molecule, thus benefiting from entropic contributions.
Figure 9.
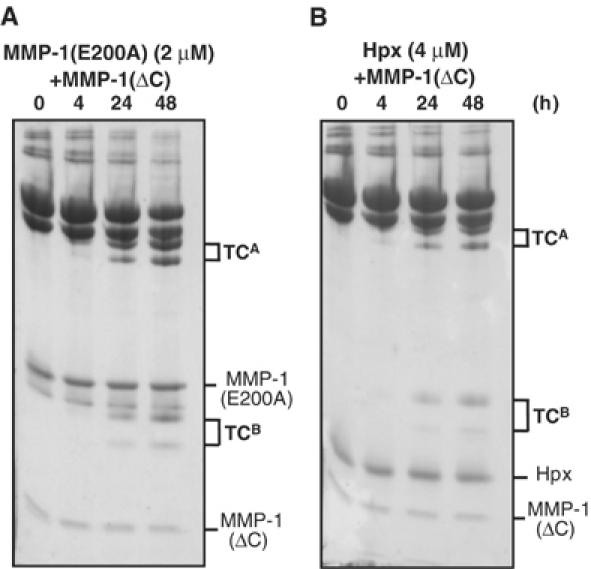
Collagenolytic activity expressed by reassociation of the catalytic domain and the Hpx domain. Collagen I (30 μg) was made to react with (A) 2μM MMP-1(E200A) and 0.8 μM MMP-1(ΔC), and (B) 4 μM HpxMMP-1 and 0.8 μM MMP-1(ΔC) at 25°C for the indicated period of time. The reactions were terminated with 20 mM EDTA and the products were analyzed as in Figure 2.
Cleavage of a single α chain during collagenolysis
We investigated the steps involved in collagenolysis by taking advantage of the unwinding ability of MMP-1(E200A) and the peptide hydrolysis activity of MMP-1(ΔC). We employed nonpepsin-treated collagen I, which retains β components consisting of two crosslinked α chains (α1(I)–α1(I) and α1(I)–α2(I) crosslinks in a 1:2 ratio) and γ components consisting of three crosslinked α chains (α1(I)–α1(I)–α2(I) crosslinks) through the noncollagenous telopeptide regions. Cleavage of one or two of these chains results in intermediate products containing cleaved α chains and intact α chains. When the nonpepsin-treated collagen I was incubated with MMP-1 and products were analyzed by SDS–PAGE with 5% total acrylamide, the crosslinked 3/4 fragments βA and γA were observed, and there were no obvious intermediates (Figure 10A). On the other hand, digestion with MMP-1(ΔC) or MMP-3(ΔC) in the presence of MMP-1(E200A) did clearly show intermediate products (I1, I2, I3 and I4) that are larger than the βA and γA products (Figure 10B). Intermediate I3 is likely to be a cleaved α1(I) chain crosslinked to an intact α1(I) through N-terminal telopeptides, and I4 is a product resulting from cleavage of either α1(I) or α2(I) chain of the α1(I)–α2(I) crosslinks. Intermediates I1 and I2 of γ chains are products resulting from cleavage of one and two of the three α chains, respectively. The detection of these intermediates was possible, probably because simultaneous binding of an unwinder and a cutter is required to cleave collagen and the dissociation of either one of these from the collagen interrupts the process. The active MMP-1, on the other hand, possesses both components in one molecule and therefore the three chains are cleaved more effectively at a rate faster than the detection method used to identify the intermediate products.
Figure 10.
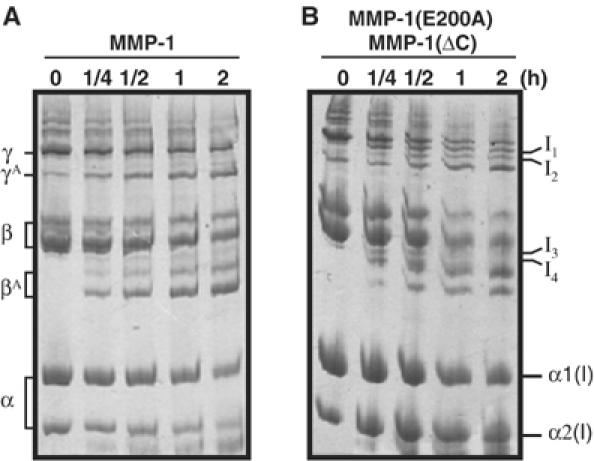
Detection of intermediate products during collagenolysis. Nonpepsin-treated collagen I was made to react with (A) 0.1 μM full-length collagenase or (B) 0.8 μM MMP-1(ΔC) and 6 μM MMP-1(E200A). The reactions were stopped with 20 mM EDTA and the products were analyzed by SDS–PAGE with 5% total acrylamide under reducing conditions. γA and βA are the 3/4 fragments of γ and β chains generated by MMP-1. I1, I2, I3 and I4 indicate intermediate products.
Discussion
Substrate specificities of proteinases are normally dictated by a short stretch of amino-acid sequence and local secondary and tertiary structures of protein substrates. The regions susceptible to proteinases are usually exposed on the surface of molecules and they are often flexible, so that the scissile bond can readily be accommodated within the active site of the enzyme. Such flexibility may be an intrinsic property of the protein or influenced by post-translational modification, such as oxidation (Mehlhase and Grune, 2002), phosphorylation (Hershko and Ciechanover, 1998), ubiquitination (Hershko and Ciechanover, 1998) and proteolysis (Murphy et al, 1985; Nagase et al, 1990; Rawson, 2003; Haass, 2004). The interstitial collagens are long triple-helical structures consisting of three left-handed poly-Pro II-like helices stabilized by hydrogen bonds formed among the backbones of three α chains and they are highly resistant to most proteinases. In this report, we have demonstrated that in order for a collagenase to cleave collagen it first binds and then unwinds the rigid triple-helical substrate before it cleaves the Gly775–Ile776 and Gly775–Leu776 bonds of collagen I. As far as we are aware, this is the first demonstration that a single polypeptide proteinase induces significant structural changes in the substrate prior to peptide bond hydrolysis. For the large multisubunit 26S proteasome, unfolding and ubiquitination of proteins are considered to be essential to digest proteins as the entrance of this complex is restricted (Voges et al, 1999). This reaction is accompanied by ATP hydrolysis (Voges et al, 1999; Goldberg, 2003), whereas collagenase activity does not require ATP. Owing to the structural constraint between collagenase and the collagen substrate, several hypotheses have been proposed to explain how collagenase may act on triple-helical collagens (Bode, 1995; de Souza et al, 1996; Gomis-Rüth et al, 1996; Ottl et al, 2000; Overall, 2002). This includes: the ‘proline zipper' model by de Souza et al (1996), proposing that the proline-rich linker region of collagenases interacts with and unwinds the triple-helical collagen, and a ‘collagen-trapping' model in which the Hpx domain folds over the catalytic site sandwiching collagen (Bode, 1995; Gomis-Rüth et al, 1996). However, the intact linker region may not be necessary as the catalytic domain and the Hpx domain added together can cleave collagen. The collagen-trapping model is also inconsistent with our observation that noncollagenolytic proteinases can cleave α1(I) and α2(I) chains in the presence of MMP-1(E200A), whereas in the model they would be protected by the Hpx domain. We also considered the following two other possible mechanisms: (i) collagenase stabilizes the partially unwound state of collagen that may occur spontaneously around the collagenase-susceptible region; and (ii) conformational changes occur within the collagenase molecule in such a way that it accommodates the triple-helical collagen in the active site. These two possibilities are also inconsistent with our results. If collagen underwent spontaneous unwinding, then such a conformational state would be recognized and cleaved by MMP-1(ΔC), and even by MMP-3(ΔC) or HLE, although the rate of these reactions could be much slower compared to full-length MMP-1. However, even with a very high concentration (3–7 μM) of MMP-1(ΔC) or MMP-3(ΔC), no collagenolytic activity was detected. It is also unlikely that the induction of a conformational change in the collagenase by collagen is the mechanism. If this were the case, MMP-1(ΔC), MMP-3(ΔC) and HLE would not be able to cleave collagen I in the presence of MMP-1(E200A), because the triple-helical collagen would be in the native conformation. Our experimental results support a mechanism in which collagenase locally unwinds the triple-helical collagen before it hydrolyzes the three peptide bonds as illustrated in Figure 11A.
Figure 11.
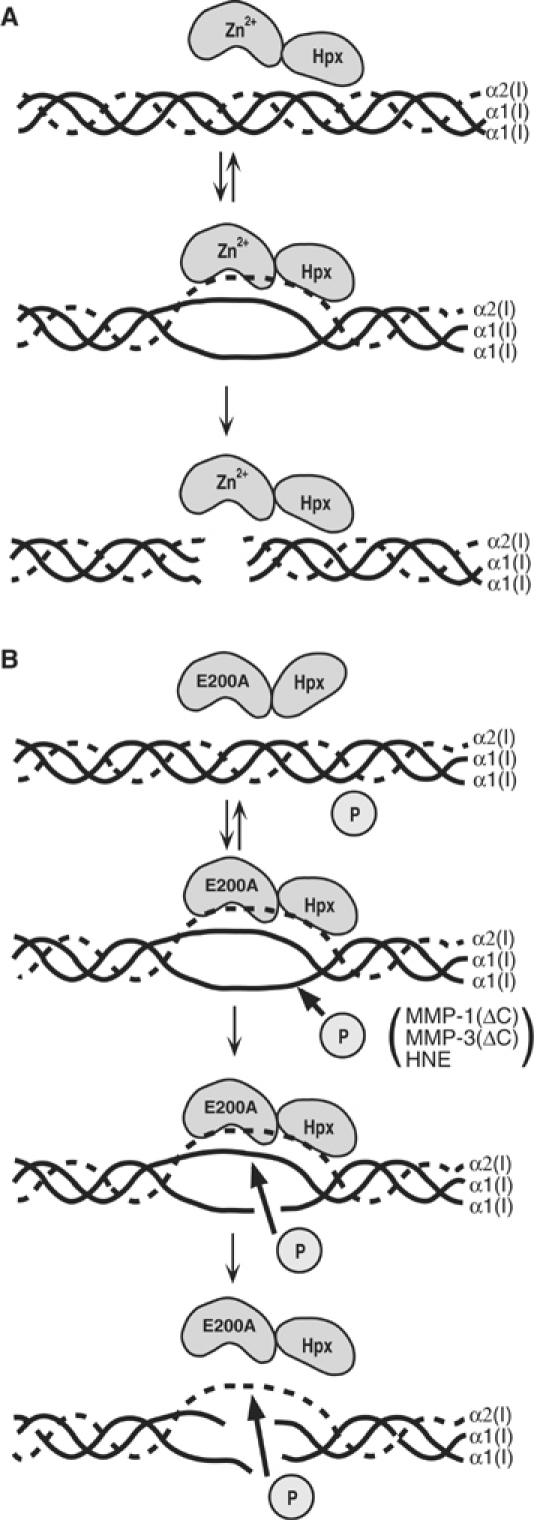
Steps involved in collagenolysis. (A) Collagenase binds to and locally unwinds collagen before it cleaves the triple-helical interstitial collagen. (B) MMP-1(E200A) binds preferentially to the α2(I) chain and unwinds the triple-helical collagen, but is unable to cleave collagen. The unwound collagen becomes susceptible to noncollagenolytic proteinases indicated as ‘P' (e.g., MMP-1(ΔC), MMP-3(ΔC) and HLE), and the α1(I) chains are initially cleaved. This reaction requires the trimolecular complex formation of the unwinder (MMP-1(E200A)), a cutter proteinase and collagen.
Studies with synthetic substrates based on the collagen sequence have shown that the primary structure dictates the catalytic activity of MMP-1 (Nagase and Fields, 1996), but it is less likely so for collagenolysis, firstly because the synthetic substrates are cleaved also by other noncollagenolytic MMPs, such as MMP-3 and MMP-9, and secondly because the Km values for these substrates are two to three orders of magnitude higher than those of collagen I (Welgus et al, 1981; Fields, 1991). The critical aspects of the collagenolytic specificity rely on the structural changes in collagen, induced by interacting with collagenase. This is evident from temperature-dependent collagenolysis versus gelatinolysis, as shown in Figure 2. At 37°C the rate of cleavage of collagen is much faster than that of gelatin, but at 10°C little or no collagenolytic activity of MMP-1 was observed compared with the gelatinolytic activity. Recent studies of Leikina et al (2002) indicated that human collagen I monomers denature at 37°C within a couple of days. Therefore, at 37°C collagen is unwound more readily by collagenase, and the configuration induced in the α chain by interacting with collagenase probably fits the active site and the specificity subsites of the enzyme optimally. Gross and Nagai (1965) postulated local structural unstability of the triple helix around the collagenase cleavage site, and Brown et al (1977) proposed that it was due to a poor imino-acid content. Fields (1991) indicated that the collagenase-recognition sites in interstitial collagens are preceded by a tightly helical conformation with a high imino-acid content (>33%) and a low side-chain molar volume, followed by a loose triple-helical region with a low imino-acid content (<17%) and a low charged residue content for the entire 25-residue cleavage site region. A recent study of a synthetic heterotrimeric triple-helical peptide containing the collagen I collagenase cleavage site has also indicated that the structure around the collagenase cleavage site is less ordered (Fiori et al, 2002). Thus, it is most likely that the collagenase cleavage sites in interstitial collagens are more susceptible to changes in conformation. The cleavage of Val783–Gly784 bond of the α1(I) chain near the collagenase cleavage site by HLE with the help of MMP-1(E200A) clearly supports this notion. It is also notable that mutation in the mouse α1(I) chain around the collagenase cleavage site makes all three α chains resistant to collagenolysis (Wu et al, 1990). In those mutants, Ile776 (P1′ subsite), or both Gln774 (P2 subsite) and Ala777 (P2′ subsite), was replaced with Pro. Addition of one or two prolines in the α1(I) chains around the collagenase cleavage site might have made collagen resistant to unwinding as the wild-type α2(I) chain was also not cleaved by collagenase (Wu et al, 1990). Nonetheless, the Tm of the wild-type mouse collagen I and those of mutants were also indistinguishable, indicating that these structural changes are too small to observe changes in the overall Tm of collagen I. These results agree well with our observation that binding and unwinding collagen I by MMP-1(E200A) did not alter the Tm. These observations support the conclusion that the structural changes induced in collagen by MMP-1 are local. On the other hand, structures of gelatin are probably not optimal for the substrate–enzyme interaction in the case of collagenase, which may be evident from the Km values of collagen I for human MMP-1 which are about 1 μM, whereas those of gelatin I are 4–7 μM at 37°C (Welgus et al, 1985).
In our experiments with MMP-1(E200A), a relatively high concentration of an unwinder protein and a cutter proteinase are required, and the efficiency of collagenolysis in this system is extremely low. This is probably because all three components (i.e., an unwinder, a cutter proteinase and the collagen substrate) need to assemble simultaneously (see Figure 11B). Our observation that the combination of HpxMMP-1 and MMP-1(ΔC) cleaved collagen I was unexpected, as our earlier work indicated that the collagenolytic activity was lost when MMP-1 is cleaved into the catalytic domain and the Hpx domain (K Suzuki and H Nagase, unpublished work). A similar loss of collagenolysis was reported for MMP-8 (Knäuper et al, 1993). When the catalytic domain and the Hpx domain are combined, they express collagenolytic activity, but a high concentration of these components is required to form a productive trimolecular complex. Such trimolecular interaction is apparently transient, explaining the low efficiency of the activity. This concentration-dependent association would also explain the earlier observation that MMP-1 and MMP-8 lost collagenolytic activity when the catalytic domain and the Hpx domain were split. When the two components are linked by the linker, it is a far more effective enzyme, as the interaction with collagen would be increased analogous to an increased free energy of binding when two low-affinity binding molecules are linked together (Jencks, 1981).
Our studies have also revealed that collagenase cleaves the three α chains one by one. Early studies of Sunada and Nagai (1983) showed that one or two α chains of collagen III were cleaved by collagenase. In the case of collagen I, we show that collagenase preferentially interacts with the α2(I) chain and this is the first chain to be cleaved. This conclusion is based on the observation that the α1(I) chain is much more readily cleaved than the α2(I) chain by MMP-1(ΔC), MMP-3(ΔC) or HLE in the presence of MMP-1(E200A). An initial cleavage of α2(I) chain by MMP-8 was reported also with a synthetic heterotrimeric triple-helical peptide (Muller et al, 2000). However, it is not known which part of the collagenase molecule makes the first contacts with collagen. Our molecular modelling suggests that one possible site is formed by the catalytic domain and the Hpx domain together, in a region rather distal to the active site of the enzyme (R Visse and H Nagase, unpublished results). This possibility and how unwinding of the triple helix occurs are currently being investigated by mapping the critical sites involved in unwinding activity by mutagenesis studies. It appears that several subsites of MMP-1 are involved in this activity. Our current hypothesis is that these sites are located both in the catalytic and Hpx domains of MMP-1 and they make contacts with the triple-helical collagen cooperatively in succession. One of the sites is the active site of the enzyme, as demonstrated here with an active site-directed synthetic inhibitor. Once these regions are clarified, it may provide new insights into generating inhibitors that specifically block unregulated collagenolysis.
Materials and methods
Materials
Chemicals were from the following sources: 4-aminophenylmercuric acetate (APMA) from ICN Biochemicals, Cleveland, OH; chymotrypsin and phenylmethylsulfonyl fluoride from Sigma, St Louis, MO; human leukocyte elastase from Athens Research, Athens, GA. The hydroxamate inhibitor GM6001X (HONHCOH2CH (isobutyl) CO-Tyr(OMe)-NHMe (Grobelny et al, 1992) was a gift from Dr J Oloksyszyn of OsteoArthritic Sciences Inc. Type I collagen was purified from guinea-pig skin (Glimcher et al, 1964). Pepsin-treated collagen I was prepared according to Trentham et al (1977). Type I gelatin was formed by heating type I collagen to 65°C for 20 min. Restriction enzymes and T4 DNA ligase were from New England Biolabs and Pfu polymerase was from Stratagene. Escherichia coli BL21(DE3) and the pET3a expression vector were from Novagen. Recombinant MMP-3(ΔC) was expressed in E. coli as an active form and purified by using a hydroxamate affinity column as described by Moore and Spilburg (1986).
Construction of MMP-1(E200A) and MMP-3(E202A)
Cloning of wild-type proMMP-1 and proMMP-3 cDNA into the expression vector pET3a was carried out as described previously (Chung et al, 2000). To generate pro-MMP-1(E200A) mutant, the pET3a-proMMP-1 vector, whose BamHI site in the original MMP-1 cDNA was removed by mutating GGATCC to GGACTT without affecting the codon for Asp 389, was used as a template for polymerase chain reaction (PCR) with a sense pET3a primer (5′-ACTTTAAGAAGGAGATATACATATG-3′), which includes the NdeI site (underlined), and an antisense primer with a mutation changing Glu200 to Ala (5′-AGAATGGCCGAGTGCATGAGCCGCAAC-3′). A second PCR was carried out with a sense primer containing the codon E200A mutation (5′-GTTGCGGCTCATGCACTCGGCCATTCT-3′) and an antisense pET3a primer (5′-GCTTTGTTAGCAGCCGGATCC-3′) containing the BamHI site (underlined). The two PCR fragments were combined and a third PCR carried out with the sense and antisense pET3a primers. The resulting fragment was digested with NdeI and BamHI and ligated into pET3a. For all PCRs, Pfu polymerase was used, and the sequence was confirmed by DNA sequencing. To generate MMP-3(E202A), the same strategy was used but with MMP-3-specific sense (5′-GTTGCTGCTCATGCAATTGGCCACTCC-3′) and antisense (5′-GGAGTGGCCAATTGCATGAGCAGCAAC-3′) primers where Glu202 was mutated to Ala. The resulting PCR fragment was digested with NdeI, and ligated into the NdeI-cleaved pET3a-proMMP-3 vector. The orientation of the fragment was checked with PCR, and the sequence was confirmed by DNA sequencing.
Expression of mutant proteins
The pET3a vector harboring wild-type or mutant MMP cDNA was transformed into E. coli BL21(DE3) cells and protein synthesis was induced by the addition of 0.4 mM isopropyl-β-D-thiogalactoside at 37°C for 4 h. Inclusion bodies were harvested as described previously (Chung et al, 2000) and dissolved in 8 M urea, 20 mM Tris–HCl (pH 8.6), 20 mM dithiothreitol and 50 μM ZnCl2, and passed over a High Q Support anion exchange column (Bio-Rad). Recombinant proteins were diluted to <0.3 mg/ml and 20 mM cystamine was added. For folding, proteins were dialyzed against a buffer containing 50 mM Tris–HCl (pH 8.6), 0.15 M NaCl, 5 mM CaCl2, 100 μM ZnCl2, 5 mM β-mercaptoethanol, 1 mM 2-hydroxyethyl disulfide and 0.02% NaN3, and then against 50 mM Tris–HCl (pH 7.5), 0.15 M NaCl, 5 mM CaCl2, 0.02% NaN3 (TNC buffer) containing 50 μM ZnCl2 at 4°C. Folded proMMPs were purified by chromatography on a Green A Dymatrex gel column (Amicon) and a Sephacryl S-200 column (Pharmacia). ProMMP-1, proMMP-1(E200A) and proMMP-3(E202A) were activated by incubating with 1 mM APMA and one-tenth molar amount of MMP-3(ΔC) at 37°C for 2 h. ProMMP-3(ΔC) lacking residues 248–460 and proMMP-3 were activated by the addition of 1 mM APMA at 37°C for 2 h. The activated MMPs and MMP mutants were applied to a Sephacryl S-200 column to remove MMP-3(ΔC), APMA and the cleaved propeptide. The hemopexin domain of MMP-1 (HpxMMP-1) and the Hpx of MMP-3(HpxMMP-3) were made by incubating either proMMP-1 or proMMP-3 with 1 mM APMA and one-tenth molar amount of MMP-3(ΔC) at 37°C for at least 16 h. The protein was run over a hydroxamate affinity column to remove the full-length MMP, and the catalytic domain of MMP and the unbound material containing the Hpx domain was collected.
Digestion of collagen and gelatin
Proteins and enzymes of indicated concentrations were used to digest 30 μg pepsin-treated type I collagen or gelatin in TNC buffer at 25°C for up to 48 h, unless otherwise specified. Reactions were stopped by the addition of SDS–PAGE loading buffer containing 20 mM EDTA. Products were analyzed by SDS–PAGE with 7.5% total acrylamide under reducing conditions, and the gels were stained with Coomassie brilliant blue R-250. Collagen digestion was examined for the generation of 3/4 and 1/4 fragments. Nonpepsin-treated type I collagen was used to detect intermediate products generated during collagenolysis and analyzed on SDS–PAGE with 5% total acrylamide under reducing conditions.
N-terminal sequencing
The 1/4 fragments generated by MMP-1(ΔC), MMP-3(ΔC) and HLE in the presence of MMP-1(E200A) were separated by SDS–PAGE, and transferred to a poly(vinylidene difluoride)-Millipore Immobilon transfer membrane. The proteins were located by staining with Coomassie brilliant blue R-250 and the bands of interest were excised and sequenced by an Applied Biosystem 447A pulse liquid sequenator.
Measurement of collagen melting temperature Tm
Pepsin-treated guinea-pig type I collagen (3 μM) was incubated with or without an equimolar concentration of MMP-1(E200A) or MMP-3(E200A) in TNC buffer. Thermal transition curves were obtained with a JASCO J-600 circular dichroism spectrometer with 0.1 cm path length quartz cell, by recording the molar ellipticity at wavelength 222 nm, while the temperature was continuously increased from 5 to 70°C at a rate of 35°C/h. Temperature was controlled using the JASCO PTC-348WI temperature control unit. The inflection point of a sigmoidal melting curve was used to define the melting temperature Tm.
Determination of KD of MMP-1(E200A) for collagen I
MMP-1(E200A) at various concentrations (0–6 μM) was incubated with 4.5 μM guinea-pig collagen I and 6 μM MMP-1(ΔC). Samples removed at different incubation times up to 24 h were analyzed by SDS–PAGE (7.5% total acrylamide) for generation of 3/4 and 1/4 collagen fragments. Proteins were stained with Coomassie brilliant blue R-250, and the percentage cleavage was determined by densitometry and analyzed using Phoretics 1D (Nonlinear Dynamics), from which the initial rate of cleavage was calculated. The equilibrium dissociation constant (KD) was determined by double reciprocal plots of the rate of product formation versus the concentration of MMP-1(E200A).
Molecular modelling
The full-length crystal structure of porcine MMP-1 (1FBL; (Li et al, 1995) and the crystal structure of triple-helical peptide comprising part of the MMP-1-specific cleavage site of type III collagen (Kramer et al, 2001) were used to model the interaction of collagenase and interstitial collagen. For this purpose, the hydroxamate inhibitor that was co-crystallized with porcine MMP-1 was removed. The collagen triple-helical peptide positioning was performed manually with InsightII/Discover, and images were produced with SwissPDB viewer (Guex and Peitsch, 1997).
Acknowledgments
We thank Professor John White for critical reading of the manuscript. This work was supported by the Wellcome Trust grant no. 057508 and National Institutes of Health grants AR39189 to HN, and CA77402 and CA98799 to GBF. JLL-F was supported by a Glenn/American Foundation for Aging Research Scholarship.
References
- Aimes RT, Quigley JP (1995) Matrix metalloproteinase-2 is an interstitial collagenase. Inhibitor-free enzyme catalyzes the cleavage of collagen fibrils and soluble native type I collagen generating the specific 3/4- and 1/4-length fragments. J Biol Chem 270: 5872–5876 [DOI] [PubMed] [Google Scholar]
- Bode W (1995) A helping hand for collagenases: the haemopexin-like domain. Structure 3: 527–530 [DOI] [PubMed] [Google Scholar]
- Brinckerhoff CE, Matrisian LM (2002) Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol 3: 207–214 [DOI] [PubMed] [Google Scholar]
- Brown RA, Hukins DW, Weiss JB, Twose TM (1977) Do mammalian collagenases and DNA restriction endonucleases share a similar mechanism for cleavage site recognition? Biochem Biophys Res Commun 74: 1102–1108 [DOI] [PubMed] [Google Scholar]
- Cawston TE (1996) Metalloproteinase inhibitors and the prevention of connective tissue breakdown. Pharmacol Therap 70: 163–182 [DOI] [PubMed] [Google Scholar]
- Chung L, Shimokawa K, Dinakarpandian D, Grams F, Fields GB, Nagase H (2000) Identification of the (RWTNNFREY191)-R-183 region as a critical segment of matrix metalloproteinase 1 for the expression of collagenolytic activity. J Biol Chem 275: 29610–29617 [DOI] [PubMed] [Google Scholar]
- Clark IM, Cawston TE (1989) Fragments of human fibroblast collagenase. Purification and characterization. Biochem J 263: 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza SJ, Pereira HM, Jacchieri S, Brentani RR (1996) Collagen/collagenase interaction: does the enzyme mimic the conformation of its own substrate? FASEB J 10: 927–930 [DOI] [PubMed] [Google Scholar]
- Fields GB (1991) A model for interstitial collagen catabolism by mammalian collagenases. J Theor Biol 153: 585–602 [DOI] [PubMed] [Google Scholar]
- Fiori S, Sacca B, Moroder L (2002) Structural properties of a collagenous heterotrimer that mimics the collagenase cleavage site of collagen type I. J Mol Biol 319: 1235–1242 [DOI] [PubMed] [Google Scholar]
- Fraser RD, MacRae TP, Suzuki E (1979) Chain conformation in the collagen molecule. J Mol Biol 129: 463–481 [DOI] [PubMed] [Google Scholar]
- Garnero P, Borel O, Byrjalsen I, Ferreras M, Drake FH, McQueney MS, Foged NT, Delmas PD, Delaissé JM (1998) The collagenolytic activity of cathepsin K is unique among mammalian proteinases. J Biol Chem 273: 32347–32352 [DOI] [PubMed] [Google Scholar]
- Glimcher MJ, Francois CJ, Richards L, Krane SM (1964) The presence of organic phosphorus in collagens and gelatins. Biochim Biophys Acta 93: 585–602 [DOI] [PubMed] [Google Scholar]
- Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895–899 [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Gohlke U, Betz M, Knäuper V, Murphy G, López-Otín C, Bode W (1996) The helping hand of collagenase-3 (MMP-13): 2.7 Å crystal structure of its C-terminal haemopexin-like domain. J Mol Biol 264: 556–566 [DOI] [PubMed] [Google Scholar]
- Grobelny D, Poncz L, Galardy RE (1992) Inhibition of human skin fibroblast collagenase, thermolysin, and Pseudomonas aeruginosa elastase by peptide hydroxamic acids. Biochemistry 31: 7152–7154 [DOI] [PubMed] [Google Scholar]
- Gross J, Nagai Y (1965) Specific degradation of the collagen molecule by tadpole collagenolytic enzyme. Proc Natl Acad Sci USA 54: 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Haass C (2004) Take five-BACE and the gamma-secretase quartet conduct Alzheimer's amyloid beta-peptide generation. EMBO J 23: 483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Jencks WP (1981) On the attribution and additivity of binding energies. Proc Natl Acad Sci USA 78: 4046–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knäuper V, Cowell S, Smith B, López-Otín C, O'Shea M, Morris H, Zardi L, Murphy G (1997) The role of the C-terminal domain of human collagenase-3 (MMP-13) in the activation of procollagenase-3, substrate specificity, and tissue inhibitor of metalloproteinase interaction. J Biol Chem 272: 7608–7616 [DOI] [PubMed] [Google Scholar]
- Knäuper V, Osthues A, DeClerck YA, Langley KE, Blaser J, Tschesche H (1993) Fragmentation of human polymorphonuclear-leucocyte collagenase. Biochem J 291: 847–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RZ, Bella J, Brodsky B, Berman HM (2001) The crystal and molecular structure of a collagen-like peptide with a biologically relevant sequence. J Mol Biol 311: 131–147 [DOI] [PubMed] [Google Scholar]
- Lei H, Furth EE, Kalluri R, Chiou T, Tilly KI, Tilly JL, Elkon KB, Jeffrey JJ, Strauss JF III (1996) A program of cell death and extracellular matrix degradation is activated in the amnion before the onset of labor. J Clin Invest 98: 1971–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikina E, Mertts MV, Kuznetsova N, Leikin S (2002) Type I collagen is thermally unstable at body temperature. Proc Natl Acad Sci USA 99: 1314–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brick P, O'Hare MC, Skarzynski T, Lloyd LF, Curry VA, Clark IM, Bigg HF, Hazleman BL, Cawston TE, Blow DM (1995) Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed beta-propeller. Structure 3: 541–549 [DOI] [PubMed] [Google Scholar]
- McCroskery PA, Wood S Jr, Harris ED Jr (1973) Gelatin: a poor substrate for a mammalian collagenase. Science 182: 70–71 [DOI] [PubMed] [Google Scholar]
- Mehlhase J, Grune T (2002) Proteolytic response to oxidative stress in mammalian cells. Biol Chem 383: 559–567 [DOI] [PubMed] [Google Scholar]
- Moore WM, Spilburg CA (1986) Purification of human collagenases with a hydroxamic acid affinity column. Biochemistry 25: 5189–5195 [DOI] [PubMed] [Google Scholar]
- Muller JCD, Ottl J, Moroder L (2000) Heterotrimeric collagen peptides as fluorogenic collagenase substrates: synthesis, conformational properties, and enzymatic digestion. Biochemistry 39: 5111–5116 [DOI] [PubMed] [Google Scholar]
- Murphy G, Allan JA, Willenbrock F, Cockett MI, O'Connell JP, Docherty AJP (1992) The role of the C-terminal domain in collagenase and stromelysin specificity. J Biol Chem 267: 9612–9618 [PubMed] [Google Scholar]
- Murphy G, McAlpine CG, Poll CT, Reynolds JJ (1985) Purification and characterization of a bone metalloproteinase that degrades gelatin and types IV and V collagen. Biochim Biophys Acta 831: 49–58 [DOI] [PubMed] [Google Scholar]
- Nagase H, Enghild JJ, Suzuki K, Salvesen G (1990) Stepwise activation mechanisms of the precursor of matrix metalloproteinase 3 (stromelysin) by proteinases and (4-aminophenyl)mercuric acetate. Biochemistry 29: 5783–5789 [DOI] [PubMed] [Google Scholar]
- Nagase H, Fields GB (1996) Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers 40: 399–416 [DOI] [PubMed] [Google Scholar]
- Ohuchi E, Imai K, Fujii Y, Sato H, Seiki M, Okada Y (1997) Membrane type 1 matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J Biol Chem 272: 2446–2451 [DOI] [PubMed] [Google Scholar]
- Ottl J, Gabriel D, Murphy G, Knäuper V, Tominaga Y, Nagase H, Kroger M, Tschesche H, Bode W, Moroder L (2000) Recognition and catabolism of synthetic heterotrimeric collagen peptides by matrix metalloproteinases. Chem Biol 7: 119–132 [DOI] [PubMed] [Google Scholar]
- Overall CM (2002) Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol 22: 51–86 [DOI] [PubMed] [Google Scholar]
- Patterson ML, Atkinson SJ, Knäuper V, Murphy G (2001) Specific collagenolysis by gelatinase A, MMP-2, is determined by the hemopexin domain and not the fibronectin-like domain. FEBS Lett 503: 158–162 [DOI] [PubMed] [Google Scholar]
- Pilcher BK, Dumin JA, Sudbeck BD, Krane SM, Welgus HG, Parks WC (1997) The activity of collagenase-1 is required for keratinocyte migration on a type I collagen matrix. J Cell Biol 137: 1445–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran GN, Kartha G (1955) Structure of collagen. Nature 176: 593–595 [DOI] [PubMed] [Google Scholar]
- Rawson RB (2003) Control of lipid metabolism by regulated intramembrane proteolysis of sterol regulatory element binding proteins (SREBPs). Biochem Soc Symp 70: 221–231 [DOI] [PubMed] [Google Scholar]
- Rich A, Crick FH (1961) The molecular structure of collagen. J Mol Biol 3: 483–506 [DOI] [PubMed] [Google Scholar]
- Sternlicht MD, Werb Z (2001) How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol 17: 463–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunada H, Nagai Y (1983) Mechanism of collagen degradation by collagenase:a transition process of the collagen molecular from collagenase-substrate to gelatinase-substrate. Biomed Res 4: 61–70 [Google Scholar]
- Trentham DE, Townes AS, Kang AH (1977) Autoimmunity to type II collagen an experimental model of arthritis. J Exp Med 146: 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visse R, Nagase H (2003) Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res 92: 827–839 [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68: 1015–1068 [DOI] [PubMed] [Google Scholar]
- Welgus HG, Grant GA, Sacchettini JC, Roswit WT, Jeffrey JJ (1985) The gelatinolytic activity of rat uterus collagenase. J Biol Chem 260: 13601–13606 [PubMed] [Google Scholar]
- Welgus HG, Jeffrey JJ, Eisen AZ (1981) The collagen substrate specificity of human skin fibroblast collagenase. J Biol Chem 256: 9511–9515 [PubMed] [Google Scholar]
- Woessner JF Jr (1998) The matrix metalloproteinase family. In Matrix Metalloproteinases, Parks WC, Mecham RP (eds), pp 1–14. San Diego: Academic Press [Google Scholar]
- Woessner JF, Nagase H (2000) Matrix Metalloproteinases and TIMPs: Protein Profile. Oxford: Oxford University Press [Google Scholar]
- Wu H, Byrne MH, Stacey A, Goldring MB, Birkhead JR, Jaenisch R, Krane SM (1990) Generation of collagenase-resistant collagen by site-directed mutagenesis of murine pro alpha 1(I) collagen gene. Proc Natl Acad Sci USA 87: 5888–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WG, Byrne MH, Boyce BF, Krane SM (1999) Bone resorption induced by parathyroid hormone is strikingly diminished in collagenase-resistant mutant mice. J Clin Invest 103: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]


